Abstract
Duchenne muscular dystrophy (DMD) is an X-linked recessive disorder resulting from lesions of the gene encoding dystrophin. These usually consist of large genomic deletions, the extents of which are not correlated with the severity of the phenotype. Out-of-frame deletions give rise to dystrophin deficiency and severe DMD phenotypes, while internal deletions that produce in-frame mRNAs encoding truncated proteins can lead to a milder myopathy known as Becker muscular dystrophy (BMD). Widespread restoration of dystrophin expression via adeno-associated virus (AAV)-mediated exon skipping has been successfully demonstrated in the mdx mouse model and in cardiac muscle after percutaneous transendocardial delivery in the golden retriever muscular dystrophy dog (GRMD) model. Here, a set of optimized U7snRNAs carrying antisense sequences designed to rescue dystrophin were delivered into GRMD skeletal muscles by AAV1 gene transfer using intramuscular injection or forelimb perfusion. We show sustained correction of the dystrophic phenotype in extended muscle areas and partial recovery of muscle strength. Muscle architecture was improved and fibers displayed the hallmarks of mature and functional units. A 5-year follow-up ruled out immune rejection drawbacks but showed a progressive decline in the number of corrected muscle fibers, likely due to the persistence of a mild dystrophic process such as occurs in BMD phenotypes. Although AAV-mediated exon skipping was shown safe and efficient to rescue a truncated dystrophin, it appears that recurrent treatments would be required to maintain therapeutic benefit ahead of the progression of the disease.
Introduction
Duchenne muscular dystrophy (DMD) is an X-linked recessive neuromuscular disorder caused by mutations in the dystrophin gene, which encodes a variety of tissue-specific protein isoforms. Among them, the full-length dystrophin (DP427), translated from a major 14-kb mRNA transcript made of 79 exons, is a large protein (427 kDa) found in skeletal and cardiac muscle fibers and neurons in particular regions of the central nervous system.1,2 In muscle, dystrophin is located on the inner side of the sarcolemma, where it associates with other proteins to form the transmembrane dystrophin–glycoprotein complex, which connects the actin cytoskeleton to the extracellular matrix. Lack of dystrophin interrupts this structural link and muscle fibers become more vulnerable to mechanical stress.3,4 As a result, dystrophic individuals display progressive weakness of skeletal muscles, which are with time replaced by adipofibrotic tissue leading to loss of ambulation, whereupon premature death can be caused by either respiratory failure or cardiomyopathy.5 In addition, about one third of DMD patients display cognitive impairment suggesting a disruption of neuronal and brain function.6
The DP 427 dystrophin is a modular protein that can retain function following loss of certain internal regions.7 This phenomenon occurs in the clinically milder disease Becker muscular dystrophy (BMD), where deletions that maintain the open reading frame lead to the synthesis of truncated semifunctional forms of dystrophin.8 Thus, it was proposed a few years ago that induction of exon skipping to remove mutated exons and/or to restore an open reading frame in partially deleted dystrophin mRNAs might be a relevant therapeutic approach for many DMD patients.9 Presently, antisense oligomers have been successfully employed for dystrophin rescue in various animal models of DMD,10,11 and clinical trials in patients are under way testing systemic delivery of both 2′-O-methyl-modified ribose oligomers with full-length phosphorothioate backbone (2OMe) and phosphorodiamidate morpholino oligomers by either recurrent subcutaneous or intravenous injections, respectively.12,13,14,15 So far, these treatments are well tolerated with no drug-related serious adverse events. Nevertheless, long-term therapeutic success requires effective delivery of the antisense to all affected tissues including the heart and the central nervous system. It would also require periodic injections throughout the patient's lifetime in order to maintain efficacy, a constraint which could be theoretically overcome by using a genetic platform such as the U7 system, for permanent delivery of the antisense. U7 is a non-spliceosomal snRNA, normally involved in the processing of the histone mRNA 3′ end, which has been engineered to deliver antisense sequences to the spliceosome.16 Proof-of-concept studies using adeno-associated virus (AAV) vectors encoding such engineered U7 have demonstrated successful exon skipping in mouse models of DMD.17,18 However, these studies have mainly focused on dystrophin rescue in the mdx murine model of DMD whose phenotype is not dramatically affected by the lack of dystrophin. Little is known about the benefit one might expect in a larger animal model exhibiting a severe dystrophic phenotype such as the golden retriever muscular dystrophy dog (GRMD), which parallels the human disease with early muscle weakness leading to locomotor, respiratory, digestive and cardiac impairments.19 The GRMD mutation consists of a single base change in the 3′ consensus splice site (A>G) of intron 6 of the dystrophin gene that provokes inaccurate mRNA processing:20 the seventh exon is skipped, which predicts a termination of the dystrophin reading frame within its N-terminal domain in exon 8. Due to exon phasing, the use of splice-switching antisenses to skip exons 6 and 8 of the dystrophin mRNA can restore a translational reading frame, thereby rescuing a truncated dystrophin, which is somewhat functional, as has been shown in recent studies using either transendocardiac delivery of exon-skipping recombinant AAV vectors21,22 or repeated systemic delivery of a cocktail of synthetic antisense oligonucleotides such as phosphorodiamidate morpholino oligomers.23
Here, our goal was to assess the muscle function recovery in GRMD after a single administration of AAV1 gene vectors expressing two engineered U7 for splice-switching of exons 6 and 8 of the mutated dystrophin mRNA. Along with dystrophin rescue, the components of the associated glycoprotein complex were expressed at the periphery of transduced muscle fibers, and the hallmarks of dystrophic pathology were lessened: utrophin was downregulated toward normal levels, the number of calcium-overloaded fibers and necrotic fibers were decreased. Likewise, NMR indices were improved and muscle strength was significantly enhanced. Finally we demonstrated the practicality of treating a forelimb by locoregional delivery of AAV1-U7 and we carried out a 5-year follow-up of the dystrophin rescue.
Results
In vitro screening of antisense sequences in GRMD primary myoblasts and design of engineered U7 for skipping exons 6 and 8
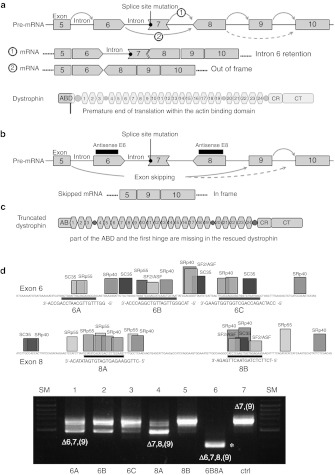
In GRMD, lack of dystrophin is due to a single base change (A to G) in the 3′ consensus splice site (splicing acceptor site—SA) of intron 6. Abolition of this SA provokes inaccurate pre-mRNA processing: either intron 6 is retained or exon 7 is skipped giving rise to a frame shift of the translational reading frame, which predicts a premature termination of dystrophin within its N-terminal domain in exon 8 (Figure 1a).20 A low level of natural skipping of multiple exons occurs spontaneously during GRMD dystrophin mRNA processing giving rise to rare revertant fibers missing exons 3 to 9 or 5 to 12.24 Due to the exon phasing around exon 7, splice-switching of exons 6 and 8 can restore the normal open reading frame (Figure 1b), rescuing a truncated dystrophin of about 403 kDa missing 158 amino-acids in the N-terminal actin-binding domain (N-ABD).
Figure 1.
The golden retriever muscular dystrophy dog (GRMD) mutation and splice-switching antisenses for dystrophin rescue. (a) The GRMD mutation consists in a single base change (A to G) in the 3′ acceptor splice site of intron 6, which abolishes dystrophin synthesis by either (a) retaining intron 6, or (b) inducing skipping of exon 7. Occasionally, exon 9 could be alternatively spliced. Subsequent mRNAs are frame-shifted and their translation is aborted at the level of the actin-binding domain (ABD). (b) Steric-blockade of RNA sequences that are recognized by the spliceosome in exons 6 and 8 would allow the synthesis of an in-frame mRNA lacking the (6 to 8)-block of exons. Succeeding dystrophin will miss part of the actin-binding domain but retain all spectrin-like repeats. (c) Shows exon-splicing enhancers (ESE) finder output for exons 6 and 8. All sites for SF2/ASF, SC35, SRp40, and SRp55 above the default thresholds are displayed as well as the location of targeted sequences by 2OMe antisense molecules (6A, 6B, 6C, 8A, 8B). 6A: 5′–GGUUUGUUGCAAUCCAGCCA-3′ 6B: 5′–UACGAGUUGAUUGUCGGACCCA-3′ 6C: 5′-CCAUCAGACCAG-CUGGUGGUGAAG-3′ 8A: 5′-CUUGGAAGAGUGAUGUGAUAUACA-3′ 8B: 5′-UCUUCUCUAGUAACUUGAGA-3′ (d) Exon skipping efficiency of selected 2OMe was assessed by transfection in GRMD primary myotubes. Subsequent mRNA extracts were analyzed by nested reverse transcription-PCR (RT-PCR) with primers in exons 3 and 10 (lanes 1 to 6: transfected GRMD myotubes; lane 7: untransfected GRMD control; SM: size marker). The major band (663-bp) detected in untreated GRMD myotubes (lane 7) corresponds to an mRNA lacking exons 7 and 9. An mRNA variant of about 792-bp, missing only exon 7, was occasionally observed. Transfection with 6A, 6B or 8A induced significant skipping of exons 6 and 8, respectively (lanes 1, 2, 4). Cotransfection of 6B and 8A resulted in a 308-bp fragment corresponding to a multi-skipped dystrophin mRNA lacking exons 6, 7, 8, and 9 (lane 6) as confirmed by DNA sequencing.
Five 2OMe (20 to 24-mer) antisense oligomers were designed against exons 6 and 8. These antisenses were chosen to anneal different putative exon-splicing enhancers of targeted exons (Figure 1c). Subsequent 2OMe were tested individually or in combination by transfection of GRMD myoblasts. Exon skipping was assessed by nested reverse transcription-PCR with primers in exons 3 and 10 of the dystrophin mRNA (Figure 1d). The three antisenses for skipping exon 6 were moderately efficient ex vivo (6B>6A>6C), whereas antisense 8A targeting exon 8 was highly efficient and antisense 8B was inefficient. Exon 9 was consistently missing from final mRNAs. It is likely that this phenomenon was not due to a general alteration of the splicing machinery in the presence of the antisenses, since it was also observed in control untreated samples. Rather, exclusion of exon 9 could result from a natural alternative splicing as it has been previously reported.23,25 The mixture of 6B and 8A antisense oligomers allowed precise multi-skipping thus giving rise to a Δ6–9 dystrophin mRNA. Consequently, these two sequences were selected to be adapted in the U7 system (Supplementary Figure S1). In order to gain specificity, both 6B and 8A were enlarged to 40 nucleotides each. Accordingly, each of the two U7 cassettes (U7E6 and U7E8) were 400-nucleotides long, comprising the U7 promoter, the appropriate antisense sequence, a smOPT (AAUUUUUGGAG) sequence to recruit the SM binding proteins of the U2 snRNA, and the terminal stem loop of U7 and its 3′ downstream sequences. U7E6 and U7E8 were cloned either independently or together within the two inverted terminal repeats of the AAV-2 genome backbone lacking all viral genes: AAV-U7E6, AAV-U7E8, and AAV-U7E6/8. Subsequent constructs were pseudotyped AAV1 by using a type 1 capsid packaging procedure as previously described.17
Evaluation of dystrophin rescue in GRMD after intramuscular delivery
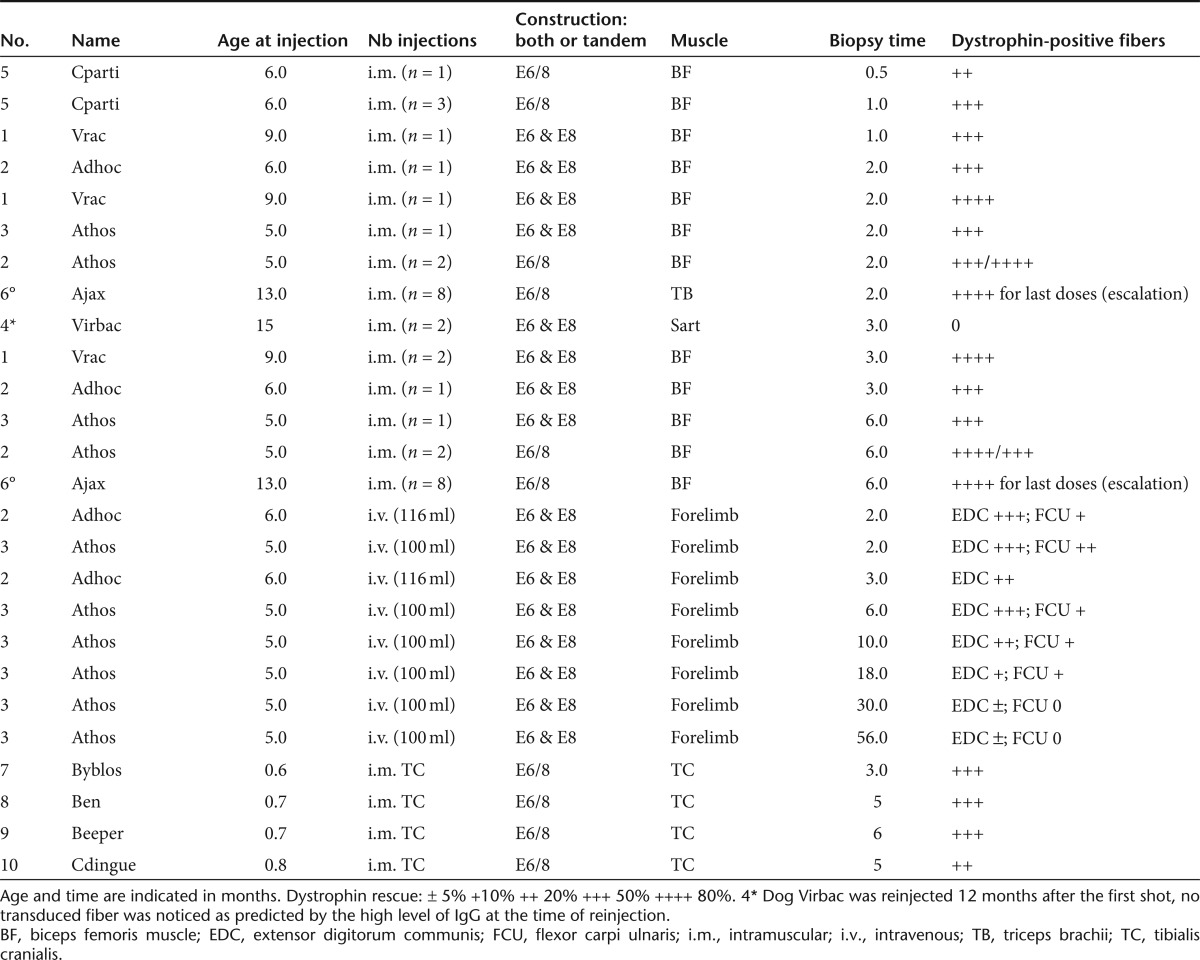
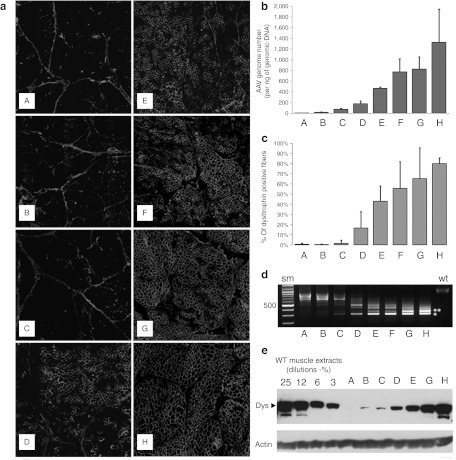
A total of six GRMD dogs were included in this study which was designed to evaluate the efficacy of the AAV1-U7E6, AAV1-U7E8, and AAV1-U7E6/8 vectors. As expected, injection of neither AAV1-U7E6 nor AAV1-U7E8 alone was sufficient to rescue the reading frame in GRMD, thus confirming that annealing a single target with U7 did not interfere with the splicing of neighbouring exons on the same pre-mRNA. This was corroborated in wild-type dog muscle, where delivery of AAV1-U7E6 or AAV1-U7E8 alone provoked the disruption of the normal dystrophin (Supplementary Figure S2). On the other hand, both conditions, a mixture of AAV1-U7E6 and AAV1-U7E8 (n = 8 biopsies) or AAV1-U7E6/8 alone (n = 22 biopsies), allowed similar dystrophin rescue after intramuscular delivery in GRMD (Table 1). Figure 2 shows a dose-range study of intramuscular injections into individual bundles of the triceps brachii muscle with increasing concentrations of AAV1-U7E6/8 [from 5 × 1010 to 3.5 × 1012 vector genomes (vg) in 500 µl phosphate-buffered saline]. Two months after injection, dystrophin-positive muscle fibers were detected by immunofluorescence staining at 2.5 × 1011 vg and higher (Figure 2a). As expected, copy numbers of AAV1-U7 present in these muscle samples increased with the dose escalation (Figure 2b). Counts on five random areas of biopsy cross-sections showed about 16% dystrophin-positive muscle fibers in fascicles injected with 2.5 × 1011 vg. This value reached 80% following injection with 3.5 × 1012 vg (Figure 2c). Consistently, reverse transcription-PCR analysis revealed that Δ6–8 and Δ6–9 in-frame multi-skipped mRNAs appeared along with increasing AAV1-U7E6/8 concentrations (Figure 2d). Western blot analysis confirmed the rescue of substantial levels of dystrophin with doses of 2.5 × 1011 vg and greater (Figure 2e). Figure 3 is representative of dystrophin rescue encompassing the whole section of a biopsy taken 3 months after intramuscular injection of both AAV1-U7E6 and AAV1-U7E8 vectors (3.5 × 1012 each). The majority of muscle fibers displayed robust immunostaining for dystrophin, which was also readily detected along longitudinal sections (Figure 3; inset b). Western blot analysis on muscle extracts confirmed highly efficient rescue of dystrophin, whose levels, in this biopsy, were improved toward normal. The skipping procedure generated a single immunoreactive protein with expected mobility of about 403 kDa without evidence for multiply deleted byproducts (Figure 3c). Results of the dose-effect at 6 months after injections of the biceps femoris muscle were similar to preceding results obtained at 2 months postinjection of the triceps brachii. However, dystrophin immunostaining was lower at 6 months that at 2 months, consistent with a reduced number of AAV-genomes in muscle biopsies when assessed by quantitative PCR, without obvious sign of immune rejection despite that AAV1-U7E6/8 generated a fast and sustained humoral response against the vector capsid (Supplementary Figure S3). AAV1 neutralizing antibodies in treated dogs appeared to peak during the first month after gene transfer, followed by a plateau. Not surprisingly, AAV-immunized dogs generated a rapid rise in antibody titers when submitted to a second injection one year later. Importantly, CD11b+ monocytes-macrophages, which typically infiltrate dystrophic muscles, were missing in areas expressing dystrophin. Moreover, neither CD4+ nor CD8+ lymphocytes were detected around dystrophin positive fibers by immunostaining, and antibodies against dystrophin were absent from sera of treated dogs, as determined by Western blot (Supplementary Figure S4 for positive controls).
Table 1. Dogs involved in the study.
Figure 2.
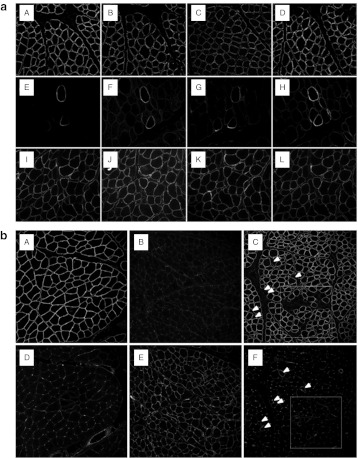
Dose-escalation study using intramuscular delivery of AAV1-U7E6E8 in golden retriever muscular dystrophy dog (GRMD). (a) Dystrophin immunostaining (NCL-DYS2 monoclonal antibody) on transverse sections of GRMD biopsies, 2 months after intramuscular injections of the tandem AAV1-U7E6E8 vector into the triceps brachii muscle. Dose-escalation: phosphate-buffered saline (PBS); 5 × 1010; 1011; 2.5 × 1011; 5 × 1011; 1012; 2 × 1012; and 3.5 × 1012 vg (A–H, respectively). (b) Quantification of the adeno-associated virus (AAV) genome copy number in samples of the dose-escalation study. (A–H) correspond to: PBS; 5 × 1010; 1011; 2.5 × 1011; 5 × 1011; 1012; 2 × 1012; and 3.5 × 1012 vg, respectively. (c) Proportion of dystrophin positive fibers in muscle samples following the dose-escalation study. For each condition, results are expressed as the mean of the percentage of dystrophin positive fibers obtained in five regions (±SD). (d) Detection of in-frame multi-skipped dystrophin mRNAs by nested reverse transcription-PCR (RT-PCR) from exon 3 to 10 in dose-escalation samples. (lane A: PBS) PCR products (792- and 663-bp fragments) correspond to out-of-frame mRNA variants lacking either exon 7 or exons 7 and 9, respectively. The RT-PCR pattern is changed accordingly to the AAV1-U7E6E8 dose. Two main fragments of 437-bp and 308-bp corresponding to in frame mRNAs lacking either exons 6 to 8 or 6 to 9 (white stars), respectively, are clearly detected at 1011 vg and above. Additional bands on top of the in frame fragments correspond to classical hetero-duplex artifacts obtained during the course of the PCR amplification. (e) Western blot of total protein (100 µg) extracted from injected GRMD muscle samples stained with NCL-DYS2. Arrow indicates the full-length dystrophin as detected in samples from normal muscle used for comparison (25, 12, 6, and 3%—diluted in GRMD to keep up total protein content). Samples were also hybridized with an actin-antibody to validate protein loadings.
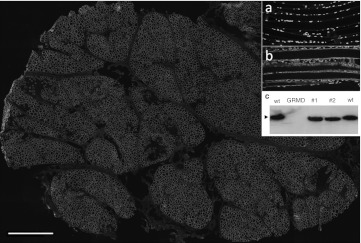
Figure 3.
Transverse section of a whole muscle biopsies taken 3 months after intramuscular injection of both AAV1-U7E6 and AAV1-U7E8 vectors (3.5 × 1012 vg each in 1 ml). Dystrophin immunostaining with NCL-DYS2 monoclonal antibody, scale bar: 1 mm. Insets (a and b) show dystrophin staining in longitudinal sections of control golden retriever muscular dystrophy dog (GRMD) and treated sample, respectively; nuclei are counterstained with Dapi. Inset (c) shows dystrophin contents appraised by western blot analysis using 40 µg of total protein extracted from normal (wt), GRMD and two treated samples (#1 and #2).
Histological features following dystrophin rescue in GRMD muscles
Figure 4a shows that along with the rescue of a truncated dystrophin missing the region encoded by exons 6–8 and/or 6–9, components of the dystrophin glycoprotein complex (DGC), including α- and β-sarcoglycans and β-dystroglycan, were readily restored at the periphery of transduced fibers, signifying that the dystrophin-dependent linkage between cytoskeleton and extracellular matrix was re-established. Further evidence of muscle repair was illustrated by assessing utrophin expression.19 The persistence of which in mature fibers is a mark of dystrophinopathy26 (Figure 4b). In treated muscle, like in normal healthy muscle, utrophin was found in capillaries and neuromuscular junctions but not in fibers expressing the rescued dystrophin. As an internal control, utrophin was still persistent in the very few fibers which did not express significant amounts of dystrophin.
Figure 4.
Restoration of the dystrophin-associated glycoprotein complex in treated golden retriever muscular dystrophy dog (GRMD) muscles and utrophin relocation. (a) Upper, middle, and lower rows show sections from biceps femoris muscles of normal dog, untreated and treated GRMD, respectively. Sections were immunoassayed for (a,e,l) dystrophin, (b,f,j) α-sarcoglycan, (c,g,k) β-sarcoglycan, and (d,h,l) β-dystroglycan. The same cluster of revertant fibers displaying dystrophin, as well as the associated glycoprotein complex, is shown on the serial sections from untreated GRMD. (b) Serial sections of (a,d) normal, (b,e) GRMD, and treated (c,f) GRMD muscles immunoassayed for (a–c) dystrophin (NCL-DYS2) and (d–f) utrophin (NCL-DRP2), respectively. Note that only the very few dystrophin-negative fibers are still stained for utrophin (arrows and highlighted square indicating a cluster of dystrophin-negative fibers).
A hallmark of the degeneration sequence that leads to fiber necrosis in dystrophic muscle is the presence of calcium-overloaded myofibers which are usually hypercontracted.27 This can be assayed with alizarin red S (ARS) staining, which is classically used to disclose intracellular calcium excess. Analysis of serial sections at injection-sites showed that dystrophin-expressing fibers were not stained by ARS, while staining was frequent in dystrophin-deficient muscle (Figure 5a–d). Consistently, the number of hypercontracted fibers decreased proportionally with the dose of injected vectors, thus confirming that the rescued dystrophin improved muscle fiber resilience (Figure 5e). However, it is worth noting that dystrophin-negative fibers in the vicinity of dystrophin-positive fibers were not protected from being damaged (Figure 5a, b).
Figure 5.
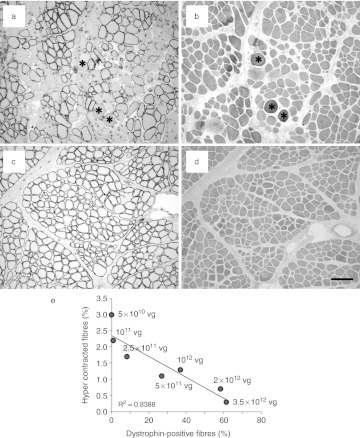
Improvement of muscle fiber resilience assessed by intracellular calcium overloading. Intracellular calcium overloading, indicative of a prenecrotic stage, was assessed by alizarin red S (ARS) staining on transverse sections of treated golden retriever muscular dystrophy dog (GRMD) muscles. (a,c) show two areas with either partial or almost complete dystrophin recue. (b,d) show corresponding serial sections stained with ARS. Stars indicate calcium-overloaded fibers in a and b (Bar = 100 µm). (e) Relationship between the percentage of dystrophin-positive fibers and prenecrotic fibers. Plotting shows a linear correlation (R2 = 0.8388) using the dose-escalation samples, 2 months after injection into the triceps brachii.
Dystrophin rescue enhanced muscle function
Muscle function was assessed for selected muscles following treatment of the hind limb in four 3-week-old GRMD dogs. To ensure standardised transduction, vectors were delivered surgically into the cranial muscles of the leg (tibialis anterior and extensor digitorum longus) through multiple intramuscular injections (n = 30) with a total dose of 3.5 × 1012 vg of the AAV1-U7E6/8. Overall dystrophin expression in transduced muscle was verified in one dog, 3 months after injection. Histological analysis revealed numerous dystrophin-positive fibers all along the entire muscle (Supplementary Figure S5). Based on this result, the three other dogs underwent, at 4-months postinjection, functional evaluation through NMR and tetanic force measurements. Muscle biopsies were taken afterwards to confirm dystrophin expression.
Overall improvement of treated muscles was appraised by NMR. A series of images were acquired before and after intravenous bolus injection of Gadolinium-DTPA.28 Two indices, one reflecting muscle tissue disorganisation (muscle signal heterogeneity on T2-weighed images), and one evaluating muscle necrosis and fibrosis (muscle maximum relative enhancement post Gadolinium-chelate i.v. injection), were found to have less variance and, on average, lower values in treated muscles than in untreated ones [heterogeneity range: (1.44–3.75) and (1.46–1.91) for untreated and treated muscles respectively, maximum relative enhancement range: (0.74–1.47) and (0.68–0.96) for untreated and treated muscles, respectively] (Figure 6). Maximum relative enhancement clearly discriminated treated muscles from untreated ones: all values of treated muscles were below those of the untreated muscles. Interestingly, anterior compartment muscle volume, normalized to body weight, was unchanged allowing direct comparison of force measurements.
Figure 6.
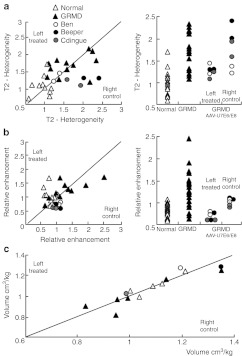
NMR assessment of cranial muscles of the leg after whole muscle transduction. (a) On T2-weighted images, muscle signal heterogeneity (H2) was calculated as follows: H2 = √(SD2T2w – SD2noise/0.655), with SDT2w representing the standard deviation of the signal in the muscle ROI and SD noise, the standard deviation of the noise in the same slice. (b) After bolus injection of 0.1 mmol/kg of Gd-D, the dynamics of the fat-saturated T1-weighted muscle intensity changes was monitored for 1 hour. Using the maximal post-gadolinium signal intensity (maxT1w signal) and the pre-gadolinium signal intensity (pre-T1w signal), muscle maximal relative enhancement was calculated as: rE = (pre-T1w signal – maxT1w signal)/pre-T1w signal. These indices were shown to be significantly higher in dystrophic muscles as compared to healthy muscles.28 As indicated by the asymmetric distribution of measurements, H2 and rE were most often lower in the treated than in the untreated muscles and all treated muscles fell within the normal range. (c) After 3D imaging, ROIs delineating the anterior compartment were drawn by hand and the anterior compartment volumes were calculated and normalized to body weight. No systematic difference was observed between the treated and the untreated sides. The relative muscle volumes were not different between healthy, dystrophic and treated dogs allowing direct comparison of force measurement.
Strength of treated cranial muscles was assessed by measuring the maximal isometric force after supramaximal tetanic stimulation of the common fibular nerve. Contralateral untreated cranial muscles were used as internal controls given that tetanic force measurements of both legs were routinely comparable in a given individual both in healthy (n = 5) and GRMD (n = 10) dogs (Supplementary Figure S6), as previously reported.29 Tetanic forces developed by treated muscles were systematically improved compared to their respective control muscles, whatever the weight and the severity of the dog's phenotype, which varied according to the GRMD genetic background. Figure 7a shows tetanic contractions of cranial muscles in a treated GRMD dog, 4 months after injection, compared to an age-matched healthy dog. Although this dog displayed the clinical hallmarks of a 5-month-old GRMD dog, the tetanic strength of its treated muscle was almost double that of its untreated contralateral muscle. Box-and-whisker plots of the measurements acquired from the three treated GRMD against untreated GRMD and healthy age-matched dogs show that relative force (N/kg) was always improved in muscles injected with AAV1-U7E6/8 compared to their untreated contralaterals (Figure 7b). A quantitative evaluation of the extent of recovery of normal characteristics can be assessed by comparing the difference between treated and untreated GRMD muscles, to the difference between normal and untreated GRMD muscles.30 Here, the recovery scores were ~25–50%.
Figure 7.
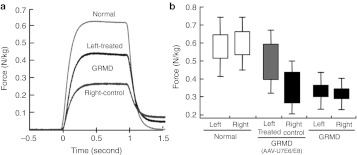
Muscle function recovery assessed by isometric tetanic contractions. (a) Example of weight-corrected maximal isometric tetanic contractions of the tibialis anterior compartment in a 4-month-old dystrophic dog (untreated and treated legs) compared to an age-matched control dog. Return to the baseline after stimulation shows a muscular memory, which is characteristic of the dystrophic muscle and is decreased in the treated compartment. (b) The box and whiskers plot shows the force, the error-type interval (height of the box, 95% confidence interval) and the standard deviation (top and bottom of the whiskers). Contraction forces of treated compartments (gray) are significantly increased compared to untreated counterparts (black) and moves toward normal (white). A Mann–Whitney U-test confirmed that force variation between left and right muscles were statistically significant (P < 0.05) for treated dogs.
Dystrophin rescue after limb perfusion with AAV1-U7E6/8
In order to assess dystrophin recovery in multiple muscles following a single injection, we performed intravenous perfusion of AAV1-U7E6 and AAV1-U7E8 into the forelimb of adult GRMD dogs (n = 2). Outflow obstruction combined with a rapid injection into the accessory cephalic vein of a large volume bolus was essential for efficient vector delivery to surrounding muscles. Iterative muscle biopsies were taken from 2 months up to 56 months after vector delivery (Table 1). Muscle to muscle variations were observed and the extensor digitorum communis muscle was usually more efficiently transduced than the flexor carpi ulnaris muscle (Table 1). As shown in Figure 8, muscle biopsies at 2 months showed numerous dystrophin positive fibers: these could be seen scattered throughout whole sections (Figure 8a for FCU muscle) or grouped alongside negative areas (Figure 8b for EDC muscle) indicating that muscle transduction was not homogenous.
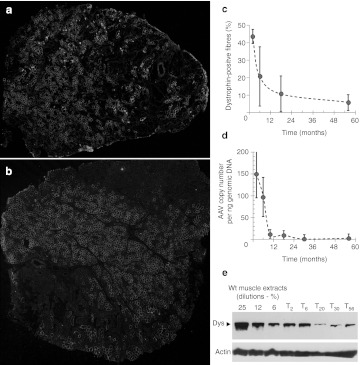
Figure 8.
Lasting effect of AAV1-U7E6E8 in golden retriever muscular dystrophy dog (GRMD) after forelimb locoregional delivery. Dystrophin immunostaining (NCL-DYS2) in transverse sections of muscles: (a) flexor carpi ulnaris (FCU), and (b) extensor digitorum communis (EDC), 2 months after high pressure intravenous delivery of adeno-associated virus (AAV) vectors into the forelimb. Amounts of rescued dystrophin were variable from one biopsy to another, sometimes: dystrophin-positive fibers were scattered throughout the biopsy (a), while a biopsy could show large domains fully transduced next to negative areas (b). (c) Percentage of dystrophin-positive fibers found in muscles biopsies taken at different time points. (d) Quantification of the AAV genome copy number in biopsies at different time points. (e) Western blot of total protein (100 µg) extracted from biopsies at different time points stained with NCL-DYS2. Arrow indicates the full-length dystrophin as detected in samples from normal muscle used for comparison (25, 12, and 6%—diluted in GRMD to keep up total protein content). Samples were also hybridized with an actin-antibody to validate protein loadings.
Dystrophin positive fibers were easily identified in all muscle biopsies taken at 2, 6, 18, and 56 months after injection. However, the number and the cluster size of dystrophin positive fibers clearly decreased with time with no obvious sign of immune rejection. Roughly, over the 5-year period of follow-up, the number of dystrophin positive fibers was decreased by at least eightfolds (Figure 8c). The overall decrease of dystrophin, as measured by Western blot, paralleled the loss of the U7 platform as measured by quantitative PCR of the AAV1-U7E6/8 genome in muscle biopsies (Figure 8d, e).
Discussion
In this work, we have designed AAV1 gene vectors encoding optimized antisense-U7 snRNA chimeras to achieve dystrophin rescue in the GRMD dog model of DMD by means of exon skipping. This approach has already been used to drive sustained restoration of dystrophin in skeletal muscle of the mdx mouse31,17 and cardiac muscle of GRMD.22 The optimized U7 gene is thought to continuously supply antisense within transduced fibers and its activity is enhanced by the U7 snRNA moiety which aids proper subcellular localization and inclusion into the pre-mRNA processing machinery.32 Here, we have examined dystrophin expression and its consequences in skeletal muscles of GRMD by using two distinct modes of delivery of AAV1 vectors: intramuscular and intravenous injections. We demonstrated rescue of dystrophin, proper localization of its associated glycoprotein complex, and the lessening of various indicators of dystrophic severity. Utrophin was downregulated toward levels found in healthy muscles. This protein is a dystrophin-related protein, which is normally expressed in capillaries and confined in adult muscle to the neuromuscular and myotendinous junctions, whereas it is found along the sarcolemma in developing muscle, and persists in dystrophic fibers in the absence of dystrophin.33 Downregulation of utrophin in transduced fibers could be seen as a step toward a recovered differentiation. Similarly, intracellular calcium leakage, which is known to coincide with progressive dystrophy and muscle weakness, was significantly reduced in a vector dose dependant manner in transduced areas, indicating that the truncated dystrophin improved muscle fiber resilience. However, this improved resilience did not preclude the possibility that, rather than a definitive escape from dystrophic fate, transduction of fibers merely postponed damage, such that it occurs over a longer time period as in the classical BMD condition compared with DMD.
In order to assess functional improvement, we performed extensive transduction of the cranial leg muscle compartment in 3-week-old GRMD puppies. These muscles were chosen because they were easily accessible and could be filled with AAV vectors by means of multiple injections. We analysed muscle function 4 months later by both NMR and tetanic force measurements. By that time, although cranial leg muscle sizes had followed the GRMD growth curve, treated muscles still displayed numerous dystrophin-positive fibers. Such maintenance was not expected since many nontransduced nuclei likely contributed to muscle growth. Nonetheless, this lack of apparent dystrophin dilution might be explained by the properties of the U7-system. Indeed, alike all spliceosomal snRNAs, engineered U7E6/8 snRNA has to be processed into U7E6/8 small nuclear ribonucleoprotein in the cytoplasm before reimport into the nucleus to participate with the spliceosome machinery. Therefore, it is possible that the U7-system supports a broadening of the extent of dystrophin rescue within growing muscle fibers as U7E6/8 small nuclear ribonucleoproteins produced by a transduced myonucleus could be conveyed to nearby nontransduced nuclei. NMR indices, although greatly improved, were not identical to healthy controls. This may have been due to the levels of dystrophin rescue, which were obviously not complete, or to the partial functionality of the rescued dystrophin, which lacked part of its actin-binding domain, or to an irreversibility of some features of the dystrophic condition. Indeed, it has been reported that dystrophins with deletions in the actin-binding domain are unstable. Patients with such in frame deletions tend to have a more severe BMD phenotype.34 As well, in the mdx mouse, delivery of an AAV encoding a truncated-dystrophin similar to the one obtained by splice-switching in GRMD gave rise to an unstable end-product, which was quickly lost in contrast to what was observed by using mini-dystrophins retaining the actin-binding domain.35 Nonetheless, subsequent force measurements confirmed the beneficial effect of AAV1-U7E6/8, and it should here be borne in mind that isometric maximal force was measured for the whole leg, so performances of individually treated cranial muscle were inevitably impacted and mitigated by the malfunctioning of untreated posterior muscles.
Hydrodynamic limb perfusion of AAV1-U7E6 and AAV1-U7E8 was carried out in order to target multiple muscles following a single injection. We analysed dystrophin recovery at different time points: whatever the mode of vector delivery (intramuscular or hydrodynamic perfusion), biopsies taken from 2 weeks after administration showed a rising of dystrophin expression culminating at 2–3 months, then declining progressively from 6 to 56 months. This longitudinal study allowed appraisal of the persistence of the AAV1-U7E6 and AAV1-U7E8 treatment in a severe dystrophic context. Biopsies showed dystrophin-positive fibers at all time points, although numbers were found to decline progressively with age. Early studies in dogs and nonhuman primates have shown both safety and persistence of AAV gene transfer in non-dystrophic skeletal muscle,36,37,38,39 while more recent reports have documented a strong immune response directed against either the transgene product or the vector capsid.40 Increased T-cell responses have been documented using different transgenes, AAV serotypes and species, leading to the loss of transduced fibers within weeks after delivery.41 Such an immune response disparity was also found in phase 1 clinical trials using intramuscular delivery of AAV vectors. A limited immune response against the vector capsid has been observed in trials for LGMD2D42 and LGMD2C,43 which did not preclude gene expression, contrasting with the rapid loss of gene expression following AAV1-LPL delivery in lipoprotein lipase deficiency44 or likely in DMD following intramuscular delivery of modified AAV-2 encoding a truncated dystrophin.45 In our study, the vanishing of dystrophin expression was likely due to the unstable nature of the rescued dystrophin rather than resulting from an immune rejection. In a recent report Mendell et al., have shown that DMD patients might display T-cell immune responses against dystrophin before gene therapy treatment46 giving an obvious explanation for the failure of the clinical trial. Although we did not carry out T-cell assays in our study, it is unlikely that such a phenomenon was involved in the present case. T-cell–mediated immune reactions usually peak around 2 weeks after administration and are gone by 4–6 weeks: recipients are then regarded as immunized. Although T-cell assays might reveal it, one can also consider that the foremost consequence of this immune response would be that transduced cells or dystrophin-expressing cells should be cleared within a few weeks. Noticeably, such clearing did not occur here and dystrophin rescue was sustained for a quite long period of time without observable sign of rejection at the histological level. Having no observable evidence of immune rejection (absence of rising antibodies against rescued dystrophin and no massive infiltration of T-cells around dystrophin positive fibers), we tend to believe that the decline we observed was due to the ongoing pathological process itself, which was not entirely stopped by expressing a truncated dystrophin that did not have the full functionality of a normal full-length dystrophin. Even though expressing such a truncated dystrophin markedly improved muscle function, we must keep in mind that it merely converted transduced GRMD fibers into BMD-like fibers, which are known to be less resilient than normal fibers as very mild BMD patients still display elevated serum CK activities. In BMD, damaged fibers are replaced by new BMD fibers, while the same degenerating progression in AAV-transduced GRMD muscles would gradually clear the U7E6/8 expression cassette, leading inexorably to the reconstitution of the original GRMD phenotype. Although this concern was theoretically expected, it needed a long-term follow-up to be noticed in a large animal model mimicking the course of DMD.
Nonetheless, we cannot extrapolate that any gene therapy treatment for DMD would follow a similar decline, since this depends upon the effectiveness of the therapeutic transgene. Appropriate mini-dystrophins as well as various exon-skipping patterns giving rise to functional proteins of high stability should allow long-lasting benefit. On the other hand, exon skipping strategies giving rise to poorly stable truncated dystrophins will mandatorily require recurrent treatments to maintain therapeutic benefit ahead of the progression of the disease. This option is not presently open to the AAV approach for the reason that a first exposure to the vector triggers at least a sustained immune response generating AAV-neutralizing antibodies, which precludes further readministrations.47 Nonetheless, one could imagine a two-step approach of long lasting clinical utility: the first step employing a starting treatment with AAV-U7, to establish an in situ factory for copious antisense production; the second step consisting in periodic injections of synthetic antisense oligomers, which are not immunogenic, to maintain exon skipping levels over time (Supplementary Figure S7). Both delivery modes are possible in the GRMD context: widespread muscle expression after a single intravenous injection of an AAV9 encoding a mini-dystrophin has been shown feasible, at least in neonate GRMD dogs,48 and recurring systemic injections of a phosphorodiamidate morpholino oligomer-cocktail for several weeks have allowed extensive dystrophin rescue throughout the whole skeletal musculature in dystrophin-deficient dogs.23
Finally, it is understood that the pre-existing immunity against dystrophin which was noticed in some DMD patients might preclude or inhibit dystrophin rescue strategies, particularly those using viral vectors that are immunogenic.46 The lack of immune concerns in clinical trials using systemic delivery of either 2OMe or phosphorodiamidate morpholino oligomers synthetic oligomers for skipping exon 5112,14 advocates for this type of personalized gene correction approach. However, this latter is only conceivable for patients with mutations for which the skipped transcript will encode a highly stable and functional dystrophin. For patients with mutations that do not meet this criteria, standard gene delivery of either a full-length dystrophin49 or deeply reshuffled mini-dystrophin46,48 is still a valuable approach provided that immunological problems are overcome both in terms of dystrophin tolerance and of gene vector readministration to achieve widespread delivery supporting clinically relevant levels of dystrophin throughout the whole life-span of the treated individual.
Materials and Methods
AAV vectors. The following antisense sequences were introduced into the U7smOPT gene as described previously:17
E6: 5′-ACATTAACCTGTGGATAATTACGAGTTGATTGTCGGACCCAG -3′
E8: 5′-ACTTGTTGAGGCAAAACTTGGAAGAGTGATGTGATATACA -3′
U7E6 and U7E8 sequences were introduced either separately or in tandem (U7E6/8) at the XbaI site of the pSMD2 AAV-2 genome. AAV1 pseudotyped vectors were prepared by transfection in 293 cells as described and vector particles were purified on cesium chloride gradients from cell lysates obtained 48 hours after transfection and titered by quantitative dot blot hybridization.
Animals. GRMD animals were part of a dog breeding colony established in France. All procedures were in accordance with the institutional and European agreement for humane treatment of animals. For all the injection procedures and the surgical biopsies, sedation was achieved with propofol (Rapinovet ND, 6.5 mg/kg i.v.) and anesthesia was maintained with 2% isoflurane. Muscle biopsies were snap-frozen in liquid nitrogen-cooled isopentane and stored at –80 °C. Table 1 summarizes the animals, the type of injection and the biopsies involved in the study.
Assessment of multi-exon skipping. Intramuscular injections were carried out in six GRMD dogs. In each subject, two to six individual bundles of proximal muscles were injected (Biceps femoris, triceps brachii, or cranial Sartorius muscles), contralateral muscles were used as control. Three dogs (Vrac, Virbac, and Adhoc) received a mixture of AAV1-U7E6 (3.5 × 1012 vg) and AAV1-U7E8 (3.5 × 1012 vg) in 1-ml phosphate-buffered saline. The three other dogs (Ajax, Athos, and Cparti) were injected with 3.5 × 1012 vg of the tandem vector AAV1-U7E6/E8. Surgical biopsies of the injected bundles were carried out at 1, 2, 3, and 6 months after injection. Each bolus of vector was delivered by using multiple injections of 50 µl by means of a Hamilton syringe.
Dose-escalation study. Eight individual bundles were injected with different doses of the combined vector AAV1-U7E6/E8) (phosphate-buffered saline, 5 × 1010 vg, 1011 vg, 2.5.1011 vg, 5.1011 vg, 1012 vg, 2.1012 vg, and 3.5.1012 vg) in 500 µl. This experiment was performed in one GRMD dog (Ajax) in two different muscles. Biopsies were taken two months after injection in triceps brachialis muscles and 6 months after injection in biceps femoris muscles. Each bolus of vector was delivered by using multiple injections of 50 µl by means of a Hamilton syringe.
Locoregional delivery. Two GRMD dogs (Athos and Adhoc, respectively aged 5 and 6 months) underwent high pressure intravenous injection in one of their forelimbs. They both received 1.4 × 1013 vg of AAV1-U7E6 mixed with the same dose of AAV1-U7E8, diluted in 8 ml of buffer per kg of body weight (i.e., Athos: 100 ml; Adhoc: 116 ml for the second one). This high volume was infused through a catheter introduced into the distal part of the accessory cephalic vein, after having tightened a tourniquet proximally to the elbow in order to transiently isolate the forearm vasculature point of view as previously described.50,51 The injection was made at a rate of 1 ml/second, meaning that the whole volume was injected within 2 minutes. Muscle biopsies (extensor digitorum communis and the flexor carpi ulnaris) were carried out at 2, 3, 6, 10, 18, 30, and 56 months after injection.
Muscle function and NMR. Four 3-week-old GRMD puppies (Byblos, Ben, Beeper, and Cdingue) were injected intramuscularly in their whole left cranial leg muscle compartment (tibialis cranialis, extensor digitorum longus), using 30 × 50 µl injections delivering a total dose of 3.5 × 1012 vg of the combined vector AAV1-U7E6/E8. One dog (Byblos) died at the age of 3 months from acute pneumonia and could not undergo functional testings. However, histological analysis was performed to confirm dystrophin recovery after AAV1-U7E6/E8 delivery.
At 4-months postinjection, the three remaining dogs underwent in vivo tetanic force measurement of the cranial muscles of both legs, using supramaximal stimulation of the fibular nerve as previously described.52 The injected leg (left side) was compared to the noninjected contralateral leg (right side). A recovery score can be defined as [(treated – untreated)/(normal – untreated)] × 100 for the maximal force; where 100% define the normal condition and 0% the GRMD condition.
At the same time, NMR imaging of the entire hind limb was performed on a whole-body 3T Trio TIM Siemens spectrometer using a birdcage transmitter-receiver coil. T1- and T2-weighed SE images were acquired with and without fat suppression. Inframillimetric (0.6 × 0.6 × 0.6) resolution 3D GE FLASH with fat-suppression was also obtained. The dynamics of muscle signal enhancement post-Gd chelate injection was monitored during 60 minutes using an inversion-recovery Turbo-FLASH sequence.
The volume of the cranial compartment (tibialis cranialis and the extensor digitorum lateralis muscles) was determined by manually tracing contours on the stacks of 3D GE images and by summing surfaces over the muscle length. Muscle structural heterogeneity was assessed on T2-weighted images. The up slopes, maximum enhancement, time to maximum enhancement and down slopes of the post Gd-chelate enhancement were determined by linear fitting. Finally, both tibialis cranialis muscles were harvested, in order to evaluate dystrophin rescue.
Histology. Serial 8-µm transverse sections, cut at 500-µm intervals over the muscle biopsies, were examined for dystrophin using the NCL-DYS2 monoclonal antibody to the C-terminal domain and for utrophin, α-dystroglycan, α- and β-sarcoglycan by immunohistochemistry (all antibodies from Novocastra, Newcastle upon Tyne, UK). Monoclonal antibodies were detected by an Alexa488 goat anti-mouse secondary antibody. The presence of calcium-overloaded muscle fibers was assessed using an Alizarin red S stain at pH 5.4, a calcium-specific stain commonly used on frozen muscle sections.27 Dried sections were stained during 5 minutes in a 2% ARS solution at pH 5.4. They were rinsed and dehydrated in acetone and acetone–xylene volume/volume, and mounted in Canada balsam. Intermediate tissue was collected for mRNA and protein analysis.
mRNA analysis. Total RNA was isolated from pooled intermediate sections using TRIZOL-reagent (Life Technologies, Saint-Aubin, France). To detect dystrophin mRNA, nested reverse transcription-PCR was carried out with 200 ng of total RNA using Acess reverse transcription-PCR system (Promega, Charbonnières, France). The first reaction was performed with the following primers for 30 cycles (94 °C/30 s; 58 °C/1 minutes; 72 °C/2 minutes).
Ex3 ext (5′-GGAAGCAGCACATAGAGAGAAC-3′) Ex10 ext (5′-TCACTTCTTCGACATCATTAG-3′) Then, 2 µl of the first reaction were amplified for 25 cycles with: Ex3 int (5′-GAGACGCCTCCTAGACCTT-3′) Ex10 int (5′-CTTCCAAAGCTGTTTGATAAC-3′)
PCR products were analyzed on 2% agarose gels, and individual bands were purified for sequence analysis.
Quantification of AAV copy number in muscle biopsies. Genomic DNA was prepared from 240 µm of muscle (TB, BF, or EDC) using the PureLink Genomic DNA Mini Kit (ref K182002; Life Technologies), according to the manufacturer's instructions. Real-time quantitative PCR was performed on Opticon2 (Bio-Rad, Marnes-la-Coquette, France) using ABsolute QPCR ROX Mix (ref AB-1139, Thermoscientific, Illkirch, France).
Primers:
AAV-22mers-F: CTCCATCACTAGGGGTTCCTTG AAV-18mers-R: GTAGATAAGTAGCATGGC TaqMan probe AAV_MGB.P: 6-FAM-TAGTTAATGATTAACCC-MGB
The AAV genome number was measured on 100 ng of genomic DNA and absolute quantification was done by using a standard curve made with different dilutions of a plasmid containing inverted terminal repeats sequences, as AAV expression cassette.
Western blot analysis. Protein extracts were obtained from pooled tissue sections treated with 4% sodium dodecyl sulfate, 125 mmol/l Tris–HCl pH 6.4, 4 mol/l urea, 10% β-mercaptoethanol, 10% glycerol, and 0.001% bromophenol blue. Forty microgram of protein were loaded onto 3–8% polyacrylamide gels, electrophoresed, blotted onto nitrocellulose membranes and probed with 1:50 NCL-DYS2, followed by incubation with a horseradish peroxidase-conjugated secondary antibody (1:1,000) and ECL Analysis System (Amersham, Courtaboeuf, France).
Detection of antibodies against AAV particles. Serum antibody titers against AAV1 capsid were determined by enzyme-linked immunosorbent assay. Briefly, 109 AAV1 particles were coated onto 96-wells plates (MaxiSorp, Nunc) overnight at 4 °C. Plates were blocked with BD Assay Diluent (BD Biosciences, Le Pont de Claix, France) at room temperature for 1 hour, washed with phosphate-buffered saline-0.05% Tween-20 and serial fivefold dilutions of canine sera diluted in BD Assay Diluent were added and incubated for 1 hour at 37 °C. Plates were washed three times as previously, and horseradish peroxidase-conjugated goat anti-dog IgG was added and incubated for 1 hour at 37 °C. Finally, plates were washed three times again, revealed with TMB Substrate reagent Set (BD Biosciences) and the OD at 450 nm was measured in the enzyme-linked immunosorbent assay plate reader.
SUPPLEMENTARY MATERIAL Figure S1. Design of the AAV-U7 vectors for skipping exons 6 and 8. Figure S2. Dystrophin disruption in normal muscle following intramuscular delivery of either AAV1-U7E6 or AAV1-U7E8 alone. Figure S3. Serum antibody titers for AAV1 capsid elicited by AAV1-U7 in GRMD dogs. Figure S4. Cellular and humoral response against full-length dystrophin after plasmid delivery in GRMD. Figure S5. Extent of the dystrophin rescue after transduction of the whole cranial muscles of the leg by using multiple injections. Figure S6. Plot of right versus left tetanic flexion measurements (cranial muscles of the legs) obtained in 4-month-old wild type and GRMD dogs. Figure S7. Expected DMD phenotype evolution following AAV-U7 treatment.
Acknowledgments
This work was supported by the Association Française contre les Myopathies, the Association Monegasque contre les Myopathies and Duchenne Parent Project France. We thank Arnaud Jollet, and Genethon and the UPR de Neurobiologie core facilities for technical assistance, as well as the Centre d'Elevage du Domaine des Souches for breeding the French GRMD colony.
Supplementary Material
Design of the AAV-U7 vectors for skipping exons 6 and 8.
Dystrophin disruption in normal muscle following intramuscular delivery of either AAV1-U7E6 or AAV1-U7E8 alone.
Serum antibody titers for AAV1 capsid elicited by AAV1-U7 in GRMD dogs.
Cellular and humoral response against full-length dystrophin after plasmid delivery in GRMD.
Extent of the dystrophin rescue after transduction of the whole cranial muscles of the leg by using multiple injections.
Plot of right versus left tetanic flexion measurements (cranial muscles of the legs) obtained in 4-month-old wild type and GRMD dogs.
Expected DMD phenotype evolution following AAV-U7 treatment.
REFERENCES
- Hoffman EP, Brown RH., Jr, and, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Chelly J, Kaplan JC, Maire P, Gautron S., and, Kahn A. Transcription of the dystrophin gene in human muscle and non-muscle tissue. Nature. 1988;333:858–860. doi: 10.1038/333858a0. [DOI] [PubMed] [Google Scholar]
- Bonilla E, Samitt CE, Miranda AF, Hays AP, Salviati G, DiMauro S.et al. (1988Duchenne muscular dystrophy: deficiency of dystrophin at the muscle cell surface Cell 54447–452. [DOI] [PubMed] [Google Scholar]
- Petrof BJ, Shrager JB, Stedman HH, Kelly AM., and, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery AEH.1993Duchenne Muscular Dystrophy2nd edn., 1–392.
- Cotton S, Voudouris NJ., and, Greenwood KM. Intelligence and Duchenne muscular dystrophy: full-scale, verbal, and performance intelligence quotients. Dev Med Child Neurol. 2001;43:497–501. doi: 10.1017/s0012162201000913. [DOI] [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C., and, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Muntoni F, Torelli S., and, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- Matsuo M. Duchenne/Becker muscular dystrophy: from molecular diagnosis to gene therapy. Brain Dev. 1996;18:167–172. doi: 10.1016/0387-7604(96)00007-1. [DOI] [PubMed] [Google Scholar]
- Lu QL, Yokota T, Takeda S, Garcia L, Muntoni F., and, Partridge T. The status of exon skipping as a therapeutic approach to duchenne muscular dystrophy. Mol Ther. 2011;19:9–15. doi: 10.1038/mt.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A. Antisense-mediated modulation of splicing: therapeutic implications for Duchenne muscular dystrophy. RNA Biol. 2010;7:453–461. doi: 10.4161/rna.7.4.12264. [DOI] [PubMed] [Google Scholar]
- Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K.et al. (2011Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study Lancet 378595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K.et al. (2011Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study Lancet 378595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C.et al. (2009Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study Lancet Neurol 8918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M.et al. (2007Local dystrophin restoration with antisense oligonucleotide PRO051 N Engl J Med 3572677–2686. [DOI] [PubMed] [Google Scholar]
- Gorman L, Suter D, Emerick V, Schümperli D., and, Kole R. Stable alteration of pre-mRNA splicing patterns by modified U7 small nuclear RNAs. Proc Natl Acad Sci USA. 1998;95:4929–4934. doi: 10.1073/pnas.95.9.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L.et al. (2004Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping Science 3061796–1799. [DOI] [PubMed] [Google Scholar]
- Goyenvalle A, Babbs A, van Ommen GJ, Garcia L., and, Davies KE. Enhanced exon-skipping induced by U7 snRNA carrying a splicing silencer sequence: Promising tool for DMD therapy. Mol Ther. 2009;17:1234–1240. doi: 10.1038/mt.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine BA, Winand NJ, Pradhan D, Moise NS, de Lahunta A, Kornegay JN.et al. (1992Canine X-linked muscular dystrophy as an animal model of Duchenne muscular dystrophy: a review Am J Med Genet 42352–356. [DOI] [PubMed] [Google Scholar]
- Sharp NJ, Kornegay JN, Van Camp SD, Herbstreith MH, Secore SL, Kettle S.et al. (1992An error in dystrophin mRNA processing in golden retriever muscular dystrophy, an animal homologue of Duchenne muscular dystrophy Genomics 13115–121. [DOI] [PubMed] [Google Scholar]
- Barbash IM, Cecchini S, Faranesh AZ, Virag T, Li L, Yang Y.et al. (2012MRI roadmap-guided transendocardial delivery of exon-skipping recombinant adeno-associated virus restores dystrophin expression in a canine model of Duchenne muscular dystrophy Gene Ther [DOI] [PMC free article] [PubMed]
- Bish LT, Sleeper MM, Forbes SC, Wang B, Reynolds C, Singletary GE.et al. (2012Long-term restoration of cardiac dystrophin expression in golden retriever muscular dystrophy following rAAV6-mediated exon skipping Mol Ther 20580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, Takeda S.et al. (2009Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs Ann Neurol 65667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatzberg SJ, Anderson LV, Wilton SD, Kornegay JN, Mann CJ, Solomon GG.et al. (1998Alternative dystrophin gene transcripts in golden retriever muscular dystrophy Muscle Nerve 21991–998. [DOI] [PubMed] [Google Scholar]
- McClorey G, Moulton HM, Iversen PL, Fletcher S., and, Wilton SD. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13:1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- Blake DJ, Tinsley JM., and, Davies KE. Utrophin: a structural and functional comparison to dystrophin. Brain Pathol. 1996;6:37–47. doi: 10.1111/j.1750-3639.1996.tb00781.x. [DOI] [PubMed] [Google Scholar]
- Valentine BA, Cooper BJ., and, Gallagher EA. Intracellular calcium in canine muscle biopsies. J Comp Pathol. 1989;100:223–230. doi: 10.1016/0021-9975(89)90099-6. [DOI] [PubMed] [Google Scholar]
- Thibaud JL, Monnet A, Bertoldi D, Barthélémy I, Blot S., and, Carlier PG. Characterization of dystrophic muscle in golden retriever muscular dystrophy dogs by nuclear magnetic resonance imaging. Neuromuscul Disord. 2007;17:575–584. doi: 10.1016/j.nmd.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Kornegay JN, Bogan DJ, Bogan JR, Childers MK, Cundiff DD, Petroski GF.et al. (1999Contraction force generated by tarsal joint flexion and extension in dogs with golden retriever muscular dystrophy J Neurol Sci 166115–121. [DOI] [PubMed] [Google Scholar]
- Deconinck N, Tinsley J, De Backer F, Fisher R, Kahn D, Phelps S.et al. (1997Expression of truncated utrophin leads to major functional improvements in dystrophin-deficient muscles of mice Nat Med 31216–1221. [DOI] [PubMed] [Google Scholar]
- Goyenvalle A, Babbs A, Wright J, Wilkins V, Powell D, Garcia L.et al. (2012Rescue of severely affected dystrophin/utrophin-deficient mice through scAAV-U7snRNA-mediated exon skipping Hum Mol Genet 212559–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümperli D., and, Pillai RS. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci. 2004;61:2560–2570. doi: 10.1007/s00018-004-4190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerk A, Morris GE, Dubowitz V, Davies KE., and, Sewry CA. Dystrophin-related protein, utrophin, in normal and dystrophic human fetal skeletal muscle. Histochem J. 1993;25:554–561. [PubMed] [Google Scholar]
- Beggs AH, Hoffman EP, Snyder JR, Arahata K, Specht L, Shapiro F.et al. (1991Exploring the molecular basis for variability among patients with Becker muscular dystrophy: dystrophin gene and protein studies Am J Hum Genet 4954–67. [PMC free article] [PubMed] [Google Scholar]
- Banks GB, Gregorevic P, Allen JM, Finn EE., and, Chamberlain JS. Functional capacity of dystrophins carrying deletions in the N-terminal actin-binding domain. Hum Mol Genet. 2007;16:2105–2113. doi: 10.1093/hmg/ddm158. [DOI] [PubMed] [Google Scholar]
- Manno CS, Chew AJ, Hutchison S, Larson PJ, Herzog RW, Arruda VR.et al. (2003AAV-mediated factor IX gene transfer to skeletal muscle in patients with severe hemophilia B Blood 1012963–2972. [DOI] [PubMed] [Google Scholar]
- Chenuaud P, Larcher T, Rabinowitz JE, Provost N, Joussemet B, Bujard H.et al. (2004Optimal design of a single recombinant adeno-associated virus derived from serotypes 1 and 2 to achieve more tightly regulated transgene expression from nonhuman primate muscle Mol Ther 9410–418. [DOI] [PubMed] [Google Scholar]
- Rivera VM, Gao GP, Grant RL, Schnell MA, Zoltick PW, Rozamus LW.et al. (2005Long-term pharmacologically regulated expression of erythropoietin in primates following AAV-mediated gene transfer Blood 1051424–1430. [DOI] [PubMed] [Google Scholar]
- Herzog RW, Hagstrom JN, Kung SH, Tai SJ, Wilson JM, Fisher KJ.et al. (1997Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus Proc Natl Acad Sci USA 945804–5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Allen JM, Riddell SR, Gregorevic P, Storb R, Tapscott SJ.et al. (2007Immunity to adeno-associated virus-mediated gene transfer in a random-bred canine model of Duchenne muscular dystrophy Hum Gene Ther 1818–26. [DOI] [PubMed] [Google Scholar]
- Wang Z, Tapscott SJ, Chamberlain JS., and, Storb R. Immunity and AAV-Mediated Gene Therapy for Muscular Dystrophies in Large Animal Models and Human Trials. Front Microbiol. 2011;2:201. doi: 10.3389/fmicb.2011.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Rodino-Klapac LR, Rosales XQ, Coley BD, Galloway G, Lewis S.et al. (2010Sustained alpha-sarcoglycan gene expression after gene transfer in limb-girdle muscular dystrophy, type 2D Ann Neurol 68629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herson S, Hentati F, Rigolet A, Behin A, Romero NB, Leturcq F.et al. (2012A phase I trial of adeno-associated virus serotype 1-?-sarcoglycan gene therapy for limb girdle muscular dystrophy type 2C Brain 135Pt 2483–492. [DOI] [PubMed] [Google Scholar]
- Mingozzi F, Meulenberg JJ, Hui DJ, Basner-Tschakarjan E, Hasbrouck NC, Edmonson SA.et al. (2009AAV-1-mediated gene transfer to skeletal muscle in humans results in dose-dependent activation of capsid-specific T cells Blood 1142077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles DE, McPhee SW, Li C, Gray SJ, Samulski JJ, Camp AS.et al. (2012Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector Mol Ther 20443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Campbell K, Rodino-Klapac L, Sahenk Z, Shilling C, Lewis S.et al. (2010Dystrophin immunity in Duchenne's muscular dystrophy N Engl J Med 3631429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorain S, Gross DA, Goyenvalle A, Danos O, Davoust J., and, Garcia L. Transient immunomodulation allows repeated injections of AAV1 and correction of muscular dystrophy in multiple muscles. Mol Ther. 2008;16:541–547. doi: 10.1038/sj.mt.6300377. [DOI] [PubMed] [Google Scholar]
- Kornegay JN, Li J, Bogan JR, Bogan DJ, Chen C, Zheng H.et al. (2010Widespread muscle expression of an AAV9 human mini-dystrophin vector after intravenous injection in neonatal dystrophin-deficient dogs Mol Ther 181501–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero NB, Braun S, Benveniste O, Leturcq F, Hogrel JY, Morris GE.et al. (2004Phase I study of dystrophin plasmid-based gene therapy in Duchenne/Becker muscular dystrophy Hum Gene Ther 151065–1076. [DOI] [PubMed] [Google Scholar]
- Hagstrom JE, Hegge J, Zhang G, Noble M, Budker V, Lewis DL.et al. (2004A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs Mol Ther 10386–398. [DOI] [PubMed] [Google Scholar]
- Thibaud JL, Huss T, Hegge J, Thioudellet C, Granger N, Gnirs K.et al. (2006Intravascular administration of dystrophin plasmid DNA in a canine model of Duchenne Muscular Dsytrophy: early side effects and long term biological efficacy J Vet Intern Med 201270–1270. [Google Scholar]
- Sampaolesi M, Blot S, D'Antona G, Granger N, Tonlorenzi R, Innocenzi A.et al. (2006Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs Nature 444574–579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Design of the AAV-U7 vectors for skipping exons 6 and 8.
Dystrophin disruption in normal muscle following intramuscular delivery of either AAV1-U7E6 or AAV1-U7E8 alone.
Serum antibody titers for AAV1 capsid elicited by AAV1-U7 in GRMD dogs.
Cellular and humoral response against full-length dystrophin after plasmid delivery in GRMD.
Extent of the dystrophin rescue after transduction of the whole cranial muscles of the leg by using multiple injections.
Plot of right versus left tetanic flexion measurements (cranial muscles of the legs) obtained in 4-month-old wild type and GRMD dogs.
Expected DMD phenotype evolution following AAV-U7 treatment.