Abstract
In quiescent tissues, minicircle DNA vectors provide at least 10 times higher sustained levels of transgene expression compared to that achieved with a canonical plasmid containing the same expression cassette. It is not known if there is a specific DNA sequence or structure that is needed for DNA silencing. To directly address this question, we substituted the bacterial plasmid DNA with various lengths of extragenic spacer DNAs between the 5′ and 3′ ends of the transgene expression cassette and determined the expression profiles using two different reporter expression cassettes. Both the human alphoid repeat (AR) and randomly generated DNA sequences of ≥1 kb in length resulted in transgene silencing while shorter spacers, ≤500 bp exhibited similar transgene expression patterns to conventional minicircle DNA vectors. In contrast, when the ≥1 kb random DNA (RD) sequences were expressed as part of the 3′-untranslated region (UTR) transgene silencing was not observed. These data suggest that the length and not the sequence or origin of the extragenic DNA flanking the expression cassette is responsible for plasmid-mediated transgene silencing. This has implications for the design of nonviral vectors for gene transfer applications as well as providing insights into how genes are regulated.
Introduction
A major limitation of nonviral plasmid vectors for gene therapy is the inability to achieve sustained therapeutic levels of transgene expression. For example, standard plasmid DNA delivery into mouse liver results in an initial high level of transgene expression that slowly declines over a period of several weeks. We have previously demonstrated that minicircle DNA vectors devoid of the plasmid DNA backbone (BB) persistently express the therapeutic transgene at tenfold to 1,000-fold higher than that of the same expression cassette from a canonical plasmid in vivo even though the amount of transfected plasmid or minicircle DNA remaining in transfected hepatocyte nuclei is the same.1 The pattern of transgene silencing and maintained expression from plasmid and minicircle, respectively is independent of the type and size of the expression cassette.1,2
In defining requirements for plasmid vector silencing in vivo, we have found that covalent attachment of the plasmid BB to the expression cassette is required for plasmid DNA silencing.2 Based on this observation, most of our current understanding of the mechanisms involved in plasmid DNA silencing have been focused specifically on the role of the plasmid-derived bacterial DNA sequence and structure. Because CpG motifs are highly enriched in the plasmid BB relative to eukaryotic expression cassettes, we previously investigated the role of CpG content and CpG DNA methylation on transgene silencing. We established that neither the CpG motifs or CpG methylation status significantly affected transgene silencing.3 In contrast, different enrichment patterns for specific modified histones from isolated minicircle and plasmid chromatinized DNA isolated from transfected mouse liver have been observed.4
Minicircle DNA vectors can be generated by different methods.1,5,6,7,8,9 Recently, we have reported a new robust system for producing minicircles—a method that is similar to routine plasmid propagation.10 This greatly simplified production scheme has allowed us to rapidly produce the vectors and further define important mechanistic parameters involved in transgene silencing of plasmid DNAs.
In the process of generating minicircle DNA, the plasmid BB is eliminated from the circular DNA construct leaving a 36 bp hybrid attR between the 5′ end of promoter region and the 3′ end of terminator region of the transgene expression cassette. Over the years we have not been able to successfully establish the role of any individual sequence within the plasmid BB DNA in plasmid silencing. Therefore, we asked whether the spatial distance between the 5′ and 3′ ends of the transgene expression cassette is the primary parameter responsible for regulating plasmid DNA silencing in vivo. To test this hypothesis, we inserted various nonbacterial, noncoding, and non-genic sequences between the 5′ and 3′ ends of the transgene expression cassette in our conventional minicircle DNA vectors and compared their relative levels of expression after transfection into mouse liver.
Results
A human alphoid repeat sequence (≥1 kb) can silence transgene expression from minicircle DNA in vivo
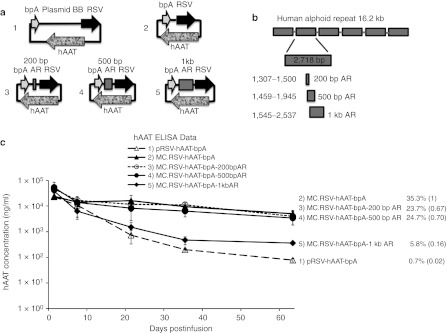
Although our previous studies have shown that covalent linkage of plasmid BB DNA to the transgene expression cassette is required for transgene silencing,2 we wanted to establish if non-plasmid, non-prokaryotic DNA sequences placed between the 3′ end of the expression cassette and 5′ end of the enhancer/promoter would also silence the expression cassette. As shown in Figure 1a, the promoter and terminator regions of the expression cassette are less than 50 bp apart in a minicircle vector. However, in plasmid DNA, the promoter and the terminator regions of gene expression cassette are separated by a plasmid BB of several kb in length. As shown in Figure 1a 1–5, multiple minicircle vectors were generated containing different lengths of exogenous DNA derived from a 16.2 kb human alphoid repeat (AR) sequence. In the first set of studies, the human AR DNA fragment was chosen as the nonbacterial, noncoding, and nongenic spacer. This segment has previously been used as an inert stuffer sequence in viral–gene deleted recombinant adenoviruses.11 The 16.2 kb human AR DNA is a centromeric sequence isolated from human chromosome 17. This centromeric sequence consists of six 2.7 kb repetitive DNA fragments and contributes to but does not define centromeric function.
Figure 1.
RSV-hAAT expression cassette constructs and transgene expression in mice. (a) Schematic of DNA constructs containing the alphoid repeats (ARs) used for injections. (b) The schematic structure of human AR and spacers generated from this sequence. The 16.2 kb human AR DNA consists of six 2.7 kb repetitive DNA fragments. A 200 bp (from 1,307 to 1,500), a 500 bp (from 1,459 to 1,945), and a 1 kb (from 1,545 to 2,537) DNA spacers were generated from a single repetitive fragment of human AR (Supplementary Figure S1 for sequence details). These three spacer sequences partially overlap. However, within each spacer, no sequences are repetitive. These AR spacers were inserted between the 5′ and the 3′ ends of expression cassette in multiple DNA constructs pictured in a. (c) DNA constructs pictured in a were injected into C57BL/6 mice respectively (n = 5 per group). Serum hAAT levels were determined over time. Two values are presented for each experimental group. The decay of hAAT expression is the level at the last time point/7-day level. The ratio of decay of expression for each transfection group/decay of expression for the corresponding minicircle transfection group (bracketed). If this ratio is <0.5, the transgene expression from a transfection group is defined as silenced. BB, backbone, ELISA, enzyme-linked immunosorbent assay; hAAT, human α1-antitrypsin.
The first expression system tested was the human α1-antitrypsin (hAAT) cDNA driven by RSV-LTR promoter. We followed transgene expression levels over time using 7 days as the early time point to allow for steady state expression and eliminate variations in early expression due to the injection procedure. We calculate the decay of expression by calculating a ratio: transgene product level at any time point after day 7/day 7 level. Transgene silencing of a plasmid vector is defined when the decay of expression (at any time point after 7 days) for a plasmid vector/minicircle vector is <0.5. The minicircle, MC.RSV-hAAT-bpA minicircle and pRSV-hAAT-bpA plasmid (including a 3 kb bacterial BB sequence) were used as the non-silenced and silenced controls, respectively. As shown in Figure 1b, 200 bp, 500 bp, and 1 kb spacers (Supplementary Figure S1 for sequence details) derived from the human AR DNA fragment were inserted in between the 5′ end of RSV-LTR promoter and the 3′ end of bovine poly A in MC.RSV-hAAT-bpA minicircle, respectively as marked in Figure 1a 3–5. These minicircle constructs, MC.RSV-hAAT-bpA-200 bp AR, MC.RSV-hAAT-bpA-500 bp AR, and MC.RSV-hAAT-bpA-1 kb AR, and control DNAs were transfected into the liver of 6–8-week-old C57BL/6 female mice through a hydrodynamic tail vein injection. The transgene expression was quantified by enzyme-linked immunosorbent assay measurement of serum hAAT protein at various time points encompassing a 2-month interval (Figure 1c). As previously shown, pRSV-hAAT-bpA was capable of expressing high levels of hAAT shortly after infusion, yet expression declined to very low levels during the next 2–3 weeks, while expression from MC.RSV-hAAT-bpA was more consistent throughout the duration of the experiment. The presence of 200 bp AR spacer and 500 bp AR spacer resulted in expression profiles similar to the minicircle whereas the 1 kb AR spacer resulted in high serum levels of hAAT signal at early time points, which rapidly declined providing an expression pattern similar to the silenced plasmid sequence (Figure 1c).
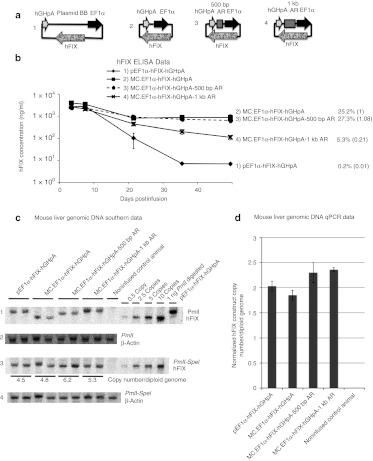
To determine if these results were consistent with different expression cassettes, we utilized a second unrelated expression cassette, a human factor IX (hFIX) producing minicircle driven by the EF1-α promoter, MC.EF1α-hFIX-hGHpA, into which we inserted the 500 bp or 1,000 bp human AR spacers (Figure 2a).
Figure 2.
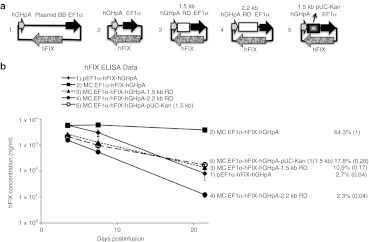
EF1α-hFIX expression cassette constructs and transgene expression in mice. (a) Schematic representation of the DNA constructs containing the alphoid repeat (AR) sequences used in the studies. (b) The DNA constructs pictured in a were injected into C57BL/6 mice (n = 5 per group). Plasma hFIX levels were determined over time. The provided values are as described in Figure 1. (c) Mouse liver genomic DNA Southern blots from 49-day postinfusion liver genomic DNA samples (n = 2 per group). Twenty microgram of each genomic DNA sample was digested with PmlI (row 1 and 2), or PmlI and SpeI (row 3 and 4) restriction enzymes to achieve single or double digestion of the infused DNA constructs, respectively; 0.5, 2.5, 5, and 10 copies of PmlI-SpeI double digested 4.6 kb MC.EF1α-hFIX-hGHpA minicircle DNA were loaded together with 20 µg noninfused control genomic DNA as copy number control. Rows 1 and 3 were probed with [P-32] dCTP-labeled 1.4 kb hFIX cDNA. Rows 2 and 4 were probed with a [P-32] dCTP-labeled 300 bp β-actin. Row 1, PmlI digestion linearized 8 kb pEF1α-hFIX-hGHpA, 4.6 kb MC.EF1α-hFIX-hGHpA, 5.1 kb MC.EF1α-hFIX-hGHpA-500 bp AR, and 5.6 kb MC.EF1α-hFIX-hGHpA-1 kb AR. Row 3, PmlI-SpeI double digestion released a 4.6 kb fragment in all groups. The corresponding DNA vector copy number per diploid genome was indicated under each band. (d) Copy number of each construct per diploid genome in 49-day postinfusion liver samples were determined by quantitative real-time PCR. Standard deviations were based on two biological samples each performed in duplicate experiments (n = 4). BB, backbone; ELISA, enzyme-linked immunosorbent assay; hFIX, human factor IX; qPCR, quantitative PCR.
As seen in Figure 2b, animals infused with the controls, MC.EF1α-hFIX-hGHpA and pEF1α-hFIX-hGHpA (including a 3.5 kb bacterial BB sequence), had persistent and transient expression profiles, respectively. The MC.EF1α-hFIX-hGHpA-500 bp AR vector produced high and prolonged plasma hFIX levels similar to that observed with the minicircle vector, while the MC.EF1α-hFIX-hGHpA-1 kb AR was incapable of maintaining high level transgene expression in vivo. Therefore, the gene expression profiles from unrelated transgenes using the same nonbacterial spacers were concordant.
In our previous studies,1,12 we determined that plasmid/episomal DNA vectors remain episomal and that the relative concentrations of minincircle and plasmid DNAs remain similar over time. To confirm that the results obtained in these studies were similar to our previous studies and not due to differential loss of episomal DNA vectors, we performed both quantitative Southern blot analysis (Figure 2c) and quantitative PCR (qPCR) assays (Figure 2d) from the livers of treated animals 7 weeks after vector administration. Restriction enzyme digest that cut the vector once established that the bulk of the vector persisted as a monocircular episome, while a two-cut vector digest was used to quantify vector copy number (Figure 2c). For all the plasmids tested, the vector copy number per diploid genome varied from 4.5 to 6.2 copies. qPCR estimated vector genome copies that ranged from 1.9 to 2.3 copies per diploid genome. These data establish that differences in vector copy are similar regardless of the vector tested and cannot explain the difference in transgene expression.
Random DNA sequences (≥1 kb) are capable of silencing transgene expression in vivo
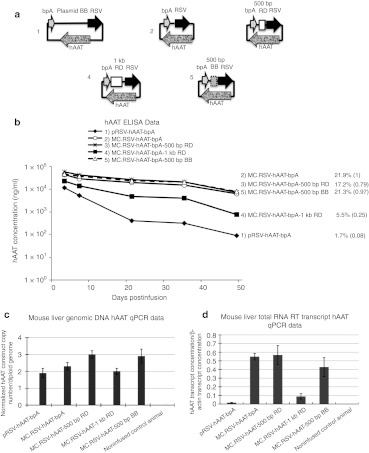
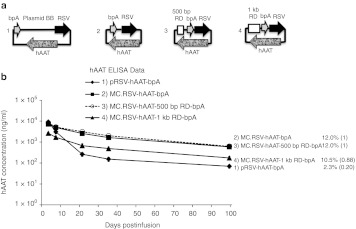
DNA sequences obtained from the genome have evolved to serve specific functions. Thus, it is not possible to exclude that the nonarbitrary nature of any genome-derived sequence might have an influence on gene expression. To exclude the possibility of such bias, we used a spacer sequence, generated by a random enzymatic process (see Methods and Supplementary Figure S2). Five hundred bp and 1 kb fragments were PCR amplified from the original 6.4 kb random sequence and then inserted into minicircle as spacers in between the 5′ and 3′ ends of each of the two transgene expression cassettes. In the first experiments, MC.RSV-hAAT-bpA-500 bp RD and MC.RSV-hAAT-bpA-1 kb RD were compared with the control plasmid and minicircle DNAs. As indicated in Figure 3b, minicircle with 500 bp random DNA (RD) spacer produced a transgene expression pattern similar to minicircle without spacer. However minicircle with 1 kb RD spacer exhibited a transgene expression pattern similar to plasmid DNA. Similar expression patterns were also obtained when a second expression cassette, EF1α-hFIX-hGHpA (Figure 4) with the various spacers were compared. In the presence of the 1 kb RD spacer, MC.EF1α-hFIX-hGHpA-1 kb RD resulted in short-lived expression whereas the MC.EF1α-hFIX-hGHpA-500 bp RD treated animals maintained high levels of plasma hFIX for at least 2 months. Again to establish that the loss of trangene expression was not due to differential plasmid loss, we quantified the vector DNAs by qPCR assays (Figure 3c). Concordant with the experiments in Figure 2, the vector copy numbers were similar between groups varying by less than two times (1.8–3.0 copies per diploid genome). To determine if transgene silencing was due to differential levels of the RNA transcript, hAAT messenger RNAs were quantified by reverse transcription (RT)-PCR. As shown in (Figure 3d), the hAAT transcript levels correlated with the level of protein expression. The MC.RSV-hAAT-bpA and MC.RSV-hAAT-bpA-500 bp RD infused groups had about five times higher amounts of transcript compared to the MC.RSV-hAAT-bpA-1 kb RD infused animals while the pRSV-hAAT injection more than a 10 times lower amount of transcript compared to the minicircle or MC.RSV-hAAT-bpA-500 bp RD injected groups.
Figure 3.
RSV-hAAT expression cassette constructs and transgene expression in mice. (a) Schematic of the DNA constructs containing random DNA (RD) or bacterial backbone (BB) used as spacers. (b) DNA constructs from a were injected into C57BL/6 mice (n = 5 per group). Serum hAAT levels were determined over time. The provided values are as described in Figure 1. (c) Vector DNA copy number (per diploid genome) and (d) Normalized hAAT mRNA transcript levels in 49-day postinfusion liver samples. In c and d, standard deviations were based on two biological samples each performed in duplicate experiments (n = 4). ELISA, enzyme-linked immunosorbent assay; hAAT, human α1-antitrypsin; mRNA, messenger RNA; qPCR, quantitative PCR; RT, reverse transcription.
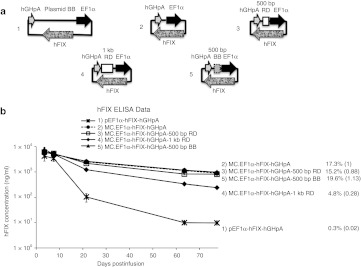
Figure 4.
EF1α-hFIX expression cassette constructs and transgene expression in mice. (a) Schematic of hFIX expressing DNA constructs with random DNA (RD) or bacterial backbone (BB) as spacer. (b) Constructs in a were injected into C57BL/6 mice (n = 5 per group). Plasma hFIX levels were determined over time. The provided values are as described in Figure 1. ELISA, enzyme-linked immunosorbent assay; hFIX, human factor IX.
We further tested whether spacers containing larger spacer inserts (>1 kb) would have stronger effects on silencing minicircle and plasmid DNA transgene expression in vivo; 1.5 kb and 2.2 kb RD fragments were used to generate MC.EF1α-hFIX-hGHpA-1.5 kb RD and MC.EF1α-hFIX-hGHpA-2.2 kb RD vectors. The 1.5 kb and 1 kb sequences only share a 400 bp stretch (Supplementary Figure S2). Mice that infused with these constructs also resulted in transgene silencing (Figure 5).
Figure 5.
EF1α-hFIX constructs and transgene expression in mice. (a) Schematic of DNA constructs containing large random DNA (RD) spacers. (b) DNA constructs in a were injected into C57BL/6 mice respectively (n = 5 per group). Plasma hFIX levels were determined over time. The provided values are as described in Figure 1. BB, backbone; ELISA, enzyme-linked immunosorbent assay; hFIX, human factor IX.
Taken together these results provide strong evidence that the extragenic DNA spacer length between the 5′ and 3′ ends of an expression cassette and not the specific DNA sequence or structure derived from a bacterial plasmid BB DNA is the important parameter dictating whether or not a plasmid DNA will be silenced.
Short plasmid BB sequences inserted as spacers are incapable of silencing trangene expression in vivo while longer plasmid BB sequences silence the transgene
Previously, we have substituted various prokaryotic antibiotic resistance genes and/or various plasmid origins of replication, and eliminated non-essential plasmid BB sequences but transgene silencing still occurred regardless of the specific substitutions. In all cases, the plasmid BB was well over 1 kb (H. Nakai and M.A. Kay, unpublished data). To further test the hypothesis that the bacterial plasmid BB DNA does not contain silencing favorable sequences, we constructed minicircles containing a 500 bp pUC-derived bacterial plasmid origin of replication in either the hFIX or hAAT expression cassette (Figures 3 and 4). As shown in Figures 3 and 4, 500 bp BB spacer failed to silence either the hAAT or hFIX transgene expression cassettes in vivo This result confirmed that generic plasmid origin sequence is not sufficient for transgene silencing in DNA plasmid vectors. On the other hand, when a fragment of a BB—a 1.5 kb BB containing pUC origin and kanamycin resistance gene was placed in between the 5′ and 3′ ends of the hFIX expression cassette (MC.EF1α-hFIX-hGHpA-pUC-Kan (1.5 kb)), transgene expression was silenced (Figure 5). This was consistent with our other results that longer extragenic spacer (≥1 kb) was sufficient to silence the transgene and supporting the idea that the length of the DNA insert and not anything specific about the bacterial plasmid DNA itself was the critical determinant responsible for transgene silencing.
DNA spacers placed into the 3′UTR do not silence minicircle transgene expression in vivo
In all of the above experiments, spacers were placed into the extragenic region of the DNA vector. We wanted to establish if the same DNA spacers placed within the transcription unit would still silence the plasmid vector. Such studies would test whether: (i) the DNA sequences could influence silencing independent of their context; (ii) establish if the total length of the vector affected silencing; (iii) establish if the length of the noncoding extragenic region affected silencing. To address these questions, 500 bp and 1 kb RD spacers were placed between the stop codon of hAAT transgene and the polyA signal, making the inserted segment a part of the 3′-untranslated region (UTR) sequences in MC.RSV-hAAT-500 bp RD-bpA and MC.RSV-hAAT-1 kb RD-bpA (Figure 6a). These constructs were tested in animals for hAAT expression in vivo. Results from animal test indicated that neither 500 bp RD and 1 kb RD when placed next to stop codon of hAAT induced transgene silencing (Figure 6b). We note that the 1 kb RD resulted in lower levels of initial transgene expression, but that this level was maintained and not silenced. Altering the 3′UTR sequence can greatly influence transgene expression with the addition of sequences generally resulting in reduced expression.13 Therefore, the low initial expression was not unexpected.
Figure 6.
RSV-hAAT expression cassette constructs containing extended 3′UTRs and transgene expression in mice. (a) Schematic of DNA constructs containing extended 3′UTRs. (b) DNA constructs in a were injected into C57BL/6 mice respectively (n = 5 per group). Serum hAAT levels were determined over time. The provided values are as described in Figure 1. BB, backbone; ELISA, enzyme-linked immunosorbent assay; hAAT, human α1-antitrypsin; UTR, untranslated region.
Particularly striking in this study was the observation that the 1 kb RD sequences that induced silencing when placed outside of the transgene expression cassette did not induce silencing when contained within the 3′UTR. This provides further support that the spacer length and not the sequence itself located between the 5′ and 3′ ends of the expression cassette as a key determinant of persistent transgene expression in vivo.
Discussion
In this study, we provide strong evidence that transgene-induced silencing is not restricted to the covalent attachment of plasmid bacterial BB sequences. Rather noncoding, nonbacterial, and nongenic DNA sequences reaching 1 kb or more in length regardless of their origin can also silence transgene expression. The sequences themselves were not inhibitory because if they were placed into the expression cassette such that it became part of the 3′UTR, silencing was not observed. The finding that extragenic spacer length rather than DNA type is the major factor influencing transgene silencing from plasmid DNA-based vectors provides important insights into mechanism.
Our results confirming that the messenger RNA and not vector DNA levels correlate with the amount of transgene protein strongly suggest that silencing is related to differences in transcription albeit the exact molecular mechanisms are not known. Naked episomal DNA vectors undergo chromatinization after transfection of mouse liver.4 Moreover, ChIP analyses performed on chromatized plasmid and minicircle DNAs isolated from mouse liver established a good correlation between specific modified histones and the degree of transgene expression.4 These experiments were only correlative and not designed to establish casual events. The nucleosomes are the basic building blocks of eukaryotic chromatin. One nucleosome core particle consists of ~147 bp of DNA wrapped in left-handed suprahelical turns around a histone octamer containing two copies of each core histones H2A, H2B, H3, and H4. Nucleosomes are connected by short stretches of linker DNA (0–80 bp) at a fixed distance between them.14 However, the inclusion of nucleosome altering sequences did affect nucleosome site formation on plasmid DNA in vivo, it did not affect transgene silencing.15 Thus the interplay between nucleosome and chromatin formation in plasmid-mediated transgene silencing is still unclear. High-resolution chromatin protein mapping might help shed additional light on the detailed molecular differences between the two types of DNA vectors. At this point, we cannot distinguish the exact step at which transgene silencing occurs albeit the current evidence points to a process affecting DNA-mediated RNA transcription.
The mammalian gene transcription mediated by RNA polymerase II involves a series of molecular steps, including transcription preinitiation complex assembly, initiation, elongation, termination and reinitiation. Studies done in Saccharomyces cerevisiae have shown that a gene promoter and terminator regions can be juxtaposed in a transcription-dependent manner and the gene loops between the promoter and terminator regions may be a common feature of gene activation by promoting efficient transcriptional elongation.16 Data collected through capturing chromosome conformation also prove that DNA looping occurs between promoter and terminator regions. Moreover the promoter is not juxtaposed to DNA downstream of the terminator, nor is the terminator juxtaposed to DNA upstream of the promoter.17 These authors further demonstrated that transcriptional activation of DNA is associated with enhanced gene loop formation and gene looping is a mechanism to promote transcription reinitiation by facilitating recycling of RNA polymerase II from the terminator to the promoter.17
A similar mechanism could explain the difference in gene expression patterns observed with minicircle and plasmid DNA. By removing bacterial BB, the 5′ end of promoter and the 3′ end of terminator regions may be more closely linked in a minicircle or plasmid with less than 1 kb of spacer DNA. More definitive studies will be required to establish the validity of this model. Nonetheless, our findings will provide new approaches for constructing episomal-based vectors that will provide robust and sustained transgene expression from quiescent tissues and cells.
Materials and Methods
RD synthesis. We prepared a population of extended double-stranded DNA molecules of arbitrary sequence as follows. A random hexanucleotide mixture (dNdNdNdNdNdN; 5′ and 3′ OH, 100 ng in 50 µl) was reacted with bovine terminal transferase (17 units; US Biochemical, Cleveland, OH) for 4 hours at 37 °C, followed by two rounds of addition of an additional aliquot of terminal transferase (17 units) and a 16 hour incubation at 37 °C. Reaction conditions were 100 mmol/l sodium cacodylate pH 6.8, 1 mmol/l cobalt chloride, 0.1 mmol/l dithiothreitol, 4 mmol/l MgCl2, 1.2 mmol/l each of dATP, dTTP, dCTP, dGTP. Twelve units of pyrophosphatase were added at the beginning of the reaction to avoid accumulation of inhibitory pyrophosphate species. This reaction led to the synthesis of single-stranded DNA with lengths ranging from several 100 to several 1,000 bases, visible upon analytical agarose gel electrophoresis of an aliquot (3 µl) in the presence of 0.3 µg/ml ethidium bromide. Following the confirmation of a DNA population on ethidium visualization, a second strand was synthesized through the action of Escherichia coli DNA polymerase I as follows: 25 µg glycogen and 340 µl of 20 mmol/l Tris-HCl pH 7.5, 1 mmol/l EDTA, 10 mmol/l MgCl2, 1 mmol/l dithioerythritol, 500 µmol/l each dTTP, dCTP, dATP, dGTP were added, followed by addition of 45 units of Escherichia coli DNA polymerase I holoenzyme, sequential incubations for 20 minutes each at 4 °C, 16 °C, 23 °C, addition of 20 additional units of Escherichia coli DNA polymerase I, and incubation at 37 °C for 2 hours. Reactions were then cleaned up by addition of 15 µl of 0.5 mol/l EDTA, 40 µl of 10 mol/l NH4OH pH 7.5, followed by extraction with phenol/CHCl3 (1:1; 400 µl), CHCl3 (400 µl) ethanol precipitation, and resuspension in 20 µl of 10 mmol/l Tris-HCl pH 7.5, 1 mmol/l EDTA. Following digestion of these samples with KpnI and XbaI, fragments were inserted into an KpnI+XbaI cut plasmid vectors and sequenced. A series of inserts were then combined using flanking sites to produce the extended random insert described. The RD sequence is listed in detail in Supplementary Figure S2. Sequences added to the vectors were examined and shown to lack a cryptic splice site using the following program, http://bioweb.uwlax.edu/genweb/molecular/seq_anal/splice_sites/splice_sites.htm. The absence of an additional polyadenylation site was confirmed by the lack of an AAUAAA sequence.
Vector construction. The pRSV-hAAT-bpA1 and pEF1α-hFIX-hGHpA2 plasmids were previously described. The hAAT and hFIX minicircle producing plasmids, pMC.RSV-hAAT-bpA and pMC.EF1α-hFIX-hGHpA, were engineered with a unique SpeI restriction enzyme site right after the polyA tail sequence. Multiple sized spacers were amplified from human AR sequence and RD sequences by PCR using primers containing the SpeI restriction enzyme digestion site at the 5′ end. The correct PCR products were digested with SpeI and ligated to SpeI digestion linearized antarctic phosphatase treated pMC.RSV-hAAT-bpA and pMC.EF1α-hFIX-hGHpA. The ligation solution was transformed into DH10B competent cells and grown on kanamycin selection agar plates at 37 °C overnight. DNA was isolated from selected colonies and retransformed into minicircle producing bacterial strain 10P3S2T and minicircle preparation.
Production of minicircle. Minicircle DNA was produced using a previously developed protocol.10 Early on day 1, cells were grown from one parental plasmid-transformed colony in 5 ml of Luria-Bertani broth containing 50 µg/ml kanamycin at 37 °C with shaking at 250 rpm. Later that evening, 100 µl of the 5 ml Luria-Bertani broth from the culture was added to 400 ml Terrific broth containing 50 µg/ml kanamycin and incubated at 37 °C with shaking at 250 rpm for 16–18 hours. The overnight culture's OD600 reading was between 3.75–4.25. The pH reading of the overnight culture was pH 6.5. The overnight culture was mixed with 400 ml fresh Luria-Bertani broth, 16 ml of 1 N NaOH and 0.4 ml of 20% L-arabinose, incubated at 32 °C with shaking at 250 rpm for 5 hours. Bacteria were pelleted and minicircles were isolated using a Qiagen mega plasmid kit (Qiagen, Valencia, CA) according to manufacturer's protocol with the exception that double the volume of P1, P2, and P3 buffers was used.
Animal studies. The animal experiments were done with approval from the Administrative Panel on Laboratory Animal Care at Stanford University and conformed to the guidelines set forth by the National Institutes of Health. Six- to 8-week-old female C57BL/6 mice purchased from Jackson Laboratory (Bar Harbor, ME) were used for DNA injection. To ensure the same molar amount of DNA was injected into each animal, 3.63 µg/kb DNA was used for various sized constructs. For example, 8 µg (3.63 µg/kb × 2.2 kb) DNA was used for 2.2 kb MC.RSV-hAAT-bpA constructs to inject each animal. Each DNA construct was diluted into 1.8 ml of 0.9% NaCl for each animal, and was delivered through hydrodynamic tail vein injection. Five animals were tested for each DNA construct in each tested experimental group. After DNA infusion, blood samples were collected periodically by a retro-orbital technique. The serum hAAT and plasma hFIX were quantitated by enzyme-linked immunosorbent assay.
Southern blot analysis of vector DNA structure in mouse liver. Liver genomic DNA was extracted through a salt-out procedure. Twenty microgram liver genomic DNA of each sample was digested with either PmlI alone or both PmlI and SpeI overnight at 37 °C. PmlI cut the expression cassette once. PmlI-SpeI double digestion cut the expression cassette twice. Various copy numbers of MC.EF1α-hFIX-hGHpA vector DNA was also digested with PmlI and SpeI, and then mixed with 20 µg noninfused mouse liver genomic DNA in each lane as copy number control. PmlI digested pEF1α-hFIX-hGHpA vector DNA was also loaded along with 20 µg noninfused mouse liver genomic DNA as a size control for 8 kb band. Vector DNA copy number was calculated based on Applied Biosystems's method, http://www6.appliedbiosystems.com/support/tutorials/pdf/quant_pcr.pdf and by using internet tool http://www.uri.edu/research/gsc/resources/cndna.html. Digested DNA samples were separated by electrophoresis in 1% agarose gel and blotted onto a nitrocellulose membrane. Southern blot membrane was hybridized with [P-32] dCTP-labeled 1.4 kb hFIX cDNA, and [P-32] dCTP-labeled 300 bp β-actin (PCR product by using forward primer 5′ACGCGTCCAATTGCCTTTCT3′ and reverse primer 5′CTCGAGGTTGAAGGTCTCAA3′). hFIX and β-actin signals were detected through phosphoimaging. The Southern blot signal strength was measured by Quantity One. hFIX signal was normalized to β-actin signal. Normalized hFIX signal was compared with copy number standard to obtain corresponding DNA vector copy number per diploid.
qPCR analysis of vector DNA structure in mouse liver. A 100 ng double-digested genomic DNA (PmlI-SpeI for Figure 2 and XbaI-SpeI for Figure 3d) from each sample was used as the template for qPCR. Or, 1 µl of RT reaction was used as template for Figure 3c. Two animal samples from each injection group were selected and two 15 µl reactions were performed for each animal sample. Various copy numbers (2 × 108 copies to 20 copies) of double-digested standard vector DNA along with 100 ng noninfused control genomic DNA per reaction was used to make copy number standard curve. Forward primer 5′ACATTGCCCTTCTGGAACTG3′ and reverse primer 5′GCTGATCTCCCTTTGTGGAA3′ oligos were used to amplify 150 bp fragment from hFIX cDNA region. Forward primer 5′AAGGCAAATGGGAGAGACCT3′ and reverse primer 5′TA CCCAGCTGGACAGCTTCT3′ oligos were used to amplify 150 bp fragment from hAAT cDNA region. Forward primer 5′TTGCTGACA GGATGCAGAAG3′ and reverse primer 5′TGATCCACATCTGCTGG AAG3′ oligos were used to amplify 150 bp fragment from β-actin as loading control. The tested transgene signal was then normalized to the β-actin signal. The mass of a single diploid copy of mouse genome is 5.88 pg. Thus 100 ng genomic DNA contains 17,007 copies of diploid genome (1 × 106 pg/5.88 pg). The average transgene copy number in 100 ng genomic DNA from each group was then divided by 17,007 to achieve the transgene construct copy number in each cell. All calculations were based on methods described by Applied Biosystems-method http://www6.appliedbiosystems.com/support/tutorials/pdf/quant_pcr.pdf and by using internet tool http://www.uri.edu/research/gsc/resources/cndna.html. qPCR was performed by using Corbett Research RG6000 PCR machine (Corbett Research, Mortlake, Australia).
RT and following qPCR analysis. Five microgram DNase I treated liver total RNA sample was used for each RT reaction with oligo(dT). The RT reaction was performed as described in the manual of SuperScript III RTS First-Strand cDNA Synthesis Kit from Invitrogen (Carlsbad, CA). Non-RT control of each sample was also performed; 1 µl RT or non-RT reaction was used as template for the following qPCR analysis. Each RT or non-RT sample was amplified by hAAT qPCR oligos and β-actin qPCR oligos as described above. The obtained hAAT signal was normalized by β-actin signal and the normalized hAAT signal from different vector infused animals were compared.
SUPPLEMENTARY MATERIAL Figure S1. Sequence of human alphoid repeat DNA spacer used in the plasmid constructs in Figures 1 and 2. Figure S2. Sequence of random DNA spacer used in the plasmid constructs in Figures 3,4,5 and 6.
Supplementary Material
Sequence of human alphoid repeat DNA spacer used in the plasmid constructs in Figures 1 and 2.
REFERENCES
- Chen ZY, He CY, Ehrhardt A., and, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- Chen ZY, He CY, Meuse L., and, Kay MA. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 2004;11:856–864. doi: 10.1038/sj.gt.3302231. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Riu E, He CY, Xu H., and, Kay MA. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol Ther. 2008;16:548–556. doi: 10.1038/sj.mt.6300399. [DOI] [PubMed] [Google Scholar]
- Riu E, Chen ZY, Xu H, He CY., and, Kay MA. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol Ther. 2007;15:1348–1355. doi: 10.1038/sj.mt.6300177. [DOI] [PubMed] [Google Scholar]
- Chen ZY, He CY., and, Kay MA. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Hum Gene Ther. 2005;16:126–131. doi: 10.1089/hum.2005.16.126. [DOI] [PubMed] [Google Scholar]
- Bigger BW, Tolmachov O, Collombet JM, Fragkos M, Palaszewski I., and, Coutelle C. An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J Biol Chem. 2001;276:23018–23027. doi: 10.1074/jbc.M010873200. [DOI] [PubMed] [Google Scholar]
- Darquet AM, Cameron B, Wils P, Scherman D., and, Crouzet J. A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Ther. 1997;4:1341–1349. doi: 10.1038/sj.gt.3300540. [DOI] [PubMed] [Google Scholar]
- Mayrhofer P, Blaesen M, Schleef M., and, Jechlinger W. Minicircle-DNA production by site specific recombination and protein-DNA interaction chromatography. J Gene Med. 2008;10:1253–1269. doi: 10.1002/jgm.1243. [DOI] [PubMed] [Google Scholar]
- Schakowski F, Gorschlüter M, Buttgereit P, Märten A, Lilienfeld-Toal MV, Junghans C.et al. (2007Minimal size MIDGE vectors improve transgene expression in vivo In Vivo 2117–23. [PubMed] [Google Scholar]
- Kay MA, He CY., and, Chen ZY. A robust system for production of minicircle DNA vectors. Nat Biotechnol. 2010;28:1287–1289. doi: 10.1038/nbt.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt A., and, Kay MA. A new adenoviral helper-dependent vector results in long-term therapeutic levels of human coagulation factor IX at low doses in vivo. Blood. 2002;99:3923–3930. doi: 10.1182/blood.v99.11.3923. [DOI] [PubMed] [Google Scholar]
- Nakai H, Yant SR, Storm TA, Fuess S, Meuse L., and, Kay MA. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J Virol. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybowska EA, Wilczynska A., and, Siedlecki JA. Regulatory functions of 3'UTRs. Biochem Biophys Res Commun. 2001;288:291–295. doi: 10.1006/bbrc.2001.5738. [DOI] [PubMed] [Google Scholar]
- Jiang C., and, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracey LE, Chen ZY, Maniar JM, Valouev A, Sidow A, Kay MA.et al. (2010An in vitro-identified high-affinity nucleosome-positioning signal is capable of transiently positioning a nucleosome in vivo Epigenetics Chromatin 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J.et al. (2004Gene loops juxtapose promoters and terminators in yeast Nat Genet 361014–1018. [DOI] [PubMed] [Google Scholar]
- Ansari A., and, Hampsey M. A role for the CPF 3'-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence of human alphoid repeat DNA spacer used in the plasmid constructs in Figures 1 and 2.