Abstract
Ubiquitin signaling plays an essential role in controlling cellular processes in eukaryotes, and the impairment of ubiquitin regulation contributes to the pathogenesis of a wide range of human diseases. During the last decade, mass spectrometry–based proteomics has emerged as an indispensable approach for identifying ubiquitinated proteins, ubiquitin modification sites, the structure of complex ubiquitin chains, as well as the interactome of ubiquitin enzymes. In particular, implementation of quantitative strategies allows the detection of dynamic changes in the ubiquitinated proteome, enhancing the ability to differentiate between function-relevant protein targets and false positives arising from biological and experimental variations. The profiling of total cell lysate and ubiquitinated proteome in the same sets of samples has become a powerful tool, revealing a subset of substrates that are modulated by specific physiological and pathological conditions, such as gene mutations in ubiquitin signaling. This strategy is equally useful for dissecting the pathways of ubiquitin-like proteins.
Keywords: ubiquitin, proteasome, E3, DUB, mass spectrometry, proteomics, SILAC
Introduction
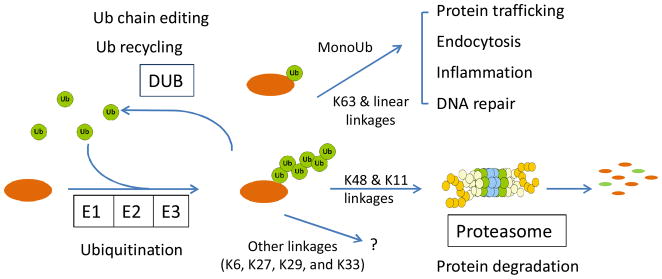
Ubiquitin (Ub) is a highly conserved essential protein of 76 amino acids that regulates almost all cellular events in eukaryotes. Although the purification of Ub was described more than 30 years ago, novel regulatory functions of Ub pathways are continuously being discovered. Ubiquitin, originally known as the ubiquitous immunopoietic polypeptide (UBIP), was first purified from the bovine thymus and then from all tested living cells (Goldstein et al., 1975; Schlesinger and Goldstein, 1975; Schlesinger et al., 1975), and it was found to form an isopeptide bond with a lysine residue in histone 2A (Goldknopf and Busch, 1977). The biological function of Ub, however, remained a mystery until 1980, in part because the monoubiquitination of histone 2A does not promote protein degradation. By accident, another protein, the ATP-dependent proteolysis factor 1 (APF-1), which facilitates protein degradation in an ATP-dependent manner, was shown to be identical to Ub (Ciehanover et al., 1978; Wilkinson et al., 1980); the physiological role of Ub in protein degradation was later shown in cells (Ciechanover et al., 1984; Finley et al., 1984). Finally, a protease complex (the 26S proteasome) was found to degrade ubiquitinated species (Hough et al., 1987). The 26S proteasome is a 2.5-Mda complex composed of a 19S regulatory particle and a 20S core particle. This 19S particle recognizes the Ub tag on substrates and then deubiquitinates, unfolds, and translocates the substrates into the 20S particle for proteolysis, whereas the 20S particle has multiple protease activities to digest the protein substrates into small peptides (Finley, 2009). These landmark findings established the concept of the ubiquitin proteasome system (UPS) in protein degradation (Hershko and Ciechanover, 1998); further studies revealed the nonproteolytic role of Ub in protein sorting, interactions, and activity modulation for a host of protein targets (Mukhopadhyay and Riezman, 2007) (Fig. 1).
Figure 1.
The chemistry and function of protein ubiquitination.
Protein ubiquitination is a reversible posttranslational process catalyzed by several enzymes. Ub molecules are mainly conjugated to the lysine residues of substrates by the catalytic action of E1 activating enzymes, E2 conjugating enzymes, and E3 ligases (Ciechanover, 2005). In some cases, non-lysine residues (e.g., N-terminal amine group and cysteine residues) can also be alternative sites for Ub conjugation (Cadwell and Coscoy, 2005; Ciechanover and Ben-Saadon, 2004). In addition to monoubiquitination, substrates can be further modified by additional Ub molecules, resulting in multi-monoubiquitination or polyUb chains. The polyUb chains are assembled through any of the 8 amine groups in the Ub sequence: the N-terminus, K6, K11, K27, K29, K33, K48, and K63 (Kirisako et al., 2006; Peng et al., 2003; Xu et al., 2009c). As Ub signaling is transduced through the interaction of Ub moieties (monoUb or polyUb) with numerous Ub receptors containing Ub-binding domains (Dikic et al., 2009; Pickart and Fushman, 2004), the 8 Ub chain linkages allow the formation of diverse chain structures (Bremm et al., 2010; Varadan et al., 2004; Virdee et al., 2010), potentially providing a basis for mediating specific downstream signaling events. Ubiquitin conjugated to substrates can be cleaved by 5 families of deubiquitinating enzymes (DUBs) that are either metalloproteases or cysteine proteases (Komander et al., 2009; Nijman et al., 2005). The DUBs function to produce Ub monomers from Ub precursors, edit polyUb chains on substrates, recycle Ub from substrates, and rescue substrates from proteasomal degradation (Reyes-Turcu et al., 2009). The specificity of Ub pathways is primarily mediated by the E3–substrate interaction, the recognition of Ub moieties (monoUb and polyUb with diverse linkages) by Ub receptors, and the selective removal of Ub modifications by DUBs. The scope of protein ubiquitination is as vast as that of protein phosphorylation. The human genome encodes 2 Ub E1 enzymes, approximately 40 different E2 enzymes, at least 600 E3 ligases, and approximately 100 DUBs (Li et al., 2008; Nijman et al., 2005; Semple, 2003), which are proposed to regulate thousands of protein substrates in cells.
Considering ubiquitin’s versatile role in cells, a link between ubiquitination dysregulation and the development of human diseases such as cancer (Voorhees and Orlowski, 2006) and neurodegenerative disorders (Ciechanover and Brundin, 2003; Goldberg, 2007) should come as no surprise. For example, the localization and degradation of the well-known tumor suppressor p53 protein are exquisitely regulated by numerous E3 ligases as well as by at least 1 known DUB (Lee and Gu, 2010). Tumorigenesis is directly associated with mutations in multiple E3 ligases, including the breast cancer protein BRCA1 (Wang, 2007), Fanconi anaemia protein complex (Wang, 2007), and Von Hippel-Lindau disease protein VHL (Kaelin, 2008). Deubiquitinating enzymes, such as the cylindromatosis protein CYLD (Bignell et al., 2000) and TNFAIP3/A20 (Musone et al., 2008), are also known as tumor suppressors. As regards to neurodegenerative disorders, some cases of early-onset Parkinson’s disease are associated with mutations in the Parkin E3 ligase (Bandopadhyay and de Belleroche, 2010). Missense mutations in a BTB-Kelch protein (KLHL7, which may function in ubiquitination through Cullin E3 ligases) cause retinitis pigmentosa, a progressive degenerative disease of rod and cone photoreceptors in the retina; mutations in another BTB-Kelch protein (gigaxonin) lead to giant axonal neuropathy (Friedman et al., 2009); and UBE3A mutations are linked to the onset of Angelman syndrome (Yi and Ehlers, 2007). Two DUB genes, UCHL1 and ATXN3, are tied genetically to Parkinson’s disease and spinal cerebral ataxia, respectively (Yi and Ehlers, 2007). In addition to genetic evidence, Ub-positive inclusion staining is a hallmark of pathology in various neurodegenerative disorders, suggesting the role of Ub in disease development (Ciechanover and Brundin, 2003; Yi and Ehlers, 2007).
In addition to Ub, a growing family of proteins referred to as ubiquitin-like proteins (Ubls) shares similar protein modification mechanisms and regulates a broad range of cellular functions in eukaryotes and even in prokaryotes (Hochstrasser, 2009; Kerscher et al., 2006). Of the more than 10 Ubls identified so far, 9 can covalently modify other proteins, such as small ubiquitin-like modifier (SUMO), neural precursor cell-expressed developmentally downregulated protein 8 (Nedd8), and interferon-stimulated 15-kDa protein (ISG15). Despite limited sequence similarity, Ubls exhibit a common globular 3D structure of the β-grasp fold or ubiquitin fold (Hochstrasser, 2000). Ubl modifications alter the chemical properties and tertiary structure of substrates and are able to compete with modification of Ub and other molecules (Jeram et al., 2009).
In summary, Ub and Ubls are key regulators of cellular events, and failure of the Ub and Ubl pathways plays a significant role in the pathogenesis of human diseases. In most cases, however, the molecular mechanisms underlying pathogenesis are far from clear, partly because of the limitation in analytic tools. Current developments in modern mass spectrometry (MS) have enabled biochemical characterization of proteins in the femtomolar or even sub-femtomolar range (Choudhary and Mann, 2010; Cravatt et al., 2007; Gstaiger and Aebersold, 2009; Peng and Gygi, 2001), providing unprecedented opportunities for dissecting ubiquitin pathways. In this review, we present the strategies for applying MS to unravel the functions of Ub or Ubl modifications, with a focus on protein ubiquitination.
Mass spectrometry (MS) analysis of the proteome modified by ubiquitin (Ub) or ubiquitin-like proteins (Ubls)
Ubiquitin-modified proteins are usually analyzed by capillary liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Choudhary and Mann, 2010; Cravatt et al., 2007; Gstaiger and Aebersold, 2009). The protein sample is first digested with a protease (usually trypsin) and the resulting peptides are then fractionated by liquid chromatography (e.g., using a C18 reverse-phase column). These peptides are ionized and transferred to a mass spectrometer, where they are further separated on the basis of mass-to-charge (m/z) ratio. It should be emphasized that tandem mass spectrometry itself is a high-resolution separation tool to isolate one ion among many coeluting ions. The isolated peptide ion is then fragmented to generate a specific MS/MS spectrum containing its sequence information. Next, a bioinformatics software such as SEQUEST (Eng et al., 1994) or Mascot (Perkins et al., 1999) is used to match the experimental MS/MS spectrum with theoretical (computer-generated) peptide spectra from a database. As current MS instruments can acquire MS/MS spectra at a rate of up to 10 Hz, this procedure allows the identification of a large number of peptide or protein sequences from complex mixtures. For example, a single run on an optimized LC-MS/MS platform identified approximately 1000 proteins from a total yeast lysate (Xu et al., 2009b), and with further modifications, identified more than 2400 proteins from mouse pancreatic islets (Waanders et al., 2009). To increase the separation power before LC-MS/MS analysis, it is common to resolve protein samples by one-dimensional SDS gel electrophoresis or fractionate peptide mixtures by strong cation exchange chromatography or isoelectric focusing (de Godoy et al., 2008). Details of these techniques are available in other review papers (Motoyama and Yates, 2008; Peng and Gygi, 2001; Steen and Mann, 2004).
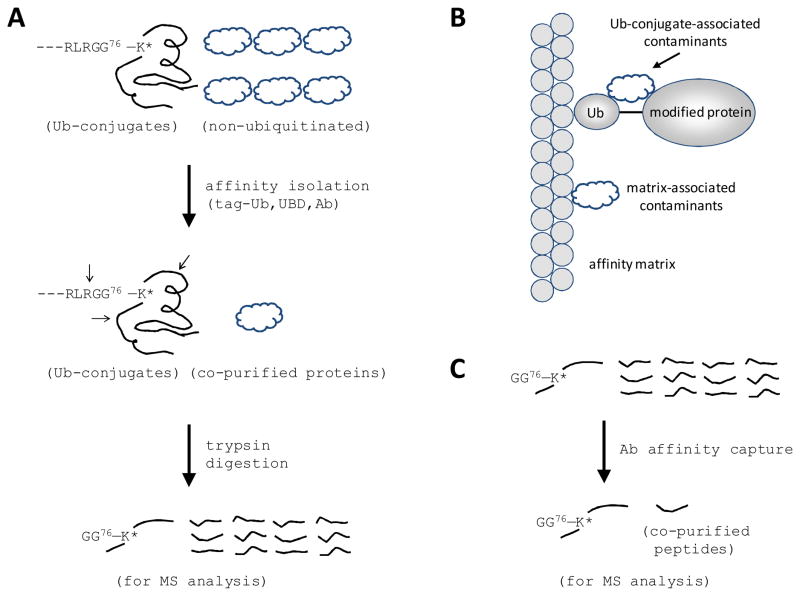
As ubiquitinated proteins are often sparse in cells, it is recommended that Ub-conjugates be pre-enriched by affinity approaches (Fig. 2) such as Ub antibodies (Matsumoto et al., 2005; Vasilescu et al., 2005), Ub-binding proteins (Bennett et al., 2007; Layfield et al., 2001; Maor et al., 2007; Weekes et al., 2003), or epitope-tagged Ub (e.g., FLAG, HA, myc, His, and biotin tag) (Kirkpatrick et al., 2005; Wang et al., 2007; Xu and Peng, 2006). Isolated Ub-conjugates are then analyzed by MS for protein identification and quantification. Most analyses have been compared in previous reviews (Wang et al., 2007; Xu and Peng, 2006); in this review, we focus on a few key experiments and recent developments. The first large-scale study used a His-tagged Ub purification method to isolate ubiquitinated proteins, followed by LC/LC-MS/MS to identify these ubiquitinated proteins and modification sites from yeast (Peng et al., 2003). In this study, 1075 proteins were identified as ubiquitinated candidates and 110 ubiquitinated sites were detected. A critical step was the use of a highly denatured condition (8M urea) to minimize associated proteins and inhibit Ub protease activities that rapidly disassemble Ub-conjugates during purification. When applied to mammalian cell cultures and even animal models (Jeon et al., 2007), however, the His tag strategy has had limited success due at least in part to the large number of His-rich native proteins in mammalian cells and the low-level expression of tagged Ub (Peng J, unpublished data). Further development of a tandem His-biotin tag strategy enabled the analysis of 669 potential ubiquitinated proteins and 44 ubiquitinated sites in HeLa cells (Meierhofer et al., 2008). More recently, a shortened biotin-tagged Ub was expressed under a neuron-specific promoter in fly, which allows the study of cell type–specific protein ubiquitination in vivo (Franco et al., 2010). To improve the expression of epitope-tagged ubiquitin in higher organisms, suppression of endogenous Ub by tetracycline-inducible RNAi strategy (Xu et al., 2009a) may be useful. In contrast to tagged Ub methods that require genetic engineering, other high-affinity reagents have been tested for their ability to capture endogenous ubiquitinated species. One promising approach is the use of tandem-repeated Ub-binding entities to increase Ub binding and to protect Ub-conjugates from degradation by DUBs (Hjerpe et al., 2009; Hjerpe and Rodriguez, 2008). A recent study used a specific antibody to enrich GG-tagged peptides and reveal 236 ubiquitinated proteins and 374 modified sites from HEK293 cells (Xu et al., 2010).
Figure 2.
Strategies for isolating ubiquitinated proteins/peptides from complex mixtures. (A) Schematic representation of the purification strategies based on tagged Ub (e.g. HA, FLAG, His and biotin), Ubiquitin-binding domains (UBDs) or Ub antibodies (Ab). Arrows indicate tryptic sites surrounding a lysine modification residue in Ub-conjugates. Digestion of the Ub moiety generates a small Gly-Gly tag on the lysine residue. (B) Purified Ub-conjugate samples contain 2 classes of contaminated proteins that are not ubiquitinated, one class associated with affinity matrix nonspecifically and the other class interacting more specifically with Ub-conjugates. (C) Immunocapture of GG-tagged peptides to enhance the capacity for identifying ubiquitination sites.
Regardless of the method employed, nonubiquitinated species are always copurified in different concentrations, depending on experimental conditions and personal expertise (Fig. 2). Lower concentrations of contaminants are expected under denaturing conditions than under native conditions. Therefore, it is essential to confirm whether proteins identified in Ub-conjugate–enriched fractions are truly modified by Ub. Three approaches may be used to confirm modification by Ub: (i) identification of ubiquitinated sites in some of these ubiquitinated proteins (Peng et al., 2003); (ii) quantitative comparison of samples with different levels of ubiquitination (Meierhofer et al., 2008); and (iii) immunoprecipitation and Western blotting to validate large mass shift caused by ubiquitination. As it is impractical to apply this method to every identified candidate, a virtual Western blotting approach was developed based on one-dimensional SDS gel and LC-MS/MS, enabling the large-scale examination of mass shift in these proteins (Seyfried et al., 2008).
The concept of using MS to identify ubiquitinated sites relies on the specificity of trypsin, which cleaves proteins at the carboxyl side of lysine or arginine unless followed by proline residues. Trypsinization of the C-terminal part of the Ub moiety produces a di-glycine (GG) that is still attached to the modified residues in ubiquitinated substrates, and the GG-modified lysine residues are resistant to trypsin digestion (Goldknopf and Busch, 1977). The GG-tagged, miscleaved lysine residues ensure a monoisotopic mass addition of 114.0429 Da and allow identification by MS (Marotti et al., 2002; Peng and Gygi, 2001; Peng et al., 2003). The same principle is applied to determine amino acid residues modified by Ubls (Table 1). As Nedd8 or ISG15 modification leads to the generation of the same GG tag as ubiquitination does, a purification procedure before MS analysis may be needed to differentiate these conjugates. Alternatively, digestion of the Ub-conjugates with other proteases such as LysC can produce specific, but long, signature peptides. A long peptide tag is also produced when trypsinizing SUMO on modified proteins. These long peptide tags, however, greatly increase the size of modified peptides, lower the detection signal on a typical LC-MS/MS platform, and increase the complexity of MS/MS spectra during fragmentation. These spectra are often difficult to interpret in database searches, but a special software such as “SUMmOn” (Pedrioli et al., 2006) or ChopNSpice (Hsiao et al., 2009) may facilitate data analysis. A combination of digestion by different proteases, database searching, and specific software analysis may improve the method of determining Ubl-conjugates (Jeram et al., 2009). Genetically shortening the tryptic SUMO peptide tag to 2 or 5 residues can enhance the mapping of SUMOylated sites (Knuesel et al., 2005; Matic et al., 2010).
Table 1.
Strategy for identifying amino acid residues modified by ubiquitin and Ubls. The conjugated proteins are usually digested by trypsin to generate a specific tag (e.g., GG for ubiquitin) on the modified residues, resulting in a mass shift that is detectable during MS and MS/MS scans. The mass of human Fat10 tag may change because of natural variations in sequence (UniProtKB/Swiss-Prot O15205).
| Human Ub or UBL | Remnant on modified peptides after trypsin digestion | Mass change in MS and MS/MS | Problems | Solutions |
|---|---|---|---|---|
| Ub |
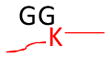
|
Monoisotopic mass shift of 114.0429 Da in MS and MS/MS | Ub, Nedd8, and ISG15 modifications lead to the same mass change after trypsin digestion | Pre-enrichment of modified proteins prior totrypsin digestion |
| Nedd8 |
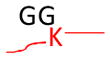
|
|||
| ISG15 |
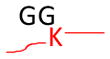
|
|||
| SUMO-1 |
|
Large mass shift in MS and complex production patterns in MS/MS | Weak signal in MS and complex MS/MS spectra that are difficult to interpret |
|
| SUMO-2/3 |
|
|||
| Fat10 |
|
During the database search of MS/MS spectra, false-positive identification of ubiquitination sites often occurs (Peng, 2008; Seyfried et al., 2008; Shi et al., 2011). For example, GG modification could be assigned to the C-terminal lysine in matched peptides by computer algorithms (Denis et al., 2007; Xu et al., 2010), resulting in a questionable conclusion that trypsin may cleave at the C-terminus of GG-modified Lys residues. However, an experiment to trypsinize a reported synthetic GG-peptide was not successful (Seyfried et al., 2008). None of the C-terminal ubiquitination events survive during manual interpretation of MS/MS matches (Seyfried et al., 2008; Shi et al., 2011). Another issue is the reliability of a reported longer tag (LRGG) due to miscleavage at Arg74 in Ub (Warren et al., 2005). The LRGG tag may be generated under certain digestion conditions but can rarely be identified. The Arg74 residue is usually cleaved at high efficiency under native or denaturing conditions (Wang et al., 2006; Xu et al., 2006). Thus, acceptance of either C-terminal ubiquitination or LRGG-modified peptides is not recommended. In addition, iodoacetamide used during purification to inhibit DUB activities by alkylating cysteine residues may also modify lysine residues twice to produce a monoisotopic mass tag of 114.0429 Da, the same mass as the GG tag (Nielsen et al., 2008). The suggested chloroacetamide (Nielsen et al., 2008) might also produce this artifact tag at high temperature, although at a much lower level than iodoacetamide (Xu et al., 2009c). However, at a low temperature (room temperature or lower) or low dosage, this side reaction is essentially eliminated (Xu et al., 2009c). As the most abundant ubiquitinated peptide in cells (K48-GG Ub peptide) can be distinguished from its iodoacetamide-modified artifact on the basis of LC retention time and a specific neutral loss in MS/MS pattern, checking for the presence of this artifact peptide in samples can be a quality control measure. Alternatively, other alkylation reagents (e.g., N-ethylmaleimide) may be used to avoid the formation of such an artifact tag. The peptide coverage of identified proteins is another parameter to evaluate false-positive matches (Shi et al., 2011). Ubiquitination sites identified on proteins with low peptide coverage need to be examined with caution. Finally, recent developments in MS such as faster scanning rates (Olsen et al., 2009) and the acquisition of high-resolution MS/MS spectra (Olsen et al., 2007) have substantially improved the analysis of protein ubiquitination (Danielsen et al., 2011; Shi et al., 2010). For example, the Nielsen group used Strep-HA tagged Ub to identify 753 ubiquitination lysine residues and more than 5700 putative Ub-conjugates, and found an approximately 20% overlap between protein ubiquitination and acetylation sites in the proteins analyzed, indicating a cross-talk between the 2 modifications (Danielsen et al., 2011).
Quantitative profiling of Ub-regulated proteome
Quantitative proteomics analysis of wild-type versus mutant cells may be an effective strategy to uncover specific substrates of Ub enzymes (e.g., E3 ligases or DUBs) (Kirkpatrick et al., 2005). The quantity of proteins can be evaluated by a number of MS approaches such as label-free quantification based on spectral counting (SC) and extracted ion current (XIC) or stable isotope labeling methods. Spectral counting uses the total number of MS/MS spectra identifying a single protein, which increases almost linearly with protein abundance after normalizing for protein size (Liu et al., 2004). This method works reasonably well for proteins with high spectral counts, but its reliability decreases significantly for proteins with low spectral counts (Zhang et al., 2006; Zhou et al., 2010). The abundance of peptides in samples can also be compared by the extracted ion current of corresponding ions (Radulovic et al., 2004; Wang et al., 2003). Because the ionization efficiency of peptides may vary among different LC runs, largely due to fluctuations of the LC system and ion suppression, considerable variations need to be normalized in this label-free method. Intrinsic LC-MS/MS variations can be effectively reduced by using stable isotope-labeled peptides as internal standards (Choudhary and Mann, 2010). As internal standards and counterparts are eluted and ionized simultaneously during the LC-MS/MS runs, relative quantification can be achieved by comparing the peptide pairs before fragmentation or comparing derived product ion peaks after fragmentation. Stable isotopes can be introduced into samples by in vitro labeling methods such as iTRAQ (Ross et al., 2004). iTRAQ reagents differentially label the amine group at the N-termini and Lys residues of peptides after protein digestion; eight iTRAQ reagents are currently available for multiplex comparisons in a single experiment. Moreover, the strategy of stable isotope labeling with amino acids in cell culture (SILAC) (Ong et al., 2002) has emerged as a highly accurate in vivo labeling method for large-scale proteomics. Many of these advanced quantitative methods and even the traditional 2D gel approach have been used to investigate protein changes in Ub pathways.
Assuming that a single E3 ligase mediates the degradation of substrates, the substrates are expected to accumulate in mutated cells. For example, to identify plasma membrane receptors modified by the transmembrane E3 ligase MARCH9, the SILAC method was used to monitor protein changes in the plasma membrane isolated from wild-type and MARCH9 mutant B cells (Hor et al., 2009). The label-free quantitative method was used to identify E3 ligase (ASB2) substrates, in which proteins from inducible cell lines expressing wild-type or ASB2 mutant were compared (Burande et al., 2009). The same method was also adapted to study the parkin-related pathway underlying the loss of parkin in a Drosophila model (Xun et al., 2009). Two-dimensional fluorescence difference gel electrophoresis (2-D DIGE) was employed to compare protein lysates from lung carcinoma cells with and without expression of viral E3 ligase (E1B55K and E40rf6) (Dallaire et al., 2009). It should be noted that proteins whose levels are altered in E3 mutants may not be genuine substrates of the related E3s, but instead may represent the adaptation of cells to mutants. Additional confirmation experiments, such as mRNA-level analysis, half-time measurement, protein–protein interaction, and in vitro ubiquitination, are required to validate the enzyme–substrate relationship.
Profiling of isolated ubiquitinated proteome is more informative than analysis of the total cell lysate, indicating direct changes caused by perturbation of Ub signaling. Meierhofer et al. studied the dynamics of ubiquitinated proteins upon protease inhibition in HeLa cells by the SILAC strategy (Meierhofer et al., 2008). Ideally, both total cell lysate and ubiquitinated proteome should be profiled from the same set of cells to reveal protein targets. For instance, Xu et al. used the SILAC method to compare 2 sets of the proteome in wild-type and Ub K11R mutant yeast strains to identify protein substrates modified by K11 polyUb chains (Xu et al., 2009c). As K11 linkage modification directs proteins to proteasomal degradation, 2 substrate candidates were identified on the basis of their enrichment in the total cell lysate and reduction in the ubiquitinated proteome. The same strategy was also used to probe a subset of Ub-conjugates recognized by Rpn10, a Ub receptor of the yeast proteasome (Mayor et al., 2007; Mayor et al., 2005), and a similar method has been used to search for SUMOylated substrates in response to heat shock (Golebiowski et al., 2009).
Quantitative analysis of polyubiquitin chains
As the linkages of polyubiquitin chains may determine the functional consequences of modified substrates (Fig. 1), analyzing the type of linkages on protein targets is of great importance. Classical K48 polyUb linkages direct substrates to the proteasome for degradation (Chau et al., 1989); the functions of newly discovered polyUb linkages (K6, K11, K27, K29, and K33), however, are much less understood, and these linkages may also contribute to proteasomal targeting (Johnson et al., 1995; Kirkpatrick et al., 2006; Xu et al., 2009c). In contrast, K63 linkages and monoUb modification mainly play roles in protein sorting (Hicke and Dunn, 2003), DNA repair (Bergink and Jentsch, 2009), and inflammation (Bhoj and Chen, 2009). Finally, linear polyUb chains are formed via the Ub N-terminal alpha amino group, but whether the linear chains function in proteolysis remains controversial (Kirisako et al., 2006; Rahighi et al., 2009; Zhao and Ulrich, 2010).
To measure all the polyUb linkages, stable isotope labeling peptides have been synthesized for all 8 linkages corresponding to ubiquitinated GG peptides (Kirkpatrick et al., 2006; Xu et al., 2006). The labeled peptides that are used as internal standards are spiked into a protein mixture. The mixture is then digested with trypsin to generate native GG peptides from polyUb chains. The pairs of native peptides and internal standards are indistinguishable during reverse-phase chromatography, but are separated by a mass spectrometer. These pairs are detected by the mass spectrometer in the setting of selected reaction monitoring [SRM, also termed MRM (multiple reaction monitoring)] for quantification. By this method, Kirkpatrick et al. detected mixed chain topologies (K11, K48, and K63) on ubiquitinated cyclin B1 catalyzed by the anaphase-promoting complex in vitro, and the heterogeneous chains were capable of mediating the degradation of cyclin B1 in a reconstituted system in vitro, suggesting a more broad involvement of linkages in substrate degradation (Kirkpatrick et al., 2006). The Kirkpatrick group has refined this approach recently and compared the detection sensitivity on different MS platforms (Phu et al., 2010). Xu et al. extended the measurement to yeast and uncovered a surprisingly high level of unconventional polyUb linkages from His-tag affinity-purified Ub-conjugates: K6, 10.9%; K11, 28%; K27, 9%; K29, 3.2%; K33, 3.5%; K48, 29%; K63, 16% (Xu et al., 2009c); the measurements in mammalian cells are slightly different (Dammer et al., 2011). The analysis of linkage changes under proteasomal inhibition in cell culture and in animals (Bedford et al., 2011) suggests that all non-K63 linkages (not including linear linkage in N-terminal-tagged Ub chains) contribute to proteasomal degradation. Other studies have also implicated the non-degradation role of unconventional linkages such as K11 (Boname et al., 2010; Goto et al., 2010) and K33 (Huang et al., 2010).
During fully tryptic digestion of Ub-conjugates, some structural information on polyUb chains is lost. For example, the current Ub-SRM method does not account for forked polyUb chains (i.e., 1 Ub molecule simultaneously modified by 2 other Ub molecules at Lys29 and Lys33) (Peng et al., 2003). To address this issue, a “middle-down” technology (Garciaa et al., 2007; Wu et al., 2006) has been used to partially digest Ub polymers under native conditions and enable analysis of the forked structure (Xu and Peng, 2008). Alternatively, the development of antibodies recognizing specific ubiquitin linkages, including K-11, K-48, and K63, makes it possible to immunoprecipitate specific chains and to perform cell imaging analysis (Matsumoto et al., 2010; Newton et al., 2008). These complementary tools will permit a more comprehensive study of Ub chain structure and function.
Interactome studies of ubiquitin enzymes
Through physical interactions, protein function is regulated by associated partners during cellular signaling in response to physiological conditions. To reveal the regulatory mechanism of Ub signaling, it is necessary to identify the interactome of core Ub enzymes such as E3 ligases, DUBs, and proteasome. Interactome studies consists of 2 main steps: (i) purification of the targeted protein complex and (ii) MS analysis to identify all components in the complex. Purification of the protein complex is the most critical step and requires optimization to increase yield and reduce nonspecific binding proteins. A commonly used approach to improve specificity is tandem-affinity purification (TAP) (Li, 2010; Puig et al., 2001), although a single step of affinity purification is also widely used in laboratories. As signaling proteins often bind to targeted proteins weakly and transiently, the quantitative analysis of tandem-affinity purified cross-linked protein complexes (QTAX) was developed to increase the recovery of these interacting proteins (Guerrero et al., 2008). A protein lysate can be made from formaldehyde-fixed cells by the QTAX method, followed by TAP under denaturing conditions. QTAX can be further coupled with SILAC to distinguish specific binding proteins from contaminants. Another strategy is to use the cleavable cross-linker disuccinimidyl sulfoxide to improve the purification of weak interacting proteins (Kao et al., 2010). Alternatively, affinity purification by GST fusion proteins is also frequently used.
A few representative examples of interactome studies are discussed here. Multiple E3 ligases ( RNF8, BRCA1, and RNF168) and the E2 enzyme Ubc13 are critical regulating proteins in DNA repair. A strep tag used to identify the RNF8 complex revealed that HERC2 interacts with RNF8 upon DNA damage–inducible phosphorylation of HERC2 (Bekker-Jensen et al., 2010). The TAP tag strategy was used to define the interaction of BRCA1 and NBA1, which is required to maintain the BRCA1 complex and to recruit BRCA1 to chromosomal damage sites (Wang et al., 2009). A recent immunoprecipitation–MS study found that OTUB1, a DUB associated with Ubc13, suppresses RNF168-dependent ubiquitination and inhibits the DNA damage response (Nakada et al., 2010). Alternatively, GST affinity purification has been used to pull down interacting proteins of mind bomb 1, a membrane-associated E3 ligase required for Notch signaling. MS analysis revealed that Mib1 primarily interacts with membrane trafficking proteins, cell adhesion components, several DUBs, and a number of kinases. Further study showed that the p35/CDK5 kinase downregulates the protein level of Mib1 and antagonizes Mib1 activity during neuronal development (Choe et al., 2007). Sowa et al. also used a global proteomic analysis strategy to identify the proteins associated with DUBs. They developed an unbiased comparative approach that integrates parallel epitope-tagged purification with LC-MS/MS analysis (Sowa et al., 2009). With this platform, 774 high-confidence candidate proteins were found to interact with 75 human DUBs. Many of the DUBs were associated with protein complexes, and protein network analysis linked the DUBs to diverse functional processes. Recently, a similar study of DUB interactome and localization was performed in fission yeast (Kouranti et al., 2010).
In addition to E3s and DUBs, the interactome of a proteasome has been under intense scrutiny in recent years. Traditional purification of a functional proteasome uses high-salt buffer for washing, making the detection of weakly associated protein impossible. In contrast, by using epitope tag and near-physiological washing conditions during purification, a large number of proteasome-interacting proteins have been found (Finley, 2009). Guerrero et al. used the QTAX method to identify transient interacting components of the proteasome, disclosing 42 novel proteins from mammalian cells (Guerrero et al., 2008). So far, proteomic analyses have characterized proteasome complexes from different tissues such as brain, heart, kidney, liver, lung, thymus, spleen, and intestines (Bingol et al., 2010; Ducoux-Petit et al., 2008; Gorbea et al.; Tai et al. 2010). These studies identified a large number of proteasome-associated proteins, many of which play important regulatory roles. For example, the proteasome can regulate Ub chains of substrates dynamically through Hul5 (a proteasome-associated E3) and Ubp6/Usp14 protease. It is of clinical importance that drug targeting to proteasome-associated proteins may represent a new way to modulate proteasomal function for disease therapy. Along with this line, Lee et al. identified a small-molecule inhibitor of Usp14 and showed that inhibition of Usp14 catalytic activity increases the turnover of several proteins such as tau, ataxin-3, and TDP-43 (Lee et al., 2010), which are involved in neurodegenerative diseases. Furthermore, the localization and activity of the proteasome can be mediated through associated proteins in cell signaling pathways. For example, phosphorylation of CaMKIIα enhances both its association to and recruitment of the proteasome to the dendritic spine in hippocampal neurons (Bingol et al. 2010). The translocation of proteasome to the spine may play an important role in synapse function and memory. Therefore, proteasome-associated proteins regulate proteasome activity and localization, and are potential targets for disease treatment.
Conclusions
Although advances in MS have enabled the sensitive analysis of the ubiquitinated proteome and partners that interact with Ub pathway enzymes, Ub proteomics still lags far behind phosphoproteomics. The development of affinity capture methods as well as MS technology is crucial for biological and medical research on Ub signaling. Nevertheless, existing technologies have played a major role in deepening our understanding of protein ubiquitination, especially in the discovery of complex Ub chain structures and in the exploration of proteins associated with Ub enzymes. With the continuous development of MS and Ub affinity capture reagents, the future holds more extensive applications of MS-based proteomics for this field.
Acknowledgments
This work was partially supported by National Institutes of Health grants (RR025822, and NS055077) and an American Cancer Society grant (RSG-09-181). We thank C. Strauss for editing the manuscript.
References
- Bandopadhyay R, de Belleroche J. Pathogenesis of Parkinson’s disease: emerging role of molecular chaperones. Trends Mol Med. 2010;16:27–36. doi: 10.1016/j.molmed.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Bedford L, Layfield R, Mayer RJ, Peng J, Xu P. Diverse polyubiquitin chains accumulate following 26S proteasomal dysfunction in mammalian neurones. Neuroscience letters. 2011;491:44–47. doi: 10.1016/j.neulet.2010.12.064. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nature cell biology. 2010;12:80–86. doi: 10.1038/ncb2008. sup pp 81–12. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nature genetics. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- Boname JM, Thomas M, Stagg HR, Xu P, Peng J, Lehner PJ. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic. 2010;11:210–220. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nature structural & molecular biology. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burande CF, Heuze ML, Lamsoul I, Monsarrat B, Uttenweiler-Joseph S, Lutz PG. A label-free quantitative proteomics strategy to identify E3 ubiquitin ligase substrates targeted to proteasome degradation. Mol Cell Proteomics. 2009;8:1719–1727. doi: 10.1074/mcp.M800410-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Choe EA, Liao L, Zhou JY, Cheng D, Duong DM, Jin P, Tsai LH, Peng J. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J Neurosci. 2007;27:9503–9512. doi: 10.1523/JNEUROSCI.1408-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends in cell biology. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochemical and biophysical research communications. 1978;81:1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- Dallaire F, Blanchette P, Branton PE. A proteomic approach to identify candidate substrates of human adenovirus E4orf6-E1B55K and other viral cullin-based E3 ubiquitin ligases. Journal of virology. 2009;83:12172–12184. doi: 10.1128/JVI.01169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest etiology of Alzheimer disease. Journal of biological chemistry. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, Jensen LJ, Mailand N, Nielsen ML. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics. 2011;10:M110 003590. doi: 10.1074/mcp.M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- Denis NJ, Vasilescu J, Lambert JP, Smith JC, Figeys D. Tryptic digestion of ubiquitin standards reveals an improved strategy for identifying ubiquitinated proteins by mass spectrometry. Proteomics. 2007;7:868–874. doi: 10.1002/pmic.200600410. [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducoux-Petit M, Uttenweiler-Joseph S, Brichory F, Bousquet-Dubouch MP, Burlet-Schiltz O, Haeuw JF, Monsarrat B. Scaled-down purification protocol to access proteomic analysis of 20S proteasome from human tissue samples: comparison of normal and tumor colorectal cells. Journal of proteome research. 2008;7:2852–2859. doi: 10.1021/pr8000749. [DOI] [PubMed] [Google Scholar]
- Eng J, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annual review of biochemistry. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Ciechanover A, Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984;37:43–55. doi: 10.1016/0092-8674(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Franco M, Seyfried NT, Brand AH, Peng J, Mayor U. A novel strategy to isolate ubiquitin conjugates reveals wide role of ubiquitination during neural development. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M110.002188. [Epub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JS, Ray JW, Waseem N, Johnson K, Brooks MJ, Hugosson T, Breuer D, Branham KE, Krauth DS, Bowne SJ, et al. Mutations in a BTB-Kelch protein, KLHL7, cause autosomal-dominant retinitis pigmentosa. Am J Hum Genet. 2009;84:792–800. doi: 10.1016/j.ajhg.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciaa BA, Siutib N, Thomasb CE, Mizzena CA, Kelleher NL. Characterization of neurohistone variants and post-translational modifications by electron capture dissociation mass spectrometry. Int J Mass Spectr. 2007;259:184–196. [Google Scholar]
- Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- Goldknopf IL, Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proceedings of the National Academy of Sciences of the United States of America. 1977;74:864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- Gorbea C, Pratt G, Ustrell V, Bell R, Sahasrabudhe S, Hughes RE, Rechsteiner M. A protein interaction network for ECM29 links the 26S proteasome to molecular motors and endosomal components. Journal of biological chemistry. 2010;285:31616–31633. doi: 10.1074/jbc.M110.154120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto E, Yamanaka Y, Ishikawa A, Aoki-Kawasumi M, Mito-Yoshida M, Ohmura-Hoshino M, Matsuki Y, Kajikawa M, Hirano H, Ishido S. Contribution of lysine 11-linked ubiquitination to MIR2-mediated major histocompatibility complex class I internalization. Journal of biological chemistry. 2010;285:35311–35319. doi: 10.1074/jbc.M110.112763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat Rev Genet. 2009;10:617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annual review of biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annual review of cell and developmental biology. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Rodriguez MS. Efficient approaches for characterizing ubiquitinated proteins. Biochem Soc Trans. 2008;36:823–827. doi: 10.1042/BST0360823. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nature cell biology. 2000;2:E153–157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor S, Ziv T, Admon A, Lehner PJ. Stable isotope labeling by amino acids in cell culture and differential plasma membrane proteome quantitation identify new substrates for the MARCH9 transmembrane E3 ligase. Mol Cell Proteomics. 2009;8:1959–1971. doi: 10.1074/mcp.M900174-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough R, Pratt G, Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. The Journal of biological chemistry. 1987;262:8303–8313. [PubMed] [Google Scholar]
- Hsiao HH, Meulmeester E, Frank BTC, Melchior F, Urlaub H. “ChopNSpice,” a Mass Spectrometric Approach That Allows Identification of Endogenous Small Ubiquitin-like Modifier-conjugated Peptides. Molecular & Cellular Proteomics. 2009;8:2664–2675. doi: 10.1074/mcp.M900087-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Jeon MS, Liao L, Yang C, Elly C, Yates JR, 3rd, Liu YC. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity. 2010;33:60–70. doi: 10.1016/j.immuni.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon HB, Choi ES, Yoon JH, Hwang JH, Chang JW, Lee EK, Choi HW, Park ZY, Yoo YJ. A proteomics approach to identify the ubiquitinated proteins in mouse heart. Biochemical and biophysical research communications. 2007;357:731–736. doi: 10.1016/j.bbrc.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Jeram SM, Srikumar T, Pedrioli PG, Raught B. Using mass spectrometry to identify ubiquitin and ubiquitin-like protein conjugation sites. Proteomics. 2009;9:922–934. doi: 10.1002/pmic.200800666. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- Kao A, Chiu CL, Vellucci D, Yang Y, Patel VR, Guan S, Randall A, Baldi P, Rychnovsky SD, Huang L. Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol Cell Proteomics. 2011;10:M110.002212. doi: 10.1074/mcp.M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nature cell biology. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Knuesel M, Cheung HT, Hamady M, Barthel KK, Liu X. A method of mapping protein sumoylation sites by mass spectrometry using a modified small ubiquitin-like modifier 1 (SUMO-1) and a computational program. Mol Cell Proteomics. 2005;4:1626–1636. doi: 10.1074/mcp.T500011-MCP200. [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Kouranti I, McLean JR, Feoktistova A, Liang P, Johnson AE, Roberts-Galbraith RH, Gould KL. A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 2010;8:e1000471. doi: 10.1371/journal.pbio.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layfield R, Tooth D, Landon M, Dawson S, Mayer J, Alban A. Purification of poly-ubiquitinated proteins by S5a-affinity chromatography. Proteomics. 2001;1:773–777. doi: 10.1002/1615-9861(200106)1:6<773::AID-PROT773>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2010;10:R110.003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Commonly used tag combinations for tandem affinity purification. Biotechnology and applied biochemistry. 2010;55:73–83. doi: 10.1042/BA20090273. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Maor R, Jones A, Nuhse TS, Studholme DJ, Peck SC, Shirasu K. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol Cell Proteomics. 2007;6:601–610. doi: 10.1074/mcp.M600408-MCP200. [DOI] [PubMed] [Google Scholar]
- Marotti LA, Jr, Newitt R, Wang Y, Aebersold R, Dohlman HG. Direct identification of a G protein ubiquitination site by mass spectrometry. Biochemistry. 2002;41:5067–5074. doi: 10.1021/bi015940q. [DOI] [PubMed] [Google Scholar]
- Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, Gnad F, Mann M, Vertegaal AC. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Molecular cell. 2010;39:641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI. Large-scale analysis of the human ubiquitin-related proteome. Proteomics. 2005;5:4145–4151. doi: 10.1002/pmic.200401280. [DOI] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Molecular cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol Cell Proteomics. 2007;6:1885–1895. doi: 10.1074/mcp.M700264-MCP200. [DOI] [PubMed] [Google Scholar]
- Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol Cell Proteomics. 2005;4:741–751. doi: 10.1074/mcp.M400220-MCP200. [DOI] [PubMed] [Google Scholar]
- Meierhofer D, Wang X, Huang L, Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res. 2008;7:4566–4576. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama A, Yates JR., 3rd Multidimensional LC separations in shotgun proteomics. Anal Chem. 2008;80:7187–7193. doi: 10.1021/ac8013669. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, Shifrin N, Petri MA, Kamboh MI, Manzi S, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nature genetics. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, O’Donnell L, Kumakubo A, Munro M, Sicheri F, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466:941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Nielsen ML, Vermeulen M, Bonaldi T, Cox J, Moroder L, Mann M. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat Methods. 2008;5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nature methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Pedrioli PG, Raught B, Zhang XD, Rogers R, Aitchison J, Matunis M, Aebersold R. Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nature methods. 2006;3:533–539. doi: 10.1038/nmeth891. [DOI] [PubMed] [Google Scholar]
- Peng J. Evaluation of proteomic strategies for analyzing ubiquitinated proteins. BMB Rep. 2008;41:177–183. doi: 10.5483/bmbrep.2008.41.3.177. [DOI] [PubMed] [Google Scholar]
- Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Phu L, Izrael-Tomasevic A, Matsumoto ML, Bustos DJ, Dynek JN, Fedorova AV, Bakalarski CE, Arnott D, Deshayes K, Dixit VM, et al. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol Cell Proteomics. 2010;10:M110.003756. doi: 10.1074/mcp.M110.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Radulovic D, Jelveh S, Ryu S, Hamilton TG, Foss E, Mao Y, Emili A. Informatics platform for global proteomic profiling and biomarker discovery using liquid chromatography-tandem mass spectrometry. Mol Cell Proteomics. 2004;3:984–997. doi: 10.1074/mcp.M400061-MCP200. [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual review of biochemistry. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Schlesinger DH, Goldstein G. Molecular conservation of 74 amino acid sequence of ubiquitin between cattle and man. Nature. 1975;255:42304. doi: 10.1038/255423a0. [DOI] [PubMed] [Google Scholar]
- Schlesinger DH, Goldstein G, Niall HD. The complete amino acid sequence of ubiquitin, an adenylate cyclase stimulating polypeptide probably universal in living cells. Biochemistry. 1975;14:2214–2218. doi: 10.1021/bi00681a026. [DOI] [PubMed] [Google Scholar]
- Semple CA. The comparative proteomics of ubiquitination in mouse. Genome Res. 2003;13:1389–1394. doi: 10.1101/gr.980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried NT, Xu P, Duong DM, Cheng D, Hanfelt J, Peng J. Systematic approach for validating the ubiquitinated proteome. Anal Chem. 2008;80:4161–4169. doi: 10.1021/ac702516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chan DW, Jung SY, Malovannaya A, Wang Y, Qin J. A dataset of human endogenous ubiquitination sites. Mol Cell Proteomics. 2010;10:M110.002089. doi: 10.1074/mcp.M110.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Xu P, Qin J. Ubiquitinated proteome: Ready for global? Mol Cell Proteomics. 2011 doi: 10.1074/mcp.R110.006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen H, Mann M. The abc’s (and xyz’s) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- Tai HC, Besche H, Goldberg AL, Schuman EM. Characterization of the Brain 26S Proteasome and its Interacting Proteins. Frontiers in molecular neuroscience. 2010;3:12. doi: 10.3389/fnmol.2010.00012. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiqutin chain provides clues to functional diversity of polyubiquitin signaling. Journal of biological chemistry. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- Vasilescu J, Smith JC, Ethier M, Figeys D. Proteomic analysis of ubiquitinated proteins from human MCF-7 breast cancer cells by immunoaffinity purification and mass spectrometry. Journal of proteome research. 2005;4:2192–2200. doi: 10.1021/pr050265i. [DOI] [PubMed] [Google Scholar]
- Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nature chemical biology. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- Voorhees PM, Orlowski RZ. The proteasome and proteasome inhibitors in cancer therapy. Annu Rev Pharmacol Toxicol. 2006;46:189–213. doi: 10.1146/annurev.pharmtox.46.120604.141300. [DOI] [PubMed] [Google Scholar]
- Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Quantitative proteomic analysis of single pancreatic islets. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18902–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes & development. 2009;23:729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Cheng D, Peng J, Pickart CM. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J. 2006;25:1710–1719. doi: 10.1038/sj.emboj.7601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- Wang X, Guerrero C, Kaiser P, Huang L. Proteomics of proteasome complexes and ubiquitinated proteins. Expert review of proteomics. 2007;4:649–665. doi: 10.1586/14789450.4.5.649. [DOI] [PubMed] [Google Scholar]
- Warren MR, Parker CE, Mocanu V, Klapper D, Borchers CH. Electrospray ionization tandem mass spectrometry of model peptides reveals diagnostic fragment ions for protein ubiquitination. Rapid Commun Mass Spectrom. 2005;19:429–437. doi: 10.1002/rcm.1798. [DOI] [PubMed] [Google Scholar]
- Weekes J, Morrison K, Mullen A, Wait R, Barton P, Dunn MJ. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics. 2003;3:208–216. doi: 10.1002/pmic.200390029. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD, Urban MK, Haas AL. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. The Journal of biological chemistry. 1980;255:7529–7532. [PubMed] [Google Scholar]
- Wu SL, Kim J, Bandle RW, Liotta L, Petricoin E, Karger BL. Dynamic profiling of the post-translational modifications and interaction partners of epidermal growth factor receptor signaling after stimulation by epidermal growth factor using Extended Range Proteomic Analysis (ERPA) Mol Cell Proteomics. 2006;5:1610–1627. doi: 10.1074/mcp.M600105-MCP200. [DOI] [PubMed] [Google Scholar]
- Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nature biotechnology. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Molecular cell. 2009a;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Cheng D, Duong DM, Rush J, Roelofs J, Finley D, Peng J. A proteomic strategy for quantifying polyubiquitin chain topologies. Israel J Chem. 2006;46:171–182. [Google Scholar]
- Xu P, Duong DM, Peng J. Systematical optimization of reverse-phase chromatography for shotgun proteomics. Journal of proteome research. 2009b;8:3944–3950. doi: 10.1021/pr900251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009c;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Peng J. Dissecting the ubiquitin pathway by mass spectrometry. Biochimica et biophysica acta. 2006;1764:1940–1947. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Peng J. Characterization of polyubiquitin chain structure by middle-down mass spectrometry. Anal Chem. 2008;80:3438–3444. doi: 10.1021/ac800016w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun Z, Kaufman TC, Clemmer DE. Stable isotope labeling and label-free proteomics of Drosophila parkin null mutants. Journal of proteome research. 2009;8:4500–4510. doi: 10.1021/pr9006238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacological reviews. 2007;59:14–39. doi: 10.1124/pr.59.1.4. [DOI] [PubMed] [Google Scholar]
- Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. Detecting differential and correlated protein expression in label-free shotgun proteomics. Journal of proteome research. 2006;5:2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- Zhao S, Ulrich HD. Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7704–7709. doi: 10.1073/pnas.0908764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JY, Afjehi-Sadat L, Asress S, Duong DM, Cudkowicz M, Glass JD, Peng J. Galectin-3 Is a Candidate Biomarker for Amyotrophic Lateral Sclerosis: Discovery by a Proteomics Approach. Journal of proteome research. 2010;9:5133–5141. doi: 10.1021/pr100409r. [DOI] [PMC free article] [PubMed] [Google Scholar]