Abstract
Combinational therapy with small RNA inhibitory agents against multiple viral targets allows efficient inhibition of viral production by controlling gene expression at critical time points. Here we explore combinations of different classes of therapeutic anti-HIV-1 RNAs expressed from within the context of an intronic MCM7 (minichromosome maintenance complex component-7) platform that naturally harbors 3 microRNAs (miRNAs). We replaced the endogenous miRNAs with anti-HIV small RNAs, including small interfering RNAs (siRNAs) targeting HIV-1 tat and rev messages that function to induce post-transcriptional gene silencing by the RNA interference pathway, a nucleolar-localizing RNA ribozyme that targets the conserved U5 region of HIV-1 transcripts for degradation, and finally nucleolar trans-activation response (TAR) and Rev-binding element (RBE) RNA decoys designed to sequester HIV-1 Tat and Rev proteins inside the nucleolus. We demonstrate the versatility of the MCM7 platform in expressing and efficiently processing the siRNAs as miRNA mimics along with nucleolar small RNAs. Furthermore, three of the combinatorial constructs tested potently suppressed viral replication during a 1-month HIV challenge, with greater than 5-log inhibition compared with untransduced, HIV-1-infected CEM T lymphocytes. One of the most effective constructs contains an anti-HIV siRNA combined with a nucleolar-localizing U5 ribozyme and TAR decoy. This represents the first efficacious example of combining Drosha-processed siRNAs with small nucleolar ribonucleoprotein (snoRNP)-processed nucleolar RNA chimeras from a single intron platform for effective inhibition of viral replication. Moreover, we demonstrated enrichment/selection for cells expressing levels of the antiviral RNAs that provide optimal inhibition under the selective pressure of HIV. The combinations of si/snoRNAs represent a new paradigm for combinatorial RNA-based gene therapy applications.
Chung and colleagues explore combinations of various classes of therapeutic anti-HIV-1 RNAs expressed from within the context of an intronic MCM7 (minichromosome maintenance complex component-7) platform. Endogenous MCM7 microRNAs (miRNAs) were replaced with various small RNA-inhibitory agents aimed at efficiently inhibiting viral production by controlling viral gene expression at critical time points. The MCM7 platform successfully allowed for expression and processing of small interfering RNAs (siRNAs), miRNA mimics, and nucleolar small RNAs. Furthermore, three of the combinatorial constructs tested potently suppressed viral replication during a 1-month HIV challenge, with greater than 5-log inhibition compared with untransduced, HIV-1-infected cells.
Introduction
HIV gene expression is a highly regulated process that involves alternative splicing from the full-length 9-kb RNA genome to generate various viral proteins. Early in the replication cycle, transcription elongation is inefficient despite having a functional viral promoter, resulting in short or early-terminated transcripts until the early regulatory protein Tat is made. Tat functions to vastly increase transcription through the trans-activation of RNA polymerase II (RNA Pol II) from the viral promoter (Kao et al., 1987; Toohey and Jones, 1989; Marciniak and Sharp, 1991; Laspia et al., 1993; Garber and Jones, 1999) via interaction with the trans-activation response (TAR) element in the 5′ untranslated region (UTR) of the viral RNAs (Kao et al., 1987). Once Tat is available, transcription becomes processive and multiply spliced transcripts are produced that encode the other regulatory proteins, including Rev. Rev facilitates the export of partially spliced and unspliced transcripts to the cytoplasm for translation into late structural proteins by interactions with the Rev response element (RRE) present on these transcripts (Felber et al., 1989, 1990; Cullen and Malim, 1991; Krug, 1993). In addition to their known functions in the nucleus, Tat and Rev exhibit nucleolar-localizing properties with poorly understood functions, hypothesized as part of the transport mechanism or temporal storage (Tat [Li, 1997; Luznik et al., 1995; Ruben et al., 1989; Siomi et al., 1990; Stauber and Pavlakis, 1998]; Rev [Nosaka et al., 1993; Dundr et al., 1995]). Similarly, full-length and partially spliced HIV transcripts have also been detected by electron microscopy in situ hybridization (Canto-Nogues et al., 2001). Taken together, these results suggest HIV-1 RNAs traffic through the nucleolus as part of the replication cycle. The importance of the nucleolus during viral replication could be more universal as transcription and replication of Borna disease virus (a negative-strand RNA virus) occur in the nucleolus (Pyper et al., 1998). Interestingly, env RNAs from the human T-lymphotropic virus were also detected in the nucleolus (Kalland et al., 1991).
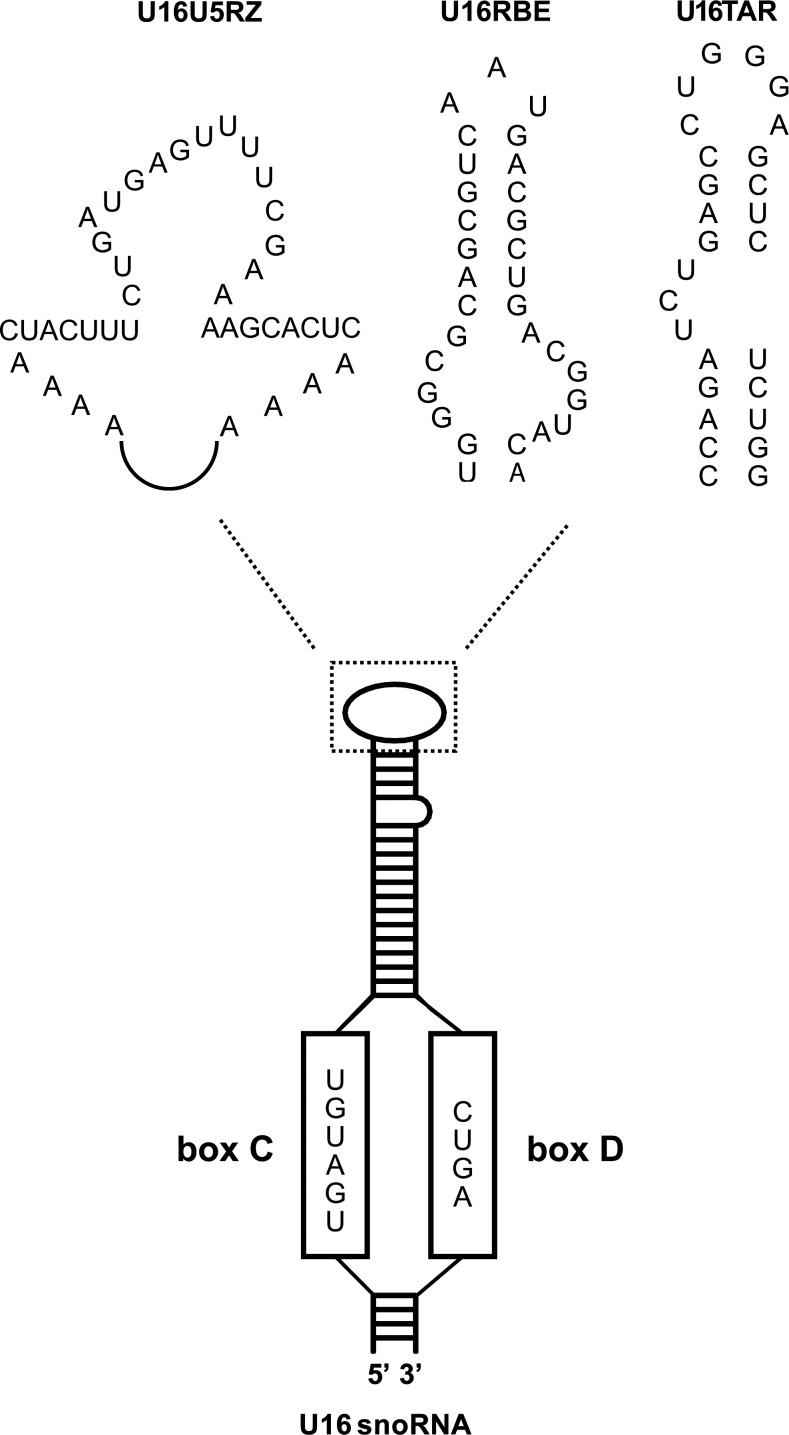
To investigate whether the nucleolus plays an important role in HIV replication, nucleolar-localizing TAR and Rev-binding element (RBE) RNA decoys (U16TAR and U16RBE, respectively) that function to trap HIV-1 Tat and Rev proteins inside the nucleolus, and a RNA ribozyme that targets a conserved U5 region of HIV-1 RNA (U16U5RZ), were created by substituting these anti-HIV small RNAs for the apical loop of the C/D box U16 small nucleolar RNA (snoRNA) (Fig. 1). The U16 chimeric RNAs were shown to localize in the nucleolus and each was shown to have strong anti-HIV-1 activity (Michienzi et al., 2002, 2006; Unwalla et al., 2008). It was also noted in these studies that the nucleolar-localizing TAR RNA decoy was a far more potent inhibitor than a nuclear-localizing counterpart. These results strengthened the importance of the nucleolar trafficking of Tat during viral replication (Michienzi et al., 2002). Taken together, these findings suggest nucleolar trapping could be a novel avenue for developing anti-HIV therapeutics and support the important functional role of Tat and Rev nucleolar localization in viral replication (Michienzi et al., 2002).
FIG. 1.
Construction of small nucleolar anti-HIV RNAs. The C/D box U16 small nucleolar RNA (snoRNA) is used as a scaffold to construct nucleolar-localizing anti-HIV small RNAs. The conserved C/D box of the U16 snoRNA is sufficient for the nucleolar-localizing property with the apical loop replaced with various anti-HIV RNAs including the U5 targeting RNA ribozymes (U16U5RZ), the Rev-binding element RNA decoy (U16RBE), and the trans-activation response RNA decoy (U16TAR).
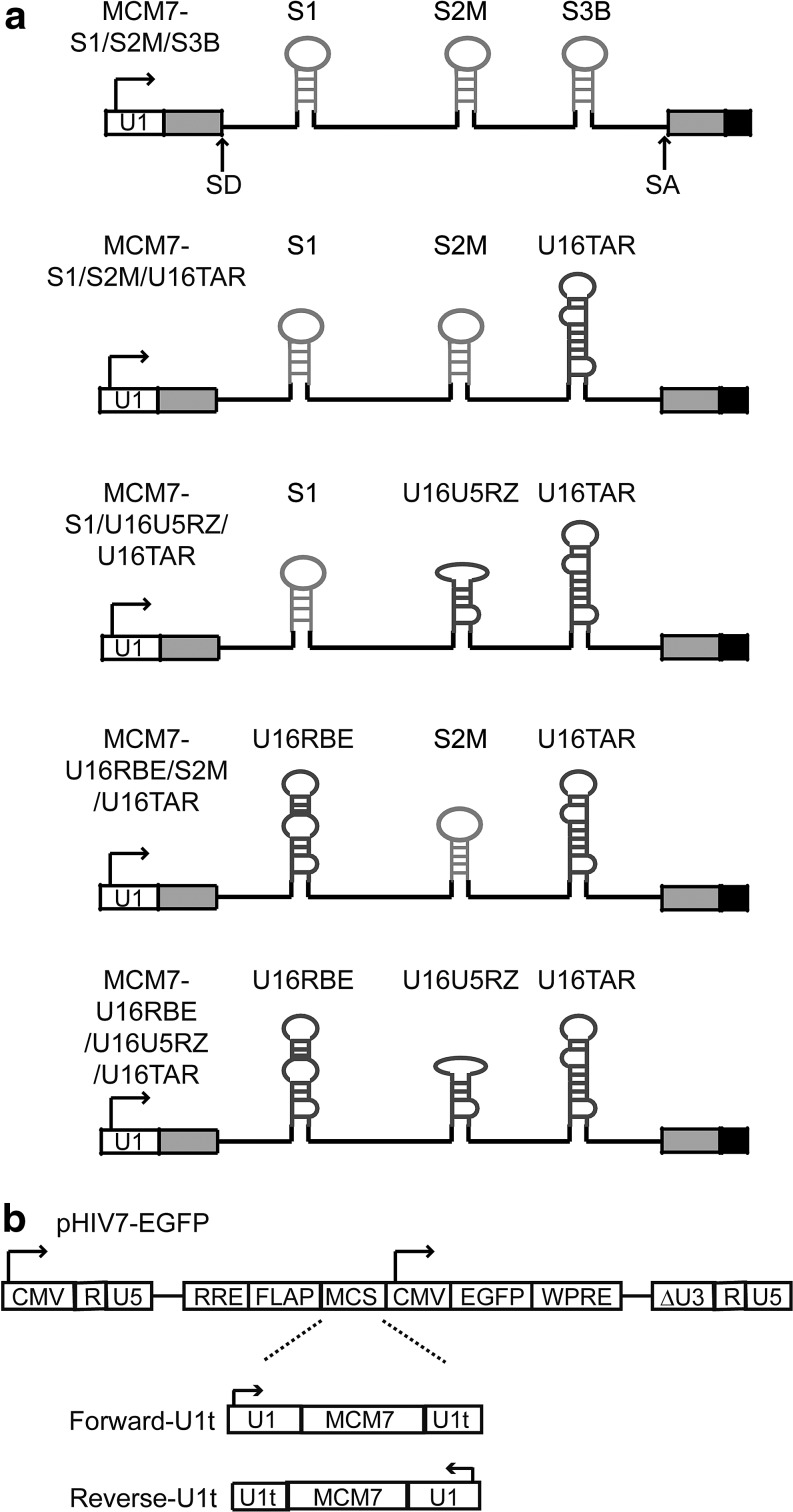
We previously chose the naturally occurring polycistronic miR-106b cluster located in intron 13 of the protein-encoding MCM7 (minichromosome maintenance complex component-7) gene on chromosome 7 as the scaffold to coexpress three anti-HIV small interfering RNAs (siRNAs) from a single RNA Pol II human U1 promoter (Aagaard et al., 2008). These studies demonstrated efficient expression and processing of three siRNAs that target the common tat/rev exon (S1), rev (S2M), and tat (S3B), respectively, as miRNA mimics (Aagaard et al., 2008). Although multiplexing siRNAs is an approach to mitigating viral escape mutants found with a single point mutation in the siRNA target site (Boden et al., 2003; Das et al., 2004; Sabariegos et al., 2006), we believe that it is also advantageous to explore the potential for combining different types of small RNA inhibitors to further reduce the likelihood of viral resistance and to exploit the potential synergy between small RNA agents within a single gene therapy construct (Li et al., 2003). The MCM7 platform offers additional advantages and flexibility over multiple small RNA agents expressed with constitutive independent Pol III promoters (e.g., Li et al., 2003) by offering opportunities to engineer tissue specificity by proper promoter choice while reducing toxicity related to overexpression. We previously demonstrated that the MCM7 platform could also be used for coexpression of the U16TAR RNA decoy by replacing the S3B subunit, as shown by the MCM7-S1/S2M/U16TAR construct (Aagaard et al., 2008). The processing of U16TAR was shown to be independent of Drosha, implicating that the snoRNA was processed independently of the siRNAs via the C/D box small nucleolar ribonucleoprotein (snoRNP) processing pathway (Aagaard et al., 2008). Here we demonstrate that multiple small nucleolar RNAs can also be incorporated in this platform, where they are effectively processed along with the siRNAs to provide combinatorial, long-term inhibition of HIV-1 replication in CEM T lymphocytes. The combinations of si/snoRNAs represent a new paradigm for combinatorial RNA-based gene therapy applications.
Materials and Methods
Generation of MCM7 snoRNA constructs
The U16RBE and U16U5RZ snoRNA molecules were amplified by PCR from pTZ/U6-U16RBE and pTZ/U6-C36U5 DNA vectors (Michienzi et al., 2006; Unwalla et al., 2008), using primer sets A and B, as XhoI–HindIII and EcoRI–BamHI fragments, respectively. The fragments were digested with appropriate enzymes followed by cloning into the pcDNA3-CMV-MCM7-S1/S2/U16TAR plasmid (Aagaard et al., 2008). The U1-specific transcriptional terminator along with a new NotI site was introduced at the terminus of the common 3′ region of the MCM7 cassette by replacing the original DNA sequence with a PCR product generated with primer set C. The cytomegalovirus (CMV) promoter was replaced by the U1 promoter flanked by MluI and KpnI sites generated by amplification from a U1 plasmid (pKS-U1; our unpublished data) with primer set D.
To generate lentiviral vectors, the U1-MCM7-U1t fragments were excised by MluI and NotI digestion and ligated into the pHIV7-EGFP lentiviral vector in both forward and reverse orientations (i.e., the U1 promoter is in the same or opposite orientation as the packaging CMV promoter, respectively), depending on the directionality of the multiple cloning site.
Primer sequences are as follows, with restriction sites underlined and U1-specific terminator in boldface:
A: Forward: 5′-CCC CCC CCTC GAG CTT GCA ATG ATG TCG TAA TTT G-3′
Reverse: 5′-CCC CAA GCT TAA AAA TTT CTT GCT CAG TAA GAA TTT-3′
B: Forward: 5′-CCC CCC CGA ATT CCT TGC AAT GAT GTC GTA ATT TG-3′
Reverse: 5′-CCC CGG ATC CAA AAA TTT CTT GCT CAG TAA GAA TTT-3′
C: Forward: 5′-ATC GAT CCG CGG ATG CTG GGG GGA GGG GGG AT-3′
Reverse: 5′-ACG TGT TAA CGC GGC CGC AGT CTA CTT TTG AAA CTC TGC CCC TTG TCT CCT AGA-3′
D: Forward: 5′-ATC GAT ACG CGT CTA AGG ACC AGC TTC TTT GGG AGA G-3′
Reverse: 5′-ATC GAT GGT ACC GAT CTT CGG GCT CTG CCC CG-3′
Lentiviral vector production
Lentiviral vectors with appropriate inserts were packaged in HEK 293T cells by calcium phosphate precipitation as previously described (Li and Rossi, 2005) with the addition of 1.5 μg of pAgo2sh plasmid (H. Soifer [MEDomics, Azusa, CA], unpublished data), which expresses a short hairpin RNA (shRNA) transcribed from the human U6 promoter to downregulate Argonaute-2 (Ago2) protein expression to reduce post-transcriptional gene silencing induced by anti-HIV siRNA within constructs during packaging. The viral supernatants were collected 48 hours after transfection, concentrated via ultracentrifugation, and stored at −80°C until use. Viral titers were determined by transduction of HT1080 cells and analyzed for enhanced green fluorescent protein (EGFP) expression by flow cytometry.
Cell culture and vector transduction
HEK 293T and HT1080 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in high-glucose (4.5 g/liter) Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mM glutamine and 10% fetal bovine serum (FBS). Human CEM T lymphocytes was cultured in RPMI 1640 medium supplemented with 10% FBS.
CEM T lymphocytes were transduced with lentiviral vectors as previously described (Li et al., 2005), with the exception of the multiplicity of infection (MOI) used. At an MOI of 50, almost 100% of the cells were EGFP+ as determined by flow cytometry and were used for subsequent experiments without sorting.
Northern blotting analysis
Total RNA was isolated from stably transduced CEM T lymphocytes with STAT-60 reagent (Tel-Test, Friendswood, TX) according to the manufacturer's protocol and resuspended in nuclease-free water. For each construct, 20 μg of total RNA was separated in an 8% polyacrylamide gel containing 8 M urea and then electroblotted onto a Hybond-N nylon membrane (Amersham, Arlington Heights, IL) and hybridized with 32P-labeled DNA probes complementary to the individual small RNAs. The U6 small nuclear RNA was used as a loading control.
The small RNA probe sequences are as follows:
S1: 5′-GCG GAG ACA GCG ACG AAG AGC-3′
S2M: 5′-GCC TGT GCC TCT TCA GCT ACC-3′
S3B: 5′-CAT CTC CTA TGG CAG GAA GAA-3′
U16RBE: 5′-CGT CAG CGT CAT TGA CGC TGC GCC CA-3′
U16U5RZ: 5′-GAG TGC TTT TCG AAA ACT CAT CAG AA-3′
U16TAR: 5′-CCA GAG AGC TCC CAG GCT CAG-3′
U6: 5′-TAT GGA ACG CTT CTC GAA TT-3′
HIV-1 challenge and p24 antigen assays
One million untransduced or stably transduced CEM T lymphocytes were infected in triplicate with the NL4-3 strain of HIV-1 at an MOI of 0.01. After overnight incubation, cells were washed three times with Hanks' balanced salts solution and cultured in RPMI 1640 with 10% FBS. At designated time points, culture supernatants were collected and analyzed for HIV-1 replication by a p24 ELISA (PerkinElmer, Waltham, MA) according to the manufacturer's protocol.
Real-time quantitative RT-PCR to quantify anti-HIV RNA expression
Total RNA from stably transduced CEM T lymphocytes challenged with HIV-1 was extracted with STAT-60 reagent (Tel-Test, Friendswood, TX) according to the manufacturer's instructions and then resuspended in nuclease-free water. Residual DNA was digested with Ambion TURBO DNase (Life Technologies, Carlsbad, CA) with 1 μg of total RNA in a 10-μl reaction, in accordance with the manufacturer's instructions. Both S1 siRNA and U16TAR RNA decoy expression were analyzed by real-time qRT-PCR with the CFX96 real-time detection system (Bio-Rad, Hercules, CA), and expression levels were normalized to the U6 small nuclear RNA. S1 siRNA was reverse transcribed into cDNA, using a TaqMan microRNA reverse transcription kit (Applied Biosystems, Foster City, CA) with 100 ng of DNase-treated total RNA and stem–loop RT primer according to the manufacturer's instructions. Real-time PCR was carried out with 1.3 μl of RT reaction, 0.2 μM S1-specific probe, 1.5 μM forward primer, and 0.7 μM reverse primer in TaqMan universal PCR master mix (Applied Biosystems, Foster City, CA) diluted to 1×concentration in a final volume of 20 μl. PCR conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 30 sec, 64°C for 30 sec, and 72°C for 30 sec (DiGiusto et al., 2010). The exact copy number of S1 siRNA was determined by comparison with a standard curve constructed with known concentrations of synthetic S1 RNA oligonucleotide (Integrated DNA Technologies, Coralville, IA).
The U16TAR RNA decoy and the internal control small nuclear U6 RNA were reverse transcribed, using 200 ng of DNase-treated total RNA with 50 ng of random primers (Invitrogen, Carlsbad, CA) and Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA) in a 20-μl reaction according to the manufacturer's instructions. Real-time PCR for the U16TAR RNA decoy was carried out with 1 μl of the RT reaction, 0.2 μM TAR-specific probe, and a 0.5 μM concentration of each U16-specific forward and reverse primer in TaqMan universal PCR master mix (Applied Biosystems, Foster City, CA) diluted to 1×concentration in a final volume of 20 μl. The PCR conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 30 sec and 64°C for 1 min. The exact copy number of RNA molecules was determined by comparison with a standard curve constructed with known concentrations of U16TAR plasmid. Quantification of the U6 internal control was accomplished with 2 μl of the RT reaction with a 0.4 μM concentration of each U6-specific forward and reverse primer, using iQ SYBR green supermix (Bio-Rad, Hercules, CA) in a final volume of 25 μl. The PCR conditions were 95°C for 5 min, followed by 40 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec. A standard curve with known amounts of total RNA input was used to determine the precise RNA input to account for sample-to-sample variability.
Quantitative RT-PCR primer sequences are as follows:
S1: Stem–loop RT primer: 5′-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA GCG GA-3′
Probe: 5′-(6-FAM)-TCG CAC TGG ATA CGA CAG CGG AGA CA-(BHQ1)-3′
Forward: 5′-GCC TCT TCG TCG CTG TCT-3′
Reverse: 5′-GTG CAG GGT CCG AGG T-3′
U16TAR: Probe: 5′-(6-FAM)-ATC TGA GCC TGG GAG CTC TCT GGC T-(BHQ1)-3′
Forward: 5′-TGC GTC TTA CTC TGT TCT CAG CGA-3′
Reverse: 5′-CGT CAA CCT TCT GTA CCA GCT TAC-3′
U6: Forward: 5′-GCT CGC TTC GGC AGC ACA TAT ACT AA-3′
Reverse: 5′-ACG AAT TTG CGT GTC ATC CTT GCG-3′
Statistical analyses
The average and standard deviation for S1 siRNA and U16TAR RNA decoy expression were derived from three independent measurements. Data were analyzed with the statistical software Prism version 5.01 (GraphPad Software, La Jolla, CA), using one-way analysis of variance followed by Bonferroni's multiple comparison test. Values of p less than or equal to 0.05 were considered statistically significant compared with cells cultured under identical conditions in the absence of HIV (i.e., day 0).
Results
Generation of MCM7 snoRNA constructs
Previously, we engineered and optimized a polycistronic miRNA cluster located in an intron of the protein-encoding gene MCM7 as an siRNA multiplexing platform (Aagaard et al., 2008). This platform, which we refer to as MCM7, was engineered to simultaneously express three anti-HIV siRNAs targeted to the common exon shared by tat/rev (S1), rev (S2M), and tat (S3B), respectively (MCM7-S1/S2M/S3B in Fig. 2a) from a single RNA Pol II human U1 promoter. Moreover, we demonstrated the potential versatility of this platform by coexpressing a U16TAR snoRNA inserted in the position of the S3B subunit. Given the demonstration of coexpression of siRNAs and snoRNAs, we hypothesized that it should be possible to insert the chimeric snoRNAs into any of the three miRNA positions to obtain processing of these small RNAs. If such was the case, we could then examine various combinations of chimeric snoRNAs and siRNAs coexpressed in the same transcript. To test this hypothesis we inserted the U16RBE and U16U5RZ snoRNAs into the MCM7-S1/S2M/U16TAR construct by replacing either the S1 or S2M unit. This resulted in the creation of three novel constructs harboring multiple snoRNA chimeras with different targets and mechanisms of action, as shown in Fig. 2a.
FIG. 2.
Overview of MCM7 intron-based lentiviral vectors. The name “MCM7” refers to a naturally occurring polycistronic miRNA cluster located in an intron of the MCM7 gene. Exons and intron of the MCM7 cassette are drawn as gray boxes and black lines, respectively, with splice donor and acceptors marked as “SD” and “SA.” Promoters are denoted by white boxes with the arrow indicating directionality, and the terminators are denoted by black boxes. shRNA and U16 snoRNA chimeras are drawn in light and dark grey, respectively. (a) The MCM7 scaffold allows coexpression of three small RNAs from the single Pol II U1 promoter. S1, S2M, and S3B represent siRNAs targeting the common tat/rev exon, rev, and tat, respectively. U16U5RZ is a nucleolar-localizing ribozyme targeting a conserved U5 region present in all HIV transcripts. U16TAR is a nucleolar-localizing TAR RNA decoy. U16RBE is a nucleolar-localizing Rev-binding element RNA decoy. (b) The MCM7 cassette with the U1-specific termination sequence (U1t) was cloned into the pHIV7-EGFP lentiviral vector in the forward orientation with respect to the CMV packaging promoter, denoted as “Forward-U1t,” whereas the cassette in the opposite orientation is denoted as “Reverse-U1t.” RRE, Rev response element; MCS, multiple cloning site; EGFP, enhanced green fluorescent protein; WPRE, woodchuck hepatitis virus post-transcription regulation element; ΔU3, deleted U3 region to generate a self-inactivating lentiviral vector after integration in targeted cells.
The original MCM7-S1/S2M/S3B and MCM7-S1/S2M/U16TAR constructs were subcloned into the pHIV7-EGFP lentiviral vector (Yam et al., 2002) in the reverse orientation with respect to the packaging CMV promoter to prevent splicing of the MCM7 intron during vector packaging (Aagaard et al., 2008). Alternatively, the HIV-1 Rev protein used in the packaging process suppresses transcript splicing, suggesting we could also orient the U1-MCM7 intron in the same transcriptional direction as the CMV packaging promoter. To test these possibilities, we cloned the MCM7 transgene in both forward and reverse orientations with a U1 promoter-specific termination sequence (Fig. 2b) and compared packaging efficiencies. Interestingly, the packaging efficiencies were greater than 100-fold better in constructs with the transgene cloned in the forward orientation (Table 1). We therefore used the forward orientations for transduction of CEM T lymphocytes to produce cell lines that stably expressed the various combinations of anti-HIV RNAs.
Table 1.
Packaging Efficiencies of Lentiviral Vectors with MCM7 Transgene
| |
Forward-U1t |
Reverse-U1t |
|
||
|---|---|---|---|---|---|
| Construct | Viral titer (TU/ml) | Ratio (to pHIV7-EGFP) | Viral titer (TU/ml) | Ratio (to pHIV7-EGFP) | Ratio (forward to reverse) |
| pHIV7-EGFP (empty) | 1.41±0.66×106 | ||||
| MCM7-S1/S2M/S3B | 4.01±1.23×106 | 2.84 | 4.20±0.50×104 | 0.030 | 96 |
| MCM7-S1/S2M/U16TAR | 4.11±1.24×106 | 2.91 | 3.00±0.42×104 | 0.021 | 137 |
| MCM7-S1/U16U5RZ/U16TAR | 4.34±1.46×106 | 3.08 | 1.44±0.34×104 | 0.010 | 301 |
| MCM7-U16RBE/S2M/U16TAR | 4.66±1.72×106 | 3.30 | 2.41±0.45×104 | 0.017 | 193 |
| MCM7-U16RBE/U16U5RZ/U16TAR | 4.12±1.58×106 | 2.92 | 4.18±0.22×104 | 0.008 | 349 |
Viral titer was determined by transducing HT1080 cells with unconcentrated viral supernatant and is reported as transduction units per milliliter (TU/ml). Samples with ∼30–40% EGFP+ cells, determined by flow cytometry, were used for calculation. The values represent averages of two independent experiments.
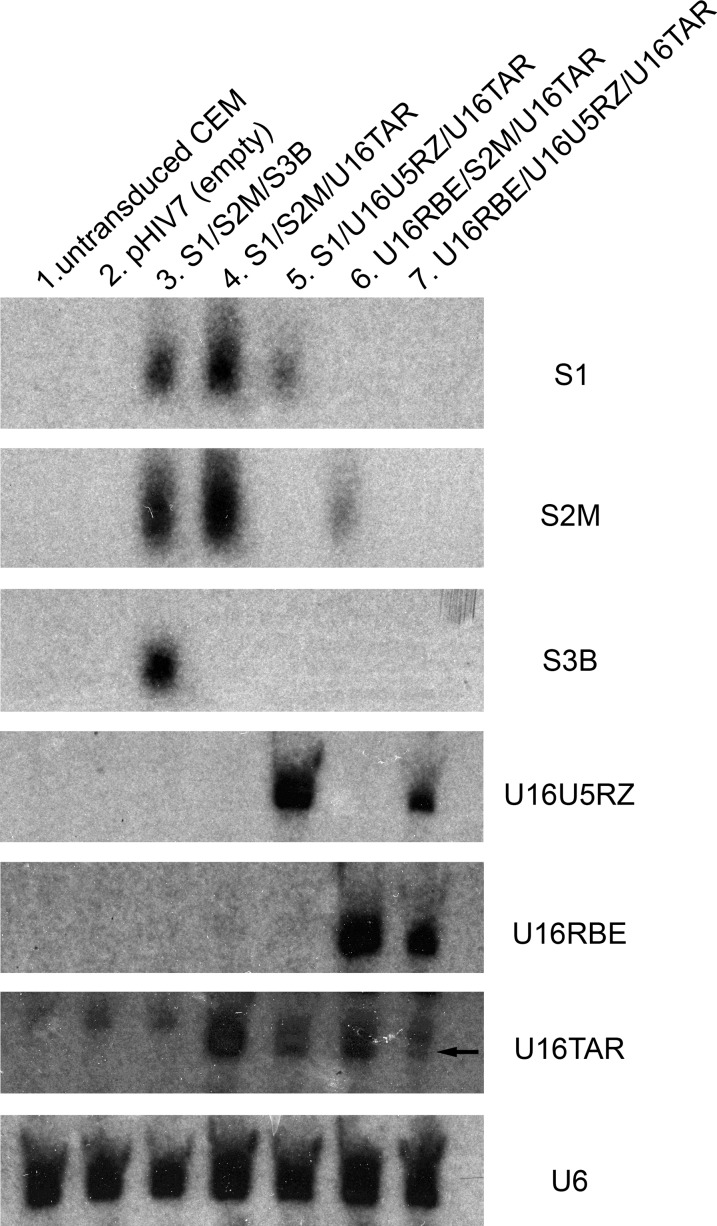
Proper processing and expression of functional small RNAs in target cells
To determine and verify whether the RNA expression of each unit within the combinatorial vector was properly transcribed and processed, Northern blotting analysis was performed on stably transduced CEM T lymphocytes (Fig. 3). After electrophoresis of the RNA samples, the blots were hybridized with probes specific for each of the RNAs. RNA expression was detected for each unit in the various constructs, with expected sizes of about 21 nucleotides for siRNAs and about 132 nucleotides for U16 chimeric snoRNAs, indicating efficient processing of the RNAs from the polycistronic transcript. It is interesting to note that certain RNA combinations express lower levels of small RNAs in this platform, implying proper processing of both si- and snoRNA within the same intron is an intricate balance between the Drosha/DGCR8 and the snoRNP pathways and furthermore may be position dependent (Hirose et al., 2003).
FIG. 3.
Expression of the small RNAs in stably transduced CEM T lymphocytes. CEM T lymphocytes were transduced with lentiviruses containing the MCM7 cassette in the forward orientation at an MOI of 50. About 20 (μg of total RNA was loaded per lane and electrophoresed in an 8% polyacrylamide gel with 8 M urea, blotted onto a nylon membrane, and hybridized with the corresponding 32P-labeled probes. RNA prepared from untransduced cells (lane 1) and cells transduced with empty pHIV7 vector (lane 2) were used as negative controls. Lanes 3 to 7 antiviral constructs as labeled. S1, S2M, and S3B siRNAs are approximately 21 nucleotides. The U16 snoRNA chimeras are approximately 132 nucleotides. U6 small nuclear RNA serves as a loading control.
Suppression of viral replication in CEM T lymphocytes expressing MCM7 intron containing anti-HIV small RNAs
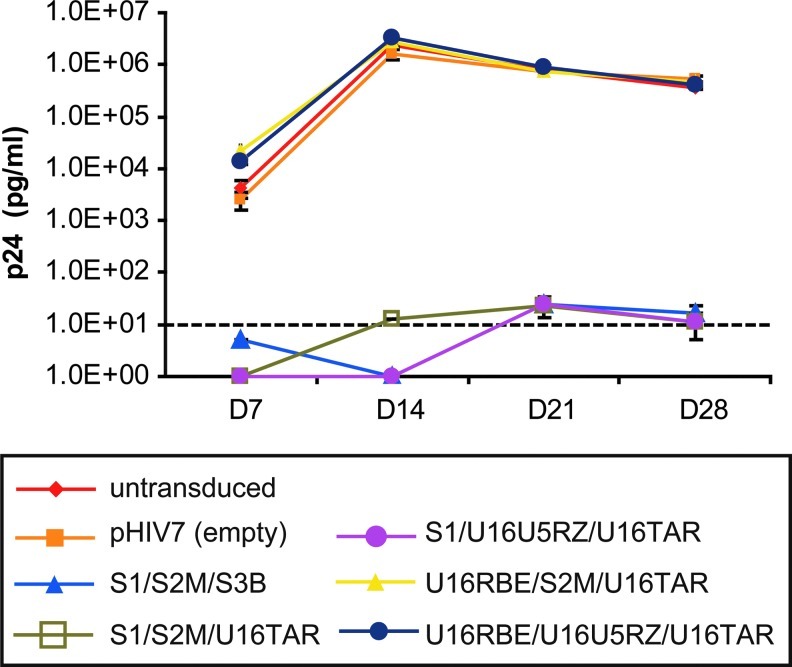
We next addressed the issue of functionality of the small RNAs as measured by antiviral activity. Long-term inhibition of viral replication in CEM T lymphocytes stably expressing the MCM7 constructs was evaluated in viral challenge assays using the NL4-3 strain of HIV-1 at an MOI of 0.01. Viral replication was monitored for 28 days by monitoring viral capsid p24 levels in the culture supernatant at the indicated time points (Fig. 4). Three of the five constructs, MCM7-S1/S2M/S3B, MCM7-S1/S2M/U16TAR, and MCM7-S1/U16U5RZ/U16TAR, showed extremely potent anti-HIV activity, providing greater than a 5-log reduction in p24 output, with almost nondetectable p24 during this 1-month challenge.
FIG. 4.
Anti-HIV activity of MCM7-based constructs. One million untransduced and stable CEM T lymphocytes were challenged in triplicate with the NL4-3 strain of HIV-1 at an MOI of 0.01, and culture supernatants were collected weekly for the HIV-1 p24 antigen ELISA to evaluate viral replication. The dashed line represents the low detection limit of the p24 assay. Three constructs, MCM7-S1/S2M/S3B, MCM7-S1/S2M/U16TAR, and MCM7-S1/U16U5RZ/U16TAR, showed potent antiviral activity with almost no detectable viral load during the 1-month challenge assay.
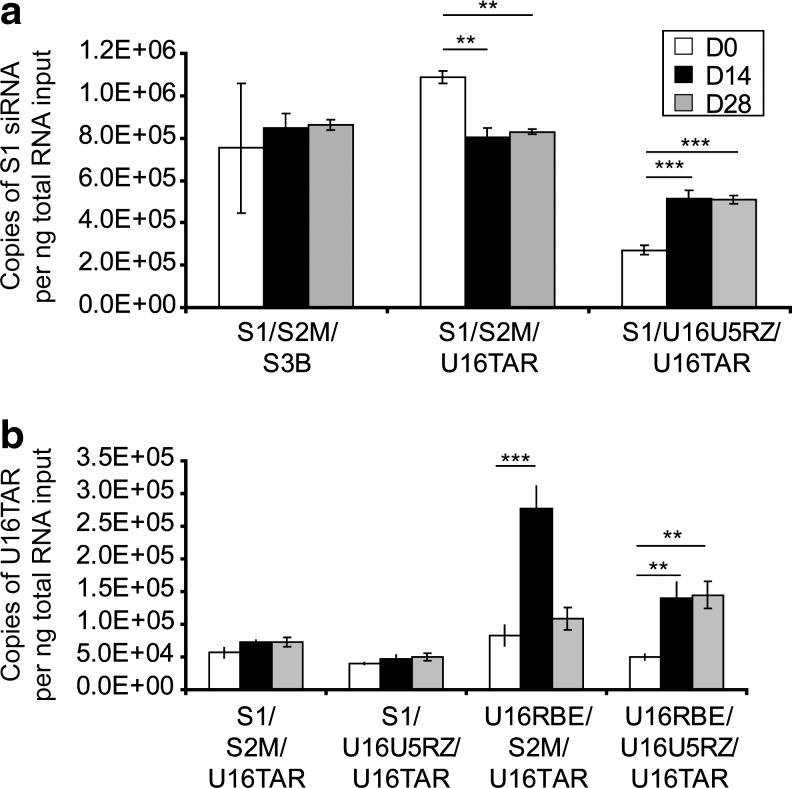
Having achieved strong suppression of viral replication during the challenge assay in CEM T lymphocytes, we were interested in correlating gene expression with functionality. We hypothesized that HIV-1 would provide selective pressure to enrich for transduced cells with levels of anti-HIV RNA gene expression that effectively suppress replication. If this is the case, we would expect selection for cells with optimal RNA expression levels during the time course of the HIV-1 challenge. We used qRT-PCR to measure S1 siRNA and U16TAR decoy RNA expression (Fig. 5a and b, respectively). Cells transduced with the MCM7-S1/U16U5RZ/U16TAR construct had a significant 2-fold enrichment for S1 siRNA expression (p<0.001), whereas interestingly, cells transduced with the MCM7-S1/S2M/U16TAR construct had a significant 20% reduction for S1 siRNA expression (p<0.01). Significant enrichment for the U16 TAR RNA decoy was observed only in constructs that did not inhibit HIV. These data are consistent with the mechanism of action for each of the small RNAs, with siRNA being catalytic and the decoys being stoichiometric in sequestering their targets. Taken together, these data suggest that under the selective pressure of HIV, there is an enrichment/selection for cells expressing levels of the anti-viral RNAs that provide optimal inhibition in the absence of toxicity.
FIG. 5.
In vitro virus-mediated selection of transduced CEM T lymphocytes with optimal anti-HIV gene expression. (a) Total RNA was extracted from CEM T lymphocytes infected with HIV-1 at designated time points (days 0, 14, and 28) to evaluate RNA expression. S1 siRNA expression was evaluated by qRT-PCR and normalized to the internal control U6 small nuclear RNA. (b) Total RNA was extracted from CEM T lymphocytes infected with HIV-1 at designated time points (days 0, 14, and 28) to evaluate RNA expression. The U16 TAR RNA decoy expression was evaluated by qRT-PCR and normalized by the internal control U6 small nuclear RNA. **p<0.01, ***p<0.001, difference compared with uninfected control (day 0).
To investigate whether combinations of the various inhibitory RNAs were more efficacious as inhibitors than single antiviral RNAs, we used a dual-luciferase reporter assay in which we transiently cotransfected a replication-deficient pNL4-3 proviral DNA harboring the firefly luciferase gene in the HIV-1 nef gene (pNL4-3.Luc.R−.E−, cat. no. 3418 from NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH [Connor et al., 1995; He et al., 1995]) and the anti-HIV RNAs driven either by the U1 or U6 promoter. The pNL4-3 luciferase construct maintains targets for each of the small RNAs in all the transcripts, both spliced and unspliced, and therefore luciferase readouts can be used as quantitative readouts of viral inhibition. We observed a general trend correlating knockdown activity with small RNA expression (Supplementary Fig. S1; supplementary data are available online at www.liebertonline.com/hum), consistent with their mechanism of action by increasing target cleavage in the case of U16U5RZ or by sequestering the Tat protein in the case of U16TAR. We were surprised to see that U16RBE did not exhibit any antiviral activity in this reporter assay, possibly because of overwhelming production of luciferase-labeled viral transcripts during transient transfection and the fact that it is noncatalytic. Overall, these observations suggest that the gene expression level is one of the determinants of an efficient RNA-based therapy.
Discussion
The use of combinations of small molecule drugs in highly active antiretroviral therapy (HAART) to stop or thwart HIV propagation has had a major impact on delaying the progression from HIV-1 infection to the development of AIDS. Despite this progress, there are problems associated with lifelong use of antiviral drug therapy, including toxicity, the emergence of virus resistant to multiple drugs, and the cost of daily medication. Gene therapy of human T lymphocytes and/or hematopoietic progenitor cells can be considered as a potential replacement or supplement to the current anti-HIV-1 therapies. Similar to small molecule therapies in which combinations of drugs targeting different steps in the viral replication cycle have been most effective, we believe that therapeutic RNAs must also be used in combinations to block various stages of the viral replication cycle to mitigate viral escape. On the basis of findings that both HIV-1 viral RNA transcripts and proteins localize in the nucleolus, we have previously demonstrated that nucleolar-localizing small RNAs can be potent therapeutic agents. For example, we had previously succeeded in inhibiting HIV-1 replication by individually expressing snoRNA chimeras, including the U16TAR and U16RBE RNA decoys that sequester the HIV-1 Tat and Rev proteins in the nucleolus, respectively (Michienzi et al., 2002, 2006). In addition, we also demonstrated that a nucleolar-localizing ribozyme targeting a conserved U5 sequence present in all HIV-1 transcripts had excellent HIV-1 inhibitory function (Michienzi et al., 2002, 2006; Unwalla et al., 2008). As a combinatorial approach to incorporate anti-HIV small RNAs with different mechanisms of action and target specificity, we multiplexed the aforementioned snoRNA chimeras in addition to siRNAs that cleave tat and rev mRNAs with the goal to block all transcript production and efficiently achieve suppression of HIV-1 replication.
We chose to use a single promoter and an intron-based platform to express combinations of siRNAs and snoRNAs. Both miRNAs and snoRNAs are processed from introns, thereby providing a rationale for our approach (Hirose et al., 2003). We previously engineered and optimized the polycistronic miRNA cluster referred to as MCM7 to coexpress three anti-HIV siRNAs from a single Pol II human U1 promoter (Aagaard et al., 2008). We have now carefully examined several aspects of using this system in a lentiviral vector backbone platform. The current constructs were further optimized for packaging efficiency by cloning the MCM7 transgene in the forward direction with respect to the CMV promoter in the lentiviral pHIV7-EGFP vector with the U1-specific transcriptional termination sequence. We were surprised by the superior packaging efficiency of our constructs, especially those with the transgene in the forward orientation, compared with the empty pHIV7-EGFP vector. Expression of the anti-HIV RNAs might be expected to negatively impact transcription of the full-length viral RNA genome during packaging because our pHIV7-EGFP lentiviral vector is dependent on HIV-1 Rev for packaging. Because all of the constructs contain at least one small RNA against HIV-1 Rev, it was expected that the viral titer of the constructs might be lower or equivalent at best to that of the parental pHIV7-EGFP vector. We have previously overcome this challenge by increasing the amount of HIV Rev-expressing plasmid (Li et al., 2005) or by inclusion of a plasmid that expresses an Ago2-targeting shRNA (H. Soifer, unpublished data) to minimize the RNAi activity in cells during packaging. In the present case, we did not find an advantage to downregulating Ago2 (data not shown) because the siRNA expression levels are relatively low compared with Pol III-transcribed shRNAs, and during packaging these do not effectively downregulate the viral transcripts. We also postulate that insertion of the MCM7 cassette produces a larger viral transcript (5.4 kb) whose size is closer to that of the natural HIV-1 RNA genome (9 kb) and therefore more favorable for packaging compared with the parental empty vector (3.9 kb). Indeed, we found a 2.5-fold increase in viral titer when the parental MCM7 intron lacking anti-HIV RNAs was incorporated (data not shown), similar to our gene therapy constructs carrying anti-HIV RNAs. Second, we observed the effect of transgene directionality on packaging efficiency, with the forward orientation yielding greater than 100-fold higher production of virus. It is likely that the transgene RNA transcript, when expressed from the U1 promoter in the reverse orientation, could create an opposing transcript during packaging that negatively impacts on levels of expression from an antisense effect.
In this study we have demonstrated the versatility of the MCM7 platform for expressing multiple siRNAs as miRNA mimics as well as snoRNAs from the polycistronic transcript, and efficient processing into mature and functional small RNAs that are readily detectable through Northern blotting analysis. Long-term inhibition of viral replication was evaluated by challenging stably transduced CEM T lymphocytes with HIV-1 NL4-3. The results of these analyses demonstrated that the MCM7-S1/S2M/S3B, MCM7-S1/S2M/U16TAR, and MCM7-S1/U16U5RZ/U16TAR constructs conferred complete protection against viral replication and spread during the 1-month challenge. Interestingly, the MCM7-U16RBE/S2M/U16TAR and MCM7-U16RBE/U16U5RZ/U16TAR constructs did not significantly inhibit HIV replication despite the fact that the small RNAs were actively expressed and readily detectable by Northern blotting. The U16 chimeras in these constructs had demonstrated antiviral activity when individually expressed from the parental vector with the Pol III U6 promoter (Michienzi et al., 2002, 2006; Unwalla et al., 2008). This discrepancy is most likely related to the difference in expression levels of these RNAs in the context of the intronic MCM7 platform driven by the Pol II U1 promoter versus independently from the Pol III U6 promoter.
The importance of optimal levels of RNA expression for anti-HIV activity and cell viability is supported by the observation that there was selection for transduced CEM T lymphocytes with optimal anti-HIV RNA expression during HIV infection. This phenomenon has also been observed for transduced CEM T lymphocytes harboring a single copy of the transgene (data not shown), reflecting selection of cells with more transcriptionally active integration sites. We evaluated overall RNA expression by qRT-PCR and showed persistent expression during challenge, and therefore it is likely that there is selective pressure for cells with optimal expression to provide antiviral activity in the absence of cellular toxicity.
In addition to RNA expression levels as a determinant for the effectiveness of an RNA-based gene therapy, the nature of the small RNAs should also be considered and carefully balanced between toxicity and therapeutic efficacy. Because RNA decoys act as “sponges” and therefore function in a stoichiometric fashion, the expression level needs to be sufficiently high to achieve therapeutic efficacy, whereas siRNAs and ribozymes are capable of multiple turnover by cleaving their targets in a catalytic manner and should be functional with lower copies per cell. Constructs with the most potent antiviral activities in the context of the MCM7 intron platform tended to have higher RNA expression levels and usually contained more than two RNA agents that are catalytic in nature, such as an siRNA or ribozyme. It is interesting to note that all the constructs that exhibit antiviral activity in the viral challenge assay contain the S1 siRNA that targets both the HIV-1 tat and rev messages. Although our current data cannot demonstrate whether the other two small RNAs in the constructs have additive effects in antiviral activity, the principle of combinational therapy is to reduce viral escape in a long-term setting. We have previously demonstrated that the combination of three is superior to two and better than single small RNA agents in prolonging anti-HIV protection in long-term setting in a viral challenge assay (Li et al., 2005).
In summary, these studies represent the first example of incorporating combinations of snoRNA-based agents with siRNA-based agents within a single expression platform driven by a single Pol II promoter. We demonstrated the versatility of the MCM7 platform for expressing a variety of small antiviral RNAs in addition to miRNAs. We also demonstrated superior packaging of these constructs versus the parental empty pHIV7-EGFP lentiviral vector. The enhanced packaging efficiency was especially pronounced when the transgene was cloned in the forward orientation with respect to the packaging CMV promoter. Last, after HIV-1 challenges of CEM T lymphocytes transduced with the various RNA combinations, we found three small RNA combinations, MCM7-S1/S2M/S3B, MCM7-S1/S2M/U16TAR, and MCM7-S1/U16U5RZ/U16TAR, that strongly inhibited viral replication during the 1-month challenge. We also found that the pressure of HIV-1 infection resulted in selection of cells with optimal levels of anti-HIV gene expression. The two RNA combinations that contained two or more nucleolar RNAs did not significantly inhibit HIV replication, perhaps owing to the noncatalytic nature of RNA decoys versus the siRNAs and ribozyme. Our results suggest these factors should be carefully considered in designing an efficient RNA-based gene therapy.
Supplementary Material
Acknowledgments
The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3.Luc.R−.E− from Dr. Nathaniel Landau (Connor et al., 1995; He et al., 1995). This work was funded by NIH AI 42552, AI29329 to J.J.R. and CIRM TR2-01771 to D.L.D. and J.J.R. The authors thank Dr. John Burnett for helpful discussions and the members of the Rossi laboratory for their support.
Author Disclosure Statement
No competing financial interests exist.
References
- Aagaard L.A. Zhang J. von Eije K.J., et al. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008;15:1536–1549. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden D. Pusch O. Lee F., et al. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto-Nogues C. Hockley D. Grief C., et al. Ultrastructural localization of the RNA of immunodeficiency viruses using electron microscopy in situ hybridization and in vitro infected lymphocytes. Micron. 2001;32:579–589. doi: 10.1016/s0968-4328(00)00053-6. [DOI] [PubMed] [Google Scholar]
- Connor R.I. Chen B.K. Choe S. Landau N.R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Cullen B.R. Malim M.H. The HIV-1 Rev protein: Prototype of a novel class of eukaryotic post-transcriptional regulators. Trends Biochem. Sci. 1991;16:346–350. doi: 10.1016/0968-0004(91)90141-h. [DOI] [PubMed] [Google Scholar]
- Das A.T. Brummelkamp T.R. Westerhout E.M., et al. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiusto D.L. Krishnan A. Li L., et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34+ cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010;2:36–43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M. Leno G.H. Hammarskjold M.L., et al. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J. Cell Sci. 1995;108:2811–2823. doi: 10.1242/jcs.108.8.2811. [DOI] [PubMed] [Google Scholar]
- Felber B.K. Hadzopoulou-Cladaras M. Cladaras C., et al. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber B.K. Drysdale C.M. Pavlakis G.N. Feedback regulation of human immunodeficiency virus type 1 expression by the Rev protein. J. Virol. 1990;64:3734–3741. doi: 10.1128/jvi.64.8.3734-3741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M.E. Jones K.A. HIV-1 Tat: Coping with negative elongation factors. Curr. Opin. Immunol. 1999;11:460–465. doi: 10.1016/S0952-7915(99)80077-6. [DOI] [PubMed] [Google Scholar]
- He J. Choe S. Walker R., et al. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T. Shu M.D. Steitz J.A. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell. 2003;12:113–123. doi: 10.1016/s1097-2765(03)00267-3. [DOI] [PubMed] [Google Scholar]
- Kalland K.H. Langhoff E. Bos H.J., et al. Rex-dependent nucleolar accumulation of HTLV-I mRNAs. New Biol. 1991;3:389–397. [PubMed] [Google Scholar]
- Kao S.Y. Calman A.F. Luciw P.A. Peterlin B.M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Krug R.M. The regulation of export of mRNA from nucleus to cytoplasm. Curr. Opin. Cell Biol. 1993;5:944–949. doi: 10.1016/0955-0674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- Laspia M.F. Wendel P. Mathews M.B. HIV-1 Tat overcomes inefficient transcriptional elongation in vitro. J. Mol. Biol. 1993;232:732–746. doi: 10.1006/jmbi.1993.1427. [DOI] [PubMed] [Google Scholar]
- Li M.J. Rossi J.J. Lentiviral vector delivery of recombinant small interfering RNA expression cassettes. Methods Enzymol. 2005;392:218–226. doi: 10.1016/S0076-6879(04)92013-7. [DOI] [PubMed] [Google Scholar]
- Li M.J. Bauer G. Michienzi A., et al. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol. Ther. 2003;8:196–206. doi: 10.1016/s1525-0016(03)00165-5. [DOI] [PubMed] [Google Scholar]
- Li M.J. Kim J. Li S., et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol. Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- Li Y.P. Protein B23 is an important human factor for the nucleolar localization of the human immunodeficiency virus protein Tat. J. Virol. 1997;71:4098–4102. doi: 10.1128/jvi.71.5.4098-4102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luznik L. Kraus G. Guatelli J., et al. Tat-independent replication of human immunodeficiency viruses. J. Clin. Invest. 1995;95:328–332. doi: 10.1172/JCI117660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak R.A. Sharp P.A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michienzi A. Li S. Zaia J.A. Rossi J.J. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14047–14052. doi: 10.1073/pnas.212229599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michienzi A. De Angelis F.G. Bozzoni I. Rossi J.J. A nucleolar localizing Rev binding element inhibits HIV replication. AIDS Res. Ther. 2006;3:13. doi: 10.1186/1742-6405-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka T. Takamatsu T. Miyazaki Y., et al. Cytotoxic activity of Rev protein of human immunodeficiency virus type 1 by nucleolar dysfunction. Exp. Cell Res. 1993;209:89–102. doi: 10.1006/excr.1993.1289. [DOI] [PubMed] [Google Scholar]
- Pyper J.M. Clements J.E. Zink M.C. The nucleolus is the site of Borna disease virus RNA transcription and replication. J. Virol. 1998;72:7697–7702. doi: 10.1128/jvi.72.9.7697-7702.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben S. Perkins A. Purcell R., et al. Structural and functional characterization of human immunodeficiency virus Tat protein. J. Virol. 1989;63:1–8. doi: 10.1128/jvi.63.1.1-8.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabariegos R. Gimenez-Barcons M. Tapia N., et al. Sequence homology required by human immunodeficiency virus type 1 to escape from short interfering RNAs. J. Virol. 2006;80:571–577. doi: 10.1128/JVI.80.2.571-577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H. Shida H. Maki M. Hatanaka M. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J. Virol. 1990;64:1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber R.H. Pavlakis G.N. Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology. 1998;252:126–136. doi: 10.1006/viro.1998.9400. [DOI] [PubMed] [Google Scholar]
- Toohey M.G. Jones K.A. In vitro formation of short RNA polymerase II transcripts that terminate within the HIV-1 and HIV-2 promoter-proximal downstream regions. Genes Dev. 1989;3:265–282. doi: 10.1101/gad.3.3.265. [DOI] [PubMed] [Google Scholar]
- Unwalla H.J. Li H. Li S.Y., et al. Use of a U16 snoRNA-containing ribozyme library to identify ribozyme targets in HIV-1. Mol. Ther. 2008;16:1113–1119. doi: 10.1038/mt.2008.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam P.Y. Li S. Wu J., et al. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol. Ther. 2002;5:479–484. doi: 10.1006/mthe.2002.0558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.