Abstract
Dehalococcoides bacteria are the only organisms known to completely reduce chlorinated ethenes to the harmless product ethene. However, Dehalococcoides dechlorinate these chemicals more effectively and grow more robustly in mixed microbial communities than in isolation. In this study, the phylogenetic composition and gene content of a functionally stable trichloroethene-degrading microbial community was examined using metagenomic sequencing and analysis. For phylogenetic classification, contiguous sequences (contigs) longer than 2500 bp were grouped into classes according to tetranucleotide frequencies and assigned to taxa based on rRNA genes and other phylogenetic marker genes. Classes were identified for Clostridiaceae, Dehalococcoides, Desulfovibrio, Methanobacterium, Methanospirillum, as well as a Spirochete, a Synergistete, and an unknown Deltaproteobacterium. Dehalococcoides contigs were also identified based on sequence similarity to previously sequenced genomes, allowing the identification of 170 kb on contigs shorter than 2500 bp. Examination of metagenome sequences affiliated with Dehalococcoides revealed 406 genes not found in previously sequenced Dehalococcoides genomes, including 9 cobalamin biosynthesis genes related to corrin ring synthesis. This is the first time that a Dehalococcoides strain has been found to possess genes for synthesizing this cofactor critical to reductive dechlorination. Besides Dehalococcoides, several other members of this community appear to have genes for complete or near-complete cobalamin biosynthesis pathways. In all, 17 genes for putative reductive dehalogenases were identified, including 11 novel ones, all associated with Dehalococcoides. Genes for hydrogenase components (271 in total) were widespread, highlighting the importance of hydrogen metabolism in this community. PhyloChip analysis confirmed the stability of this microbial community.
Keywords: bioremediation, cobalamin, Dehalococcoides, metagenomics, reductive dehalogenation, trichloroethene
Introduction
Chlorinated ethenes are common groundwater contaminants that pose human health risks (McCarty, 1997; Moran et al., 2007; US Dept. of H&HS, 2007). PCE (tetrachloroethene) and TCE (trichloroethene) are suspected human carcinogens (US Dept. of H&HS, 2005) that have been widely used for a variety of applications, including dry cleaning and metal degreasing (Mohn and Tiedje, 1992; McCarty, 1997). The partially dechlorinated degradation products of TCE, dichloroethene (cis-DCE and trans-DCE) and VC (vinyl chloride), are also toxic, and VC is a known human carcinogen (Kielhorn et al., 2000; US Dept. of H&HS, 2005).
Although several groups of organisms can reductively dechlorinate PCE and TCE to the toxic intermediate DCE (Scholz-Muramatsu et al., 1995; Sharma and McCarty, 1996; Holliger et al., 1998; Luijten et al., 2003; Löffler et al., 2004), Dehalococcoides (Dhc) species are the only organisms known to dechlorinate these compounds completely to the harmless product ethene (Maymó-Gatell et al., 1997; Smidt and de Vos, 2004). Dhc are strictly anaerobic bacteria that use chlorinated ethenes and other chlorinated organics as electron acceptors (Maymó-Gatell et al., 1997; Smidt and de Vos, 2004). These reductive dechlorination reactions are catalyzed by membrane-associated enzymes called reductive dehalogenases (RDases) (Smidt and de Vos, 2004). Genome sequencing of several Dhc strains has revealed a large variety of putative RDase genes. The complement of RDase genes varies greatly between strains and corresponds to variation in dechlorination abilities (Kube et al., 2005; Seshadri et al., 2005; McMurdie et al., 2009; Lee et al., 2011). Further, the suite of Dhc RDase genes that have been tied to functional activity are far fewer, currently numbering six in all (pceA, tceA, vcrA, bvcA, cbrA and mbrA) (Magnuson et al., 1998, 2000; Krajmalnik-Brown et al., 2004; Müller et al., 2004; Adrian et al., 2007; Chow et al., 2010).
Dhc species have been found to grow more robustly and reduce chlorinated organics more effectively when grown in mixed communities rather than in isolation (Maymó-Gatell et al., 1997; He et al., 2007). This may be due to Dhc's stringent metabolic needs. For example, in addition to requiring chlorinated organics as terminal electron acceptors, all known Dhc species obligately use hydrogen as an electron donor and acetate as a carbon source (Maymó-Gatell et al., 1997; Adrian et al., 2000; He et al., 2003). Further, although cobalamin is a necessary cofactor for RDases (Smidt and de Vos, 2004), no Dhc strains have been reported to be capable of synthesizing cobalamin de novo (Kube et al., 2005; Seshadri et al., 2005; He et al., 2007). Previously sequenced Dhc strains have genes encoding for enzymes in the second part of the cobalamin biosynthesis pathway, lower ligand attachment and rearrangement (Maymó-Gatell et al., 1997; Kube et al., 2005; McMurdie et al., 2009), but not for the first part of the pathway, corrin ring synthesis. Even when Dhc isolates are provided sufficient hydrogen, acetate and cobalamin, they do not grow or dechlorinate as robustly as when grown in mixed communities or defined consortia (He et al., 2007; Lee et al., 2011; Men et al., 2012).
The dechlorinating community studied here is an enrichment culture that has been stably dechlorinating TCE to ethene for over 10 years. This culture was derived from sediment collected at the Alameda Naval Air Station and is referred to as ANAS (Richardson et al., 2002). The phylogenetic composition of ANAS has been studied using clone libraries (Richardson et al., 2002; Lee et al., 2006), and the Dhc strains in ANAS have been analyzed using quantitative PCR and whole-genome microarrays (Holmes et al., 2006; West et al., 2008; Lee et al., 2011). ANAS contains two Dhc strains, which have recently been isolated (Holmes et al., 2006; Lee et al., 2011). A comparative genomics analysis showed these strains to have very similar core genomes, but different RDase genes, with correspondingly different dechlorination abilities (Lee et al., 2011).
Metagenomic sequencing analysis is used in this study to examine the Dhc component in the context of the ANAS microbial community. Metagenomic data provide a broad view of the genetic composition of a community, including information about the identity and potential metabolic capabilities of community members. Metagenomic approaches have been used to study a variety of microbial communities, including those inhabiting termite guts, human intestines, wastewater treatment plants and acid mines (Tyson et al., 2004; Gill et al., 2006; Warnecke et al., 2007; Sanapareddy et al., 2009). In the case of dechlorinating communities, metagenomic data can provide insights into the organisms that support dechlorination activity (Waller, 2009).
In this study, we identify and examine DNA sequences of Dhc and other ANAS community members from metagenomic sequence data. We also focus on three categories of functional genes related to dechlorination activity: genes for RDases, genes for cobalamin biosynthesis enzymes and genes for hydrogenases. Cobalamin biosynthesis was targeted because cobalamin is a required cofactor for RDases (Smidt and de Vos, 2004). Hydrogenases, which catalyze the reversible oxidation of molecular hydrogen, were targeted because Dhc couple reductive dechlorination to hydrogen oxidation (Maymó-Gatell et al., 1997; Adrian et al., 2000; He et al., 2003).
Materials and methods
ANAS enrichment culture and DNA sample preparation
Culture conditions and maintenance procedures for ANAS have been described previously (Richardson et al., 2002). Briefly, 350 ml of culture was grown at 25–28 °C and 1.8 atm with a N2-CO2 (90:10) headspace in a 1.5 l continuously stirred semi-batch reactor. The culture was amended with 13 μl TCE and 25 mM lactate every 14 days.
Cells were collected from 30 ml culture samples by vacuum filtration onto hydrophilic Durapore membrane filters (0.22 μm pore size, 47 mm diameter (Millipores, Billerica, MA, USA)), and filters were stored in 2 ml microcentrifuge tubes at −80 °C until further processing. For PhyloChip experiments, samples were collected from the same time point (27 h) from three different 14-day cycles of the culture to achieve biological triplication. For metagenomic sequencing, samples from the same time point (27 h) from four different feeding cycles were pooled to collect enough material for sequencing. Total nucleic acids were extracted from frozen filters using a modified version of the bead beating and phenol extraction method described previously (West et al., 2008).
Metagenome sequencing, assembly and annotation
Metagenome sequencing, assembly and annotation were performed at the Department of Energy Joint Genome Institute (JGI). A combination of 454-Titanium sequencing (453 944 reads) and paired-end short-insert Sanger sequencing (76 272 mate pairs, approximate insert size 3 kb) was used. 454-Titanium sequencing reads were assembled into contiguous sequences (contigs) using Newbler (454 Life Sciences, Roche Applied Sciences, Branford, CT, USA). Those contigs were shredded to resemble overlapping Sanger sequencing reads, which were then combined into an assembly with the paired-end Sanger sequencing reads using the Paracel Genome Assembler (Paracel Inc., Pasadena, CA, USA). Similar methods have been used by other researchers to combine Sanger and 454-Titanium sequencing data (Goldberg et al., 2006; Woyke et al., 2009). The contigs resulting from this second assembly, as well as Sanger reads and Newbler contigs that could not be further assembled, were annotated through a version of the JGI microbial annotation pipeline (Mavromatis et al., 2009) adapted to metagenomes, which includes prediction of protein coding and RNA genes and product naming. Annotation was automated and no manual annotation was preformed. Data were loaded into the Integrated Microbial Genomes with Microbiome Samples (IMG/M) database (Markowitz et al., 2010) and used in the following analyses.
Analysis of metagenomic sequence data
Identification of Dhc contigs by sequence similarity (SS)
Dhc contigs were identified in a two-stage SS process. In the first stage, contigs were identified by comparison to previously sequenced Dhc reference genomes (Dhc strains 195, BAV1, CBDB1, VS and GT). Reference genome sequences were retrieved from the National Center for Biotechnology Information (NCBI) genomes database (ftp://ftp.ncbi.nlm.nih.gov/genomes), and blastn (Zhang et al., 2000) was used to compare the reference genome sequences against a database of all metagenome contig sequences. For each reference genome, the top 250 BLAST hit contigs were selected for the second-stage comparison (at this cutoff, additional contigs did not expand the useful contig set), where their identities were checked by comparison with the NCBI genomes database using megablast (Zhang et al., 2000). All contigs whose top BLAST hit (lowest expect value) in the genomes database was to Dhc were selected and expect values were checked for significance. For all contigs identified as Dhc, the expect value of the identifying BLAST hit was ⩽ 10−35.
Classification of ANAS contigs by tetranucleotide frequencies (TF)
ANAS contigs larger than 2500 bp were grouped by TF using a procedure based on one described by Dick et al., (2009) with some modifications described in the Supplementary Information.
Comparisons to reference genomes and identification of novel Dhc genes
To identify regions of similarity and difference between a set of metagenome contigs and a reference genome, each contig in the set was compared with the reference genome using blastn, with an expect value cutoff of 10−12 unless otherwise stated. Based on the results of these searches, aligning and non-aligning regions were identified in the contigs and the reference genome.
Two measures of overall similarity between contigs and reference are reported. The first, contig match, is the percentage of total bases in all contigs that are part of an alignment to the reference genome. The second, reference match, is the percentage of bases in the reference genome that are part of an alignment to some contig in the set.
To identify contig regions containing potentially novel Dhc genes, Dhc metagenome contigs were compared with five sequenced Dhc genomes (strains 195, BAV1, CBDB1, VS and GT) that were publicly available in August 2010. Contig regions that were not in alignments to any reference genomes and were over 100 bases in length were investigated further. A less stringent expect value cutoff (10−6) was used to ensure that only low similarity regions were left unaligned and were included in the analysis. All annotated genes contained in the non-aligning regions, or overlapping the regions by at least five bases, were identified as novel.
Confirmation of novel Dhc genes in Dhc isolates from ANAS
Selected novel Dhc genes identified in the metagenome were amplified and sequenced from Dhc strains previously isolated from ANAS. Primers were designed based on the metagenome gene sequence using Primer3 (Table 1). PCR reactions were performed in 0.2 ml tubes in using Qiagen Taq DNA Polymerase. The thermocycler program was as follows: 12 min at 94 °C; 40 cycles of 1 min at 94 °C, 45 s at the annealing temperature (Table 1) and 2 min at 72 °C; 12 min at 72 °C. gDNA (genomic DNA) from Dhc strains ANAS1 and ANAS2 were used as templates for separate reactions. ANAS metagenomic DNA was used as a positive control template and Dhc strain 195 gDNA was used as a negative control template. PCR products were visualized on agarose gels and purified using the QIAquick PCR Purification Kit (QIAGEN Inc., Valencia, CA, USA). Purified PCR products were sequenced by Sanger sequencing.
Table 1. PCR primers and annealing temperatures.
| Target gene name | JGI IMG gene object ID |
Primer sequences |
Annealing temperature for PCR | |
|---|---|---|---|---|
| Forward | Reverse | |||
| cbiD | 2014753801 | 5′-ACCGCCAGCCTCAGGGTTGA-3′ | 5′-ACAGCCGCCATGGCACACAG-3′ | 59 °C |
| cbiF | 2014753804 | 5′-CGCTGTCTGGAAGAAGCCGACC-3′ | 5′-TGCATGGCGGAGGCCAGATT-3′ | 57 °C |
| cbiC | 2014753814 | 5′-CGCCGTTGTCCGCCAGCTTA-3′ | 5′-TTTCACCCGCCGCTTCTGCC-3′ | 58 °C |
Abbreviations: IMG, integrated microbial genomes; JGI, Joint Genome Institute.
PhyloChip assessment of community composition
Metagenomic DNA and RNA extracted from ANAS were applied to separate PhyloChip microarrays to examine the phylogenetic composition of ANAS. Details of the methods for these experiments are in the Supplementary Information and draw on several previously published methods (Cole et al., 2004; Brodie et al., 2006; DeSantis et al., 2007; West et al., 2008).
Results
ANAS metagenome overview
ANAS metagenome sequences were assembled into 26 293 contigs, totaling 41 065 977 bp of DNA sequence. Contigs ranged in length from 78 bp to 921 258 bp, with an N50 length of 2149 bp. A total of 60 992 protein-coding genes and 565 RNA genes were identified. The annotation is available through IMG/M (http://img.jgi.doe.gov/cgi-bin/m/main.cgi) (Taxon Object ID 2014730001) (Markowitz et al., 2008b).
Dhc in ANAS
Identification of Dhc contigs
The SS method identified 301 contigs as Dhc. In the TF analysis, one class containing 45 contigs was identified as Dhc, based on the presence of 16S and 23S rRNA genes that were 100% and 99% identical to those of Dhc strain 195.
The Dhc contigs identified by SS and by TF were compared to evaluate the two methods. Of the 301 Dhc contigs (1 810 488 bp total) identified by SS, 49 (1 643 099 bp total) were sufficiently long (> 2500 bp) for classification by TF. Of those, the TF method classified 45 as Dhc (the class of 45 identified above) and one (ANASMEC_C10442) as a Synergistete, leaving three (ANASMEC_C5086, ANASMEC_C818 and ANASMEC_C10029) unclassified.
The four contigs identified by SS but not by TF were further examined to determine possible reasons for the discrepancy. The BLAST alignments identifying these contigs by SS covered 25% or less of each contig's length. In the non-aligning sequence regions, two contigs (ANASMEC_5086 and ANASMEC_C818) contained several phage-related genes and recombinases, indicating possible horizontal DNA transfer, which could explain non-Dhc TF classification of sequences from a Dhc genome. The other contigs (ANASMEC_C10029 and ANASMEC_C10442) did not contain genes that were obvious indicators of horizontal transfer, although this does not rule out that explanation. Mis-assembly may also be responsible for the presence of both Dhc and non-Dhc sequence in these contigs. Given this uncertainty, the following analyses consider contigs identified by TF and SS separately, and make special note when these four contigs are relevant to a particular analysis.
Identification of novel Dhc genes
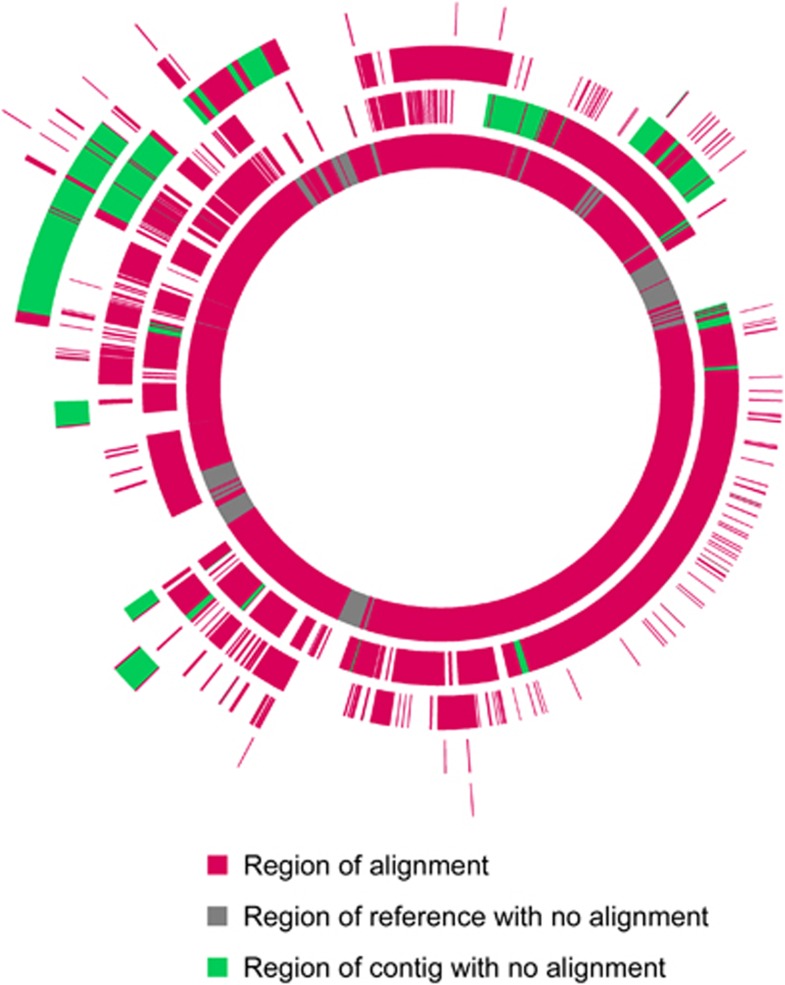
Of 406 novel genes, 184 with annotated functions (Supplementary Table S1), were identified on 26 contigs (15 identified as Dhc by both TF and SS, 4 identified by SS alone as described above and 7 that were too short for TF analysis but were identified by SS) (Figure 1). The most surprising finding was the presence of nine genes predicted to be involved in corrin ring synthesis, the first half of the cobalamin biosynthesis pathway.
Figure 1.
Alignment of ANAS metagenome Dhc contigs (identified by TF and/or SS) to the Dhc strain 195 genome. The inner circle represents the reference strain 195 genome, with the origin of replication at the top. Magenta areas indicate alignment to ANAS metagenome contigs, whereas gray areas indicate regions with no alignment. Each contiguous bar in the outer circles represents a contig, positioned based on its aligning regions, with contigs plotted on different circles to avoid overlap. Green areas indicate regions with no alignment to the reference genome, potentially containing novel Dhc genes (if they also do not align to other Dhc reference genomes).
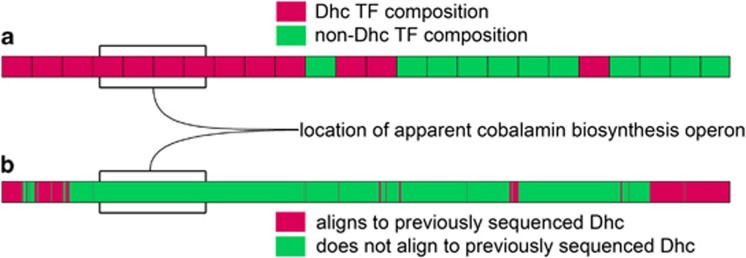
The corrin ring synthesis genes are on contig ANASEMC_C6240, which was identified as Dhc by both SS and by TF. Eight of the nine genes are oriented in the same direction and appear to be in a single operon, along with seven genes for ABC-transporter (ATP-binding cassette transporter) components (Figure 2), some specifically annotated as ABC-type cobalamin/Fe3+-siderophore transport systems components. All regions of this contig aligning to reference Dhc genomes aligned to previously identified High Plasticity Regions, which contain much of the variation between sequenced Dhc genomes (McMurdie et al., 2009). Based on the TF analysis, the region of this contig containing the cobalamin biosynthesis genes grouped with the Dhc sequences and not with any other contig class (Figure 3).
Figure 2.
Operon structure for genes for the first (corrin ring synthesis) part of the cobalamin biosynthesis pathway identified in an ANAS metagenome contig associated with Dhc. Genes in white are the corrin ring synthesis genes, labeled with the gene name. Genes with hatching are genes for ABC-transporter components.
Figure 3.
Evidence for the association of contig ANASMEC_C6240 (containing cobalamin biosynthesis genes) with Dhc. (a) The top bar shows how different segments of the contig were grouped with Dhc based on TF analysis, whereas (b) the bottom bar shows which parts of the contig aligned with previously sequenced Dhc genomes (magenta matches Dhc and green does not). The location of the apparent cobalamin biosynthesis operon is indicated and has a Dhc TF composition but does not align to previously sequenced Dhc genomes.
PCR amplification and sequencing were used to confirm the presence of three of the cobalamin biosynthesis genes in Dhc strains previously isolated from ANAS. Genes tested included two from the apparent cobalamin biosynthesis operon (cbiD and cbiF) and the one from elsewhere on the same contig (cbiC). All three genes were successfully amplified and sequenced from gDNA from Dhc strain ANAS2 as well as from ANAS metagenomic DNA but not from Dhc strain ANAS1 or strain 195. Sequences had 99.6–100% nucleotide identity with corresponding metagenome sequences. Amplification with the primers for cbiF produced products of a different size than the target sequence, when gDNA from Dhc strain ANAS1 or strain 195 was used as the template. Sequencing of PCR products confirmed that these were a different sequence from the target, the result of non-specific primer binding.
Several other groups of genes are well represented among the novel Dhc genes identified here. A total of 15 novel genes for ABC-transporter components were identified, including 13 on the same contig as the corrin ring synthesis genes. In all, 15 genes for phage proteins and 14 genes for recombinases were also present. A total of 11 novel genes for RDases were identified. However, for one RDase, the first-third of the gene matched (98% ID) the Dhc strain 195 gene DET0088, an RDase domain gene, which is approximately one-third the length of a typical RDase gene. Together with the remaining two-thirds of the RDase gene, this appears to be a full-length novel Dhc RDase gene in the ANAS metagenome.
Metagenome coverage of Dhc genes detected by microarray
Metagenome coverage of Dhc genes was assessed by comparison with results from a previous comparative genomics study performed with microarrays targeting 98.6% of annotated genes in Dhc strains 195, BAV1, CBDB1 and VS (Lee et al., 2011). Coverage was evaluated by identifying Dhc genes detected in ANAS by the microarray analysis and determining which of those genes were present in the metagenome sequences (Figure 4). Presence of Dhc genes in the metagenome was determined by blastn comparisons of the genome sequences of Dhc strains 195, BAV1, CBDB1 and VS to all metagenome contigs (expect value cutoff of 1 × 10−12).
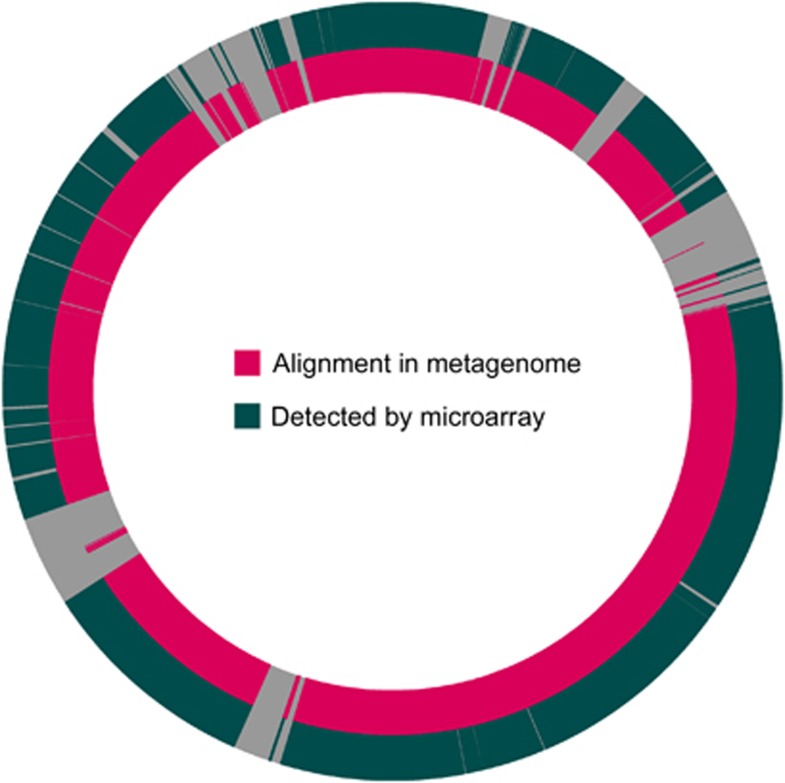
Figure 4.
Comparison of metagenomic Dhc coverage with ANAS genes detected by microarray. Although the analysis was performed for genes from Dhc strains 195, BAV1, CBDB1 and VS, only results for Dhc strain 195 are shown here for simplicity. Circles represent the Dhc strain 195 genome, with the origin of replication at the top. The inner circle shows regions with ANAS metagenome / strain 195 alignments in magenta. The outer circle shows ANAS genes detected by microarray in blue-green.
The metagenome contigs contained 96.2% (1311 of 1363) of the genes identified as present by the microarray analysis. Another 3.4% (47 genes) were partially present, overlapping the contig end. Only 5 of the 1363 Dhc genes identified by microarray were not found in any of metagenome contigs. These were all genes from the Dhc195 genome, and include fabG (DET1277), nusB (DET1278) and three genes coding for hypothetical proteins (DET0768, DET1405 and DET1406). Based on the alignment of the metagenome contigs to the Dhc 195 genome, these genes appear to fall in gaps between contigs. Blastn comparisons of these genes to the raw sequencing reads revealed that all five genes had significant alignments (expect value < 1 × 10−50) to 454-Titanium sequencing reads but not to Sanger sequencing reads, indicating that they were missed by Sanger sequencing.
Co-assembly of sequence from distinct Dhc strains
Comparisons to the previous comparative genomics microarray analysis (Lee et al., 2011) were also used to determine whether sequences from the two distinct Dhc strains were co-assembled in the metagenome. The presence of two different Dhc strains (ANAS1 and ANAS2) in the ANAS community has been established previously (Holmes et al., 2006; Lee et al., 2011), and the previous study identified 60 genes distinct to ANAS1 and 36 genes distinct to ANAS2 (Lee et al., 2011). The metagenome contigs containing these non-shared genes were identified using BLAST and identifications were confirmed by BLAST comparison of metagenome sequences to the NCBI non-redundant nucleotide database. Although all genes analyzed had significant (expect value < 10−12) alignments in the contigs, we also required alignment of at least 75% of gene length for positive identification for this analysis. Five genes distinct to ANAS1 failed this alignment length requirement and were not considered. Six contigs, representing 541 431 bp combined, were found to be co-assembled because each contained at least one gene distinct to ANAS1 and one gene distinct to ANAS2. In total, 17 contigs contained genes distinct to ANAS1, and 15 contigs contained genes distinct to ANAS2.
ANAS community structure
TF classification of metagenome contigs
TF was used to analyze all contigs longer than 2500 bp, comprising 2323 contigs representing 46% of the total sequence length of all contigs. Of these contigs, 95% were classified into 10 classes (Table 2). 141 contigs were left unclassified because they did not cluster with other contigs.
Table 2. Classification of contigs by TF and identification of contig classes by 16S and 23S BLAST comparisons.
| Classa | Number of contigs | Total sequence length (bp) | Median contig length (bp) | Average read depth | Taxa | 16S rRNA gene closest BLAST hitb (23S when 16S not present) | %ID of closest BLAST hitb |
|---|---|---|---|---|---|---|---|
| Class 1 | 13 | 2 279 508 | 39 051 | 53 | Clostridiaeceae | Clostridiaceae bacterium SH021 | 95 |
| Class 2 | 45 | 1 483 420 | 14 384 | 39 | Dehalococcoides | Dehalococcoides sp MB and Dehalococcoides ethenogenes 195 | 100 |
| Class 3 | 77 | 2 654 085 | 27 104 | 18 | Spirochaetes | Spirochaetes bacterium enrichment culture clone DhR^2/LM-B02 | 92 |
| Class 4 | 152 | 2 550 033 | 11 242 | 11 | Methanobacterium | Methanobacterium formicicum strain FCam | 99 |
| Class 5 | 382 | 2 249 123 | 4714 | 8 | Desulfovibrio | Desulfovibrio desulfuricans subsp. Desulfuricans str. ATCC 27774c | 96c |
| Class 6 | 449 | 2 295 796 | 4122 | 7 | unknown taxa | no rRNA genes | |
| Class 7 | 550 | 2 732 616 | 3821 | 7 | Synergistetes | Synergistetes bacterium enrichment culture clone DhR^2/LM-F01 | 98 |
| Class 8 | 205 | 791 052 | 3346 | 6 | Deltaproteobacterium | no rRNA genes | |
| Class 9 | 191 | 675 183 | 3112 | 6 | unknown taxa | no rRNA genes | |
| Class 10 | 118 | 421 953 | 3156 | 5 | Methanospirillum | Methanospirillum hungatei JF-1 | 99 |
| Unclassified | 141 | 864 492 | 3451 | ||||
| All classes | 2323 | 18 997 261 | 4003 |
Abbreviation: TF, tetranucleotide frequency.
Classes are ordered by average read depth.
Identity and %ID are presented for the top 16S (or 23S) rRNA gene BLAST hit in the NCBI nucleotide database that was identified beyond ‘uncultured bacterium'. BLAST searches were performed in August 2010.
23S rRNA gene was used for identification of the Desulfovibrio class because no 16S gene was present on those contigs.
Based on 16S and 23S rRNA genes present on the contigs, 7 of the 10 classes were attributed to the following taxa: a Clostridiaceae, Dhc, Desulfovibrio (23S only), Methanobacterium, Methanospirillum, a Spirochete, and a Synergistete. The remaining three classes did not contain 16S or 23S rRNA genes. Notably, an additional contig (ANASMEC_C9204) containing a set of rRNA gene sequences from a Clostridium did not cluster with any contig class, although it was more similar to the Clostridiaceae class than to other contig classes. This 8722 bp contig also contains genes for subunits of a type IIA topoisomerase and a gene for a hypothetical protein. A partial 23S rRNA gene belonging to a Bacteroides and a 16S gene belonging to a Desulfovibrio were also identified, but were on contigs smaller than the 2500 bp cutoff used for the TF analysis.
IMG/M Phylogenetic Marker clusters of orthologous group (Markowitz et al., 2008a) were used to try to identify the remaining three classes. One class was identified as a Deltaproteobacterium, likely from the Desulfovibrionales order. Marker genes in Class 6 did not give a clear identification, and Class 9 contained no marker genes.
Comparisons with previously sequenced reference genomes
Contig classes were compared with relevant reference genomes in the NCBI genomes database (accessed September 2010). Desulfovibrio and Methanospirillum contigs were compared with fully sequenced genomes from the same genus. Methanobacterium contigs were compared with genomes of members of the Methanobacteriaceae family. Clostridiaceae contigs were compared with Clostridium genomes (most similar genus based on 16S and 23S sequences). Comparisons were not performed for the Spirochete, Synergistete, or unknown Deltaproteobacterium contigs because sufficiently close relatives (same family or genus) could not be identified.
Dhc contigs had the most similarity to reference genomes, whereas Clostridiaceae and Methanobacterium contigs had < 4% contig match (percent of contig bases in alignments to reference genome) or reference match (percent of reference genome bases in alignments to contigs) (Table 3, Supplementary Table S2). For comparison, it is useful to consider what these values are for a set of contigs compared with a reference genome that is not closely related. A comparison of the Dhc contigs to seven Desulfovibrio reference genomes results in contig matches and reference matches of 0.1% to 0.2%. For Methanospirillum contigs, the disparity between contig match and reference match is probably due to poor sequencing coverage (0.5 Mbp compared with 3.5 Mbp for Methanospirillum hungatei).
Table 3. Comparison of ANAS contig classes to the most similar sequenced genomes.
| ANAS contig class | Most similar reference genome based on contig and reference match | Contig matcha (%) | Reference matchb (%) |
|---|---|---|---|
| Dehalococcoides | Dehalococcoides ethenogenes strain 195 | 81.7 | 82.2 |
| Desulfovibrio | Desulfovibrio desulfuricans ATCC 27774 | 32.9 | 26.1 |
| Clostridiaceae | Clostridium kluyveri NBRC 12016 | 1.1 | 1.1 |
| Methanospirillum | Methanospirillum hungatei | 46.5 | 6.1 |
| Methanobacterium | Methanothermobacter thermautotrophicus | 2.1 | 3.2 |
Contig match is the percentage of total bases in all contigs in the set that are part of an alignment to the reference genome.
Reference match is the percentage of total bases in the reference genome that are part of an alignment to some contig in the set.
Metabolic functions in ANAS
Metagenome gene content overview
The ANAS metagenome contains 60 992 putative protein-coding genes. Of these, 36 101 could be assigned to clusters of orthologous groups, and 32 520 of those were assigned to categories beyond general function prediction (Supplementary Table S3).
Three types of functional genes related to dechlorination (genes directly involved in dechlorination, genes involved in cobalamin biosynthesis and genes involved in hydrogen production and consumption) were selected for further analysis because they may provide insight into the dechlorinating abilities of this community and the interactions between community members that result in an efficient dechlorinating consortium.
Reductive dechlorination
A total of 15 putative RDase genes located on six contigs were identified in the JGI annotation of the ANAS metagenome contigs (Supplementary Table S4). In addition, three RDase genes identified by previous microarray analysis (Lee et al., 2011) but not annotated in the JGI annotation were found by BLAST search on an additional contig (ANASMEC_C7898) and gene identities were confirmed by comparison to the NCBI non-redundant nucleotide database. Two of these were present as full-length RDase genes. The third, matching Dhc strain 195 gene DET1535, was present as two partial RDase genes disrupted by an apparent frame-shift mutation. Of the seven contigs containing RDase genes, six were identified as Dhc by both SS and TF. The remaining contig (ANASMEC_C818), which contained only one RDase gene, was identified as Dhc by the SS method, but was left unclassified in the TF analysis. This contig had significant SS to Dhc strain 195 over approximately 25% of its length, including the RDase gene region. The non-aligning contig regions contained recombinases and phage-related genes, indicating possible horizontal transfer and perhaps accounting for the atypical tetranucleotide composition.
Of the 17 full-length RDase genes identified, 7 were matched to putative RDase genes in the NCBI non-redundant protein database (⩾98% amino acid ID). Together with the partial RDase genes mentioned above, which match DET1535 (97% amino acid ID), these correspond to the eight RDase genes identified as present in ANAS (or Dhc isolates from ANAS) by the previous microarray study (Lee et al., 2011). These include two (tceA and vcrA) that have been linked to enzymes with demonstrated RDase activity, and one (DET0088) that appears as a truncated RDase gene in Dhc strain 195, but which is extended to a full-length novel RDase gene in the ANAS metagenome as noted above. Of the other ten putative RDase genes, one had 91% amino-acid identity to another putative RDase, and the remaining nine had less than 70% identity to any sequences in the NCBI protein database as of July 2011.
Cobalamin biosynthesis
In total, 20 genes along the first (corrin ring synthesis) and second (lower ligand attachment and rearrangement) parts of the cobalamin biosynthesis pathway were targeted for analysis (Kanehisa and Goto, 2000; Warren et al., 2002). Near-complete cobalamin biosynthesis pathways appear to be present in the Dhc, Methanobacterium and Clostridiaceae classes (Table 4, Supplementary Table S5).
Table 4. Cobalamin biosynthesis genes identified in ANAS metagenome contigs.
| Genesa |
Contig classifications |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clostridiaceae | Dehalococcoides | Spirochaetes | Methanobacterium | Desulfovibrio | (class 6) | Synergistetes | Deltaproteobacterium | (class 9) | Methanospirillum | Unclassified | short contigs | |
| cbiX/cbiK(cobN)b | x | x | x | x | x | x | ||||||
| cbiL (cobI)c | x | x | x | x | x | x | ||||||
| cbiH (cobJ)c,d | x | x | x | x | x | x | ||||||
| cbiF (cobM)c,d | x | x | x | x | x | x | x | |||||
| cbiGc | x | x | x | x | x | x | ||||||
| cbiD (cobF)c | x | x | x | x | x | x | x | x | ||||
| cbiJ (cobK)c | x | x | x | x | ||||||||
| cbiE (cobL)c,d,e | x | x | x | x | x | x | x | |||||
| cbiT (cobL)c,d | x | x | x | x | x | |||||||
| cbiC (cobH)c,d | x | x | x | x | x | x | ||||||
| cbiA (cobB)d,e | x | x | x | x | x | x | x | |||||
| cobA (cobO)d,e | x | x | x | x | x | |||||||
| cbiP (cobQ)d,e | x | x | x | x | x | x | x | |||||
| cbiB (cobD)d,e | x | x | x | x | x | x | ||||||
| cobU (cobP)d,e | x | x | x | x | x | x | ||||||
| cobT (cobU)d,e | x | x | x | x | ||||||||
| cobC (cobU)e | x | |||||||||||
| cobS (cobV)d,e | x | x | x | x | x | x | ||||||
Abbreviation: Dhc, Dehalococcoides.
Gene names are given for the anaerobic (early cobalt insertion) cobalamin biosynthesis pathway, with the names for the aerobic pathway genes with the same function in parentheses.
cbiX and cbiK (and cobN) are grouped together because they code for alternative cobaltochelatases.
Genes involved in corrin ring synthesis, the first part of the cobalamin synthesis pathway.
Genes present in Hodgkinia cicadicola.
Genes present in Dhc strain 195.
Genes for incomplete biosynthesis pathways were identified in both the ANAS Desulfovibrio and Methanospirillum contigs. However, the total sequence length of these contig classes is significantly smaller than would be expected for a full genome (Desulfovibrio contigs, 2249 123 bp total, represent 43–78% of the length of sequenced Desulfovibrio genomes; Methanospirillum contigs, 421 953 bp total, represent 12% of the length of the Methanospirillum hungatei genome), indicating incomplete coverage.
Hydrogen production and consumption
Hydrogenases, enzymes that catalyze the reversible oxidation of molecular hydrogen, appear to be widespread in the ANAS community, with 271 genes annotated as hydrogenase components (Supplementary Table S6). Of those, 126 genes were present on contigs that were large enough for classification by TF, spread across all classes except the Methanospirillum class. However, this is likely a false negative result given the low coverage of this genome as described above. Methanospirillum are expected to have genes for hydrogenases used in methanogenesis (Madigan et al., 2008). Of the 126 hydrogenase genes in large contigs, the Methanobacterium class had the largest proportion (36 genes), followed by the Desulfovibrio class (26 genes) and Dhc (17 genes). The Clostridiaceae class contained only three genes for hydrogenase components.
PhyloChip analysis of ANAS community composition
PhyloChip analysis of metagenomic DNA identified 1056 bacterial and archaeal taxa in ANAS (37 bacterial phyla, 2 archaeal phyla) (Supplementary Table S7). Of these, 285 taxa were identified as highly active by detection when hybridizing RNA to the PhyloChip (29 bacterial phyla, 2 archaeal phyla) (Supplementary Table S8).
The community composition of ANAS as detected by DNA PhyloChip experiments remained stable between the three feeding cycles sampled (mean coefficient of variation for normalized signal intensity: 0.083). The greatest variation was seen for taxa with the lowest average signal intensity. A total of 11 taxa, all among the lowest 5% of average signal intensity, had coefficients of variation ⩾0.20. The highest coefficient of variation (0.48) was for a Methanosarcinaceae.
The highly active taxa (taxa detected by RNA PhyloChip experiments) were also stable between the three sampling time points (mean coefficient of variation: 0.085). Only four taxa, the four Methanobacteriaceae detected, had coefficients of variation > 0.20 for the RNA PhyloChip experiments. These had coefficients of variation ranging from 0.24 to 0.36 and fell within the lowest 15% of signal intensity.
For all contig class taxa (Clostridiaceae, Dhc, Desulfovibrio, Deltaproteobacteria, Methanobacterium, Methanospirillum, Spirochetes and Synergistetes) except for Methanospirillum, representatives of the same taxa were detected as both present and active by PhyloChip experiments using DNA and RNA, respectively. Representatives of all bacterial contig class taxa were among the highest 10% of average signal intensity in DNA PhyloChip experiments, consistent with these taxa being dominant members of the community. However, all Methanobacterium detected were among the lowest 15% of signal intensity, and Methanospirillum were not detected by the PhyloChips. In the RNA PhyloChip experiments, Dhc was the only contig class taxa among the top 10% of signal intensity, although several Clostridiales not identified as Clostridiaceae were also in this most active group. One Spirochete also appeared in the top 15% of signal intensity.
Discussion
In this study, metagenomic sequencing and analysis were used to examine the phylogenetic composition of ANAS and the genes present in the dominant community members, with a focus on Dhc. Although Dhc and non-Dhc metagenome contigs were classified based on TF, an alternative SS approach was also used to identify Dhc contigs. Both approaches have advantages: SS can identify smaller contigs, whereas TF works even when closely related reference genomes are unavailable.
Metagenomic analysis has provided some insight into the functions and interactions of different community members in the context of overall TCE dechlorination activity. The widespread presence of genes for hydrogenases emphasizes the importance of hydrogen metabolism in this community. In the ANAS bioreactor, lactate is fermented to acetate and hydrogen, which are used by Dhc and by other organisms. Because hydrogenases can catalyze both the formation and degradation of molecular hydrogen, the presence of hydrogenase genes does not differentiate the organisms that are producing hydrogen from those that are consuming it. Based on knowledge of other organisms in these taxonomic groups, however, the Clostridiaceae, the Desulfovibrio and the Spirochete are potential fermenters that produce hydrogen, although some may also be homoacetogens, consuming hydrogen and carbon dioxide to produce acetate (Leadbetter et al., 1999; Madigan et al., 2008). The methanogens likely consume hydrogen as an electron donor, competing with Dhc (Madigan et al., 2008). These different hydrogen producers and consumers (fermenters, homoacetogens, reductive dechlorinators and methanogens) have different thermodynamic requirements and different hydrogen thresholds. However, in this community, they appear to have developed working syntrophic relationships, allowing stable long-term dechlorination activity.
With respect to dechlorination reactions, although other organisms are known to reductively dechlorinate TCE to DCE in many environments (Scholz-Muramatsu et al., 1995; Sharma and McCarty 1996; Holliger et al., 1998; Löffler et al., 2004), the association of all RDase genes in the metagenome with Dhc contigs implies that Dhc is the dominant, and possibly sole dechlorinator in ANAS. Previous studies have indicated that ANAS contains two distinct Dhc strains (Holmes et al., 2006; Lee et al., 2011). Consequently, we analyzed the metagenomic data set to determine whether sequences from these strains were co-assembled. Although co-assembly at the domain level has been reported for both real and simulated metagenomic data sets, these errors are expected to be rare and easy to identify (DeLong, 2005; Mavromatis et al., 2007). Co-assembly of closely related species or strains is more common and more difficult to detect (Mavromatis et al., 2007; Kunin et al., 2008). In this study, co-assembly of sequences from the two Dhc strains was detected for at least six contigs, representing 541 431 bp. Considering the similarity between these two strains (Lee et al., 2011), this amount of co-assembly is not surprising. However, it is worth recognizing as one characteristic of this approach and highlights the importance of parallel sequencing of isolates and/or single cells to metagenome studies.
Because the medium provided for ANAS contains only 2 μgl−1 cobalamin, a lower than optimal concentration for Dhc (He et al., 2007), cobalamin synthesis in the bioreactor is likely necessary to support the observed dechlorination abilities. Several community members, including Dhc, appear to have genes for complete or near-complete cobalamin biosynthesis pathways. Although some genes appear to be missing, not all genes identified in the pathway are necessary for de novo cobalamin synthesis. For example, Hodgkinia cicadicola, an endosymbiont of cicadas with a highly streamlined genome, retains cobalamin synthesis capabilities despite its lack of several of the enzymes in the pathway (Table 4) (McCutcheon et al., 2009).
As previously sequenced Dhc do not have these genes and Dhc are assumed to obtain this cofactor from other organisms, the association of genes for corrin ring synthesis (the first part of cobalamin biosynthesis) with Dhc was unexpected (Kube et al., 2005; Seshadri et al., 2005; He et al., 2007). The contig regions containing the corrin ring synthesis genes have TF compositions that were grouped with the Dhc sequences and not with any of the other contig classes (Figure 3), implying that these genes were not recently horizontally transferred to Dhc, but have been maintained in the ANAS Dhc for some time. Given that Dhc are known to have relatively streamlined genomes (Kube et al., 2005; Seshadri et al., 2005; McMurdie et al., 2009), it is interesting that the ANAS Dhc appear to be maintaining genes for this pathway even though other community members appear to be capable of supplying this cofactor and cobalamin has been supplied in the medium, albeit at a low level, for over 10 years. As PCR amplification and sequencing have confirmed the presence of these genes in Dhc strain ANAS2, the functionality of the Dhc cobalamin biosynthesis pathway is currently being investigated in that strain.
The description of the community composition derived from metagenomic analysis is generally consistent with those of previous 16S clone library studies (Richardson et al., 2002; Lee et al., 2006) and the PhyloChip study presented here. Overall, data from the clone libraries and metagenome sequencing agreed on the most abundant bacterial taxa, which were also detected by the PhyloChip. The PhyloChip also detected many other taxa because it is more effective at detecting low abundance organisms (Brodie et al., 2006; DeSantis et al., 2007). This is because the PhyloChip is less sensitive to random sampling effects that impact sequencing-based approaches (Zhou et al., 2008, 2011). With the exception of Methanospirillum, the archaeal taxa detected in the metagenome were also detected by the PhyloChip, along with several other archaea. No archaeal clone libraries have yet been prepared for ANAS.
One notable discrepancy between the bacterial clone libraries and the metagenome was in the relative abundance of taxa detected by the two methods. Specifically, the Spirochete exhibited only low abundance (1–2% of clones) in both clone library experiments (Richardson et al., 2002; Lee et al., 2006). Based on the median contig length and average read depth of Spirochete contigs (Table 2) however, the Spirochete appears to be one of the more abundant organisms in ANAS. Studies of other Dhc-containing dechlorinating microbial communities have also detected Spirochetes (Gu et al., 2004; Macbeth et al., 2004; Duhamel and Edwards, 2006). Based on what is known of Spirochetes in general, they may be fermenters or homoacetogens in these communities (Leadbetter et al., 1999; Madigan et al., 2008). Clone libraries are known to be susceptible to PCR and cloning biases (von Wintzingerode et al., 1997), and some studies have found Spirochetes in particular to be underrepresented in some clone libraries (Campbell and Cary, 2001; Hongoh et al., 2003). However, recent studies suggest that estimates of relative abundance based on metagenomic sequencing read depth are also biased (Amend et al., 2010; Morgan et al., 2010).
The notable discrepancies between the metagenome and the PhyloChip results were with the methanogens. The PhyloChip did not detect any Methanospirillum, and although the read depth and contig length of the Methanobacterium contigs indicates that they were dominant community members, their low signal intensity using the PhyloChip suggests otherwise. Because these experiments involved amplification of 16S genes prior to PhyloChip hybridization, the low signal intensity may be due to poor amplification. Methanogens had the highest coefficients of variation in both the PhyloChip DNA and RNA results, lending weight to the explanation that the methanogen population is less stable than the rest of this microbial community.
In this study, we also compared the metagenome sequences with a previous comparative genomics study that used microarrays to detect known Dhc genes in ANAS (Lee et al., 2011). The agreement between the two approaches in detecting Dhc genes (Figure 4) confirms that the coverage of Dhc in the metagenomic sequence data was very high. Most differences between the results of the two methods are regions of the reference Dhc genomes for which no genes were detected in ANAS by microarray, but which had an alignment in the metagenome contigs. This highlights the specificity of the microarray to detect only very closely matched sequences. Alternatively, metagenomic sequencing allows the detection of somewhat more divergent versions of genes as well as unexpected or novel genes.
This analysis of metagenomic sequence data has advanced our understanding of this dechlorinating microbial community. The phylogenetic composition of ANAS described by metagenomic sequencing generally confirms the composition described by PhyloChip and previous 16S clone library studies, with a few discrepancies in the relative abundances of some taxa and possible variability in the methanogen population. More importantly, our analysis of functional genes relevant to dechlorination provides insight into the capabilities of microbial community members. Dhc appear to be the dominant reductive dechlorinators in ANAS, as all RDase genes identified were associated with Dhc. Genes related to the synthesis of cobalamin, an important cofactor for reductive dechlorination, are present in several community members, including Dhc, highlighting the importance of this cofactor in the function of ANAS. This is the first time that genes for the first part of the cobalamin biosynthesis pathway have been identified in a Dhc strain, further highlighting the unique adaptation of the ANAS strains to reductive dechlorination, but also suggesting that the non-Dhc community members likely have additional important roles beyond cobalamin biosynthesis.
Acknowledgments
This research was supported by the Strategic Environmental Research and Development Program (SERDP) through grant ER-1587 and the NIEHS Superfund Basic Research Project ES04705-19. We are grateful to staff at the DOE Joint Genome Institute for the sequencing and annotation work carried out in this project and to the Department of Energy Office of Biological and Environmental Research for funding sequencing under the JGI Community Sequencing Program. Part of this work was performed at Lawrence Berkeley National Lab supported by the Office of Science, US Department of Energy under Contract No. 470 DE-AC02-05CH11231.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adrian L, Rahnenführer J, Gobom J, Hölscher T. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl Environ Microbiol. 2007;73:7717–7724. doi: 10.1128/AEM.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian L, Szewzyk U, Wecke J, Görisch H. Bacterial dehalorespiration with chlorinated benzenes. Nature. 2000;408:580–583. doi: 10.1038/35046063. [DOI] [PubMed] [Google Scholar]
- Amend AS, Seifert KA, Bruns TD. Quantifying microbial communities with 454 pyrosequencing: does read abundance count. Mol Ecol. 2010;19:5555–5565. doi: 10.1111/j.1365-294X.2010.04898.x. [DOI] [PubMed] [Google Scholar]
- Brodie EL, DeSantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Cary SC. Characterization of a novel spirochete associated with the hydrothermal vent polychaete annelid, Alvinella pompejana. Appl Environ Microbiol. 2001;67:110–117. doi: 10.1128/AEM.67.1.110-117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow WL, Cheng D, Wang S, He J. Identification and transcriptional analysis of trans-DCE-producing reductive dehalogenases in Dehalococcoides species. ISME J. 2010;4:1020–1030. doi: 10.1038/ismej.2010.27. [DOI] [PubMed] [Google Scholar]
- Cole K, Truong V, Barone D, McGall G. Direct labeling of RNA with multiple biotins allows sensitive expression profiling of acute leukemia class predictor genes. Nucleic Acids Res. 2004;32:e86. doi: 10.1093/nar/gnh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF. Microbial community genomics in the ocean. Nat Rev Microbiol. 2005;3:459–469. doi: 10.1038/nrmicro1158. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53:371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, et al. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 2009;10:R85. doi: 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhamel M, Edwards EA. Microbial composition of chlorinated ethene-degrading cultures dominated by Dehalococcoides. FEMS Microbiol Ecol. 2006;58:538–549. doi: 10.1111/j.1574-6941.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg SMD, Johnson J, Busam D, Feldblyum T, Ferriera S, Friedman R, et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc Natl Acad Sci USA. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu AZ, Hedlund BP, Staley JT, Strand SE, Stensel HD. Analysis and comparison of the microbial community structures of two enrichment cultures capable of reductively dechlorinating TCE and cis-DCE. Environ Microbiol. 2004;6:45–54. doi: 10.1046/j.1462-2920.2003.00525.x. [DOI] [PubMed] [Google Scholar]
- He J, Holmes VF, Lee PKH, Alvarez-Cohen L. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl Environ Microbiol. 2007;73:2847–2853. doi: 10.1128/AEM.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Ritalahti KM, Yang KL, Koenigsberg SS, Löffler FE. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature. 2003;424:62–65. doi: 10.1038/nature01717. [DOI] [PubMed] [Google Scholar]
- Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, et al. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch Microbiol. 1998;169:313–321. doi: 10.1007/s002030050577. [DOI] [PubMed] [Google Scholar]
- Holmes VF, He J, Lee PKH, Alvarez-Cohen L. Discrimination of multiple Dehalococcoides strains in a trichloroethene enrichment by quantification of their reductive dehalogenase genes. Appl Environ Microbiol. 2006;72:5877–5883. doi: 10.1128/AEM.00516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongoh Y, Ohkuma M, Kudo T. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae) FEMS Microbiol Ecol. 2003;44:231–242. doi: 10.1016/S0168-6496(03)00026-6. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielhorn J, Melber C, Wahnschaffe U, Aitio A, Mangelsdorf I. Vinyl chloride: still a cause for concern. Environ Health Perspect. 2000;108:579–588. doi: 10.1289/ehp.00108579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajmalnik-Brown R, Hölscher T, Thomson IN, Saunders FM, Ritalahti KM, Löffler FE. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl Environ Microbiol. 2004;70:6347–6351. doi: 10.1128/AEM.70.10.6347-6351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kube M, Beck A, Zinder SH, Kuhl H, Reinhardt R, Adrian L. Genome sequence of the chlorinated compound respiring bacterium Dehalococcoides species strain CBDB1. Nat Biotechnol. 2005;23:1269–1273. doi: 10.1038/nbt1131. [DOI] [PubMed] [Google Scholar]
- Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P. A bioinformatician's guide to metagenomics. Microbiol Mol Biol Rev. 2008;72:557–578. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbetter JR, Schmidt TM, Graber JR, Breznak JA. Acetogenesis from H2 plus CO2 by Spirochetes from termite guts. Science. 1999;283:686–689. doi: 10.1126/science.283.5402.686. [DOI] [PubMed] [Google Scholar]
- Lee PKH, Cheng D, Hu P, West KA, Dick GJ, Brodie EL, et al. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME J. 2011;5:1014–1024. doi: 10.1038/ismej.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PKH, Johnson DR, Holmes VF, He J, Alvarez-Cohen L. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl Environ Microbiol. 2006;72:6161–6168. doi: 10.1128/AEM.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler FE, Cole JR, Ritalahti KM, Tiedje JM.2004Diversity of Dechlorinating BacteriaIn: Häggblom MM, Bossert ID (eds),Dehalogenation: Microbial Processes and Environmental Applications Kluwer Academic Publishers: New York; 53–87. [Google Scholar]
- Luijten MLGC, de Weert J, Smidt H, Boschker HTS, de Vos WM, Schraa G, et al. Description of Sulfurospirillum halorespirans sp nov., an anaerobic, tetrachloroethene-respiring bacterium, and transfer of Dehalospirillum multivorans to the genus Sulfurospirillum as Sulfurospirillum multivorans comb. nov. Int J Syst Evol Microbiol. 2003;53:787–793. doi: 10.1099/ijs.0.02417-0. [DOI] [PubMed] [Google Scholar]
- Macbeth TW, Cummings DE, Spring S, Petzke LM, Sorenson KS., Jr Molecular characterization of a dechlorinating community resulting from in situ biostimulation in a trichloroethene-contaminated deep, fractured basalt aquifer and comparison to a derivative laboratory culture. Appl Environ Microbiol. 2004;70:7329–7341. doi: 10.1128/AEM.70.12.7329-7341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan MT, Martinko JM, Dunlap PV, Clark DP.2008Brock Biology of Microorganisms12th edn,Benjamin Cummings: Boston [Google Scholar]
- Magnuson JK, Romine MF, Burris DR, Kingsley MT. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl Environ Microbiol. 2000;66:5141–5147. doi: 10.1128/aem.66.12.5141-5147.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson JK, Stern RV, Gossett JM, Zinder SH, Burris DR. Reductive dechlorination of tetrachloroethene to ethene by two-component enzyme pathway. Appl Environ Microbiol. 1998;64:1270–1275. doi: 10.1128/aem.64.4.1270-1275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2010;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Ivanova NN, Anderson I, Lykidis A, Mavromatis K, Pati A, et al. Using IMG-M: Comparative Analysis with the IMG/M System: Addendum to Using IMG. Technical Report LBNL-63614. Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory: Berkeley; 2008a. [Google Scholar]
- Markowitz VM, Ivanova NN, Szeto E, Palaniappan K, Chu K, Dalevi D, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008b;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- Mavromatis K, Ivanova NN, Chen IMA, Szeto E, Markowitz VM, Kyrpides NC. The DOE-JGI standard operating procedure for the annotations of microbial genomes. Stand Genomic Sci. 2009;1:63–67. doi: 10.4056/sigs.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maymó-Gatell X, Chien YT, Gossett JM, Zinder SH. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- McCarty PL. Breathing with chlorinated solvents. Science. 1997;276:1521–1522. doi: 10.1126/science.276.5318.1521. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Behrens SF, Müller JA, Göke J, Ritalahti KM, Wagner R, et al. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 2009;5:e1000714. doi: 10.1371/journal.pgen.1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men Y, Feil H, VerBerkmoes NC, Shah MB, Johnson DR, Lee PKH, et al. Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analysis. ISME J. 2012;6:410–421. doi: 10.1038/ismej.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn WW, Tiedje JM. Microbial reductive dehalogenation. Microbiol Rev. 1992;56:482–507. doi: 10.1128/mr.56.3.482-507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MJ, Zogorski JS, Squillace PJ. Chlorinated solvents in groundwater of the United States. Environ Sci Technol. 2007;41:74–81. doi: 10.1021/es061553y. [DOI] [PubMed] [Google Scholar]
- Morgan JL, Darling AE, Eisen JA. Metagenomic sequencing of an in vitro-simulated microbial community. PLoS One. 2010;5:e10209. doi: 10.1371/journal.pone.0010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller JA, Rosner BM, von Abendroth G, Meshulam-Simon G, McCarty PL, Spormann AM. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp strain VS and its environmental distribution. Appl Environ Microbiol. 2004;70:4880–4888. doi: 10.1128/AEM.70.8.4880-4888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RE, Bhupathiraju VK, Song DL, Goulet TA, Alvarez-Cohen L. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ Sci Technol. 2002;36:2652–2662. doi: 10.1021/es0157797. [DOI] [PubMed] [Google Scholar]
- Sanapareddy N, Hamp TJ, Gonzalez LC, Hilger HA, Fodor AA, Clinton SM. Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing. Appl Environ Microbiol. 2009;75:1688–1696. doi: 10.1128/AEM.01210-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz-Muramatsu H, Neumann A, Meßmer M, Moore E, Diekert G. Isolation and characterization of Dehalospirillum Multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch Microbiol. 1995;163:48–56. [Google Scholar]
- Seshadri R, Adrian L, Fouts DE, Eisen JA, Phillippy AM, Methe BA, et al. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science. 2005;307:105–108. doi: 10.1126/science.1102226. [DOI] [PubMed] [Google Scholar]
- Sharma PK, McCarty PL. Isolation and characterization of a facultatively aerobic bacterium that reductively dehalogenates tetrachloroethene to cis-1,2-dichloroethene. Appl Environ Microbiol. 1996;62:761–765. doi: 10.1128/aem.62.3.761-765.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt H, de Vos WM. Anaerobic microbial dehalogenation. Annu Rev Microbiol. 2004;58:43–73. doi: 10.1146/annurev.micro.58.030603.123600. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- US Department of Health and Human Services. 2005. Report on Carcinogens, 11th edition.
- US Department of Health and Human Services. 2007. 2007 CERCLA Priority List of Hazardous Substances.
- von Wintzingerode F, Göbel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- Waller AS.2009. Molecular Investigation of Chloroethene Resuctive Dehalogenation by the Mixed Microbial Community KB1. Doctor of Philosophy thesis, University of Toronto, Toronto, 2009.
- Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. The biosynthesis of adenosylcobalamin (vitamin B12) Nat Prod Rep. 2002;19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- West KA, Johnson DR, Hu P, DeSantis TZ, Brodie EL, Lee PKH, et al. Comparative genomics of ‘Dehalococcoides ethenogenes' 195 and an enrichment culture containing unsequenced ‘Dehalococcoides' strains. Appl Environ Microbiol. 2008;74:3533–3540. doi: 10.1128/AEM.01835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Xie G, Copeland A, González JM, Han C, Kiss H, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4:55299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Zhou J, Kang S, Schadt CW, Garten CT., Jr Spatial scaling of functional gene diversity across various microbial taxa. Proc Natl Acad Sci USA. 2008;105:7768–7773. doi: 10.1073/pnas.0709016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wu L, Deng Y, Zhi X, Jiang YH, Tu Q, et al. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J. 2011;5:1303–1313. doi: 10.1038/ismej.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.