Abstract
Microbes are transported in hydrological networks through many environments, but the nature and dynamics of underlying microbial metacommunities and the impact of downslope inoculation on patterns of microbial diversity across landscapes are unknown. Pyrosequencing of small subunit ribosomal RNA gene hypervariable regions to characterize microbial communities along a hydrological continuum in arctic tundra showed a pattern of decreasing diversity downslope, with highest species richness in soil waters and headwater streams, and lowest richness in lake water. In a downstream lake, 58% and 43% of the bacterial and archaeal taxa, respectively, were also detected in diverse upslope communities, including most of the numerically dominant lake taxa. In contrast, only 18% of microbial eukaryotic taxa in the lake were detected upslope. We suggest that patterns of diversity in surface waters are structured by initial inoculation from microbial reservoirs in soils followed by a species-sorting process during downslope dispersal of both common and rare microbial taxa. Our results suggest that, unlike for metazoans, a substantial portion of bacterial and archaeal diversity in surface freshwaters may originate in complex soil environments.
Keywords: Bacteria, Archaea, Eukarya, metacommunity, landscape, MIRADA-LTERS
Introduction
The chemistry and in turn the biology of freshwater and near-shore marine environments are tightly coupled to terrestrial ecosystems through hydrological networks on the landscape. However, relatively little is known about land–water transfers of microorganisms and the impacts on patterns of species diversity. Most research on microbial biogeography has focused on bacteria, and is beginning to reveal that mechanisms controlling the structure of microbial communities are very similar to those controlling the structure of metazoan communities. For example, there is ample evidence that bacterial diversity in freshwaters is strongly influenced by environmental variation that controls ecological ‘species-sorting' processes (Crump et al., 2003; Leibold et al., 2004; Judd et al., 2006; Fierer et al., 2007; Jones and McMahon, 2009). Dispersal also influences the biogeography of microbial species (Martiny et al., 2006; Telford et al., 2006), but the mechanisms of dispersal and the scales at which they are relevant are poorly understood. It is becoming clear that patterns of microbial diversity are controlled by both dispersal and environmental conditions including biological interactions, but what remains unclear is the relative importance of these two factors and how they interact on different temporal and spatial scales.
Several recent studies demonstrate the importance of dispersal at broad geographic scales by showing how specific habitats (for example, lakes, soils and ocean) share similar microbial communities at locations around the globe (Lozupone and Knight, 2007; Crump et al., 2009; Tamames et al., 2010). However, few studies have explored the effects of dispersal at local to landscape scales, and thus far most of these studies have focused on environments with short residence times such as stream water (Judd et al., 2006; Lindström et al., 2006; Crump et al., 2007). There is, however, a degree of overlap in microbial community diversity generally across soil, sediment, stream and lake habitats (Lozupone and Knight, 2007; Ramette and Tiedje, 2007; Tamames et al., 2010), suggesting that patterns of species distribution are nonrandom and instead mechanistically related across landscapes. For example, one study found consistent shifts in bacterial community composition in surface waters along a chain of streams and lakes, and suggested that this pattern was controlled by downslope inoculation of individual species and subsequent species sorting (Crump et al., 2007). Although this mechanism may help explain community composition of microbes in surface waters, the specific role of upslope habitats, such as soils, in determining diversity over landscape scales is unknown. In addition, because this one study (Crump et al., 2007) used less sensitive DNA fingerprinting techniques that ignore rare taxa with ∼<0.1% abundance (Kan et al., 2006), it remains untested whether or not the distribution and dispersal of rare bacterial taxa, or even archaeal and eukaryotic taxa, are also controlled via dispersal through hydrological connections between habitats.
The movement of microbe-sized particles within soil, groundwater and surface water is common and has been extensively studied and modeled (McDowell-Boyer et al., 1986; Bergström and Jansson, 2000), but these studies did not address the degree to which downslope transport of microbes contributes to patterns in diversity. In this study, we used pyrosequencing of small subunit rRNA (ribosomal RNA) gene V6 and V9 hypervariable regions to analyze microbial community composition of both common and rare members of Bacteria, Archaea and Eukarya in connected habitats within an arctic tundra catchment (Supplementary Figure S1). We report, for the first time as far as we know, a pattern of decreasing alpha diversity downslope along the hydrological continuum for Bacteria and Eukarya. We also discovered that the Bacteria and Archaea that dominated lake and lake-influenced environments were first observed in soil water and other upslope environments, suggesting that terrestrial environments serve as critical reservoirs and sources of microbial diversity for downslope surface waters.
Materials and Methods
Toolik Lake (1.5 km2) is a deep (maximum depth, 25 m; mean depth, 7 m) kettle lake located on the North Slope of Alaska, USA. The lake is usually ice free and thermally stratifies from late June through September. Ice cover forms in early October (O'Brien et al., 1997). The catchment of Toolik Lake (65 km2) has vegetation dominated by tussock and upland heath tundra (Whalen and Cornwell, 1985; O'Brien et al., 1997). Soils in the catchment have a maximum thaw depth of ∼0.5 m and are underlain by continuous permafrost.
Toolik Inlet is a third-order, cobble-bottomed stream that serves as the primary inlet stream to the lake. It drains 75% of the lake catchment and lies at the base of a chain of 12 smaller lakes (Kling et al., 2000). Stream flow usually begins in mid to late May and quickly reaches its peak flow rate as snow on the catchment melts (Hobbie et al., 1983). Soils sampled along the bank of Toolik Inlet stream are elevated ∼0.5–2 m above the level of the stream, and are primarily covered by riparian birch-willow tundra. Lake I-8 inlet stream is a headwater (no upstream lake influence), cobble-bottomed stream that drains a catchment consisting primarily of tussock and upland heath tundra.
Duplicate water samples were collected on 18 May 2008 and 11 July 2008 from the epilimnion (3 m) and hypolimnion (16 m) of Toolik Lake using a Van Dorn sampler (Supplementary Figure S1). May samples were collected from the same depths through a hole drilled in the ice (68.629961°N, −149.612633°W). On 11 July, duplicate samples were collected from the primary inlet streams of Lake I-8 and Toolik Lake by dipping an acid-washed amber polypropylene bottle below the surface. Duplicate samples were also collected from the shallow hyporheic zone below Toolik Inlet stream and from soil water of birch-willow tundra near the bank of the Toolik Inlet stream.
To collect hyporheic water, a steel spike with a pipe sleeve was pounded into the stream bed to a depth of ∼55 cm. The spike was removed and replaced with a nylon tube that was slotted and screened at one end and equipped with a luer-lock connector at the other. After inserting the tube, the pipe was removed and hyporheic water was drawn up through the tube with a syringe. The tube was rinsed with hyporheic water several times before collecting samples.
Soil water was collected by inserting a steel needle into the soil at several locations near the bank of Toolik Inlet stream at an elevation above the level of the stream, and withdrawing water with an attached syringe. At each site, samples were pooled from 5–10 randomly chosen locations of depths between 5 and 20 cm.
We filtered water samples onto a 0.2-μm Sterivex filter (Millipore, Billerica, MA, USA), added 2.0 ml of Puregene lysis buffer (Qiagen, Valencia, CA, USA) and stored samples at −20 °C until further processing. We extracted DNA as described previously (Amaral-Zettler et al., 2009) and stored our samples at −20 °C until amplification. Water filtration and DNA extraction protocols are available at http://amarallab.mbl.edu.
We amplified V6 hypervariable regions (Bacteria and Archaea) using primers targeting the regions between 947 and 1046 of the 16S rRNA gene (according to the E. coli numbering scheme) for bacterial targets and the 958 and 1048 regions for archaeal targets. We amplified eukaryotic V9 hypervariable regions following protocols in Amaral-Zettler et al. (2009). We multiplex-sequenced the resulting amplicons with a bar-coded primer strategy (Huber et al., 2007; Amaral-Zettler et al., 2009) on a 454 Genome Sequencer FLX (Roche, Basel, Switzerland) using the manufacturer's suggested amplicon protocol for the GS-FLX platform. The average amplicon sequence length was 65 bp for Bacteria, 61 bp for Archaea and 129 bp for Eukarya. The number of sequences per sample varied, and for most samples was between 1770 and 39 280 for bacterial and eukaryotic sequences, except for bacterial sequences from one headwater stream sample (304 sequences) and eukaryotic sequences from one soil water sample (223 sequences). Fewer sequences were recovered overall for Archaea. PCR failed for several samples (Supplementary Table S1).
We trimmed adaptor and primer sequences, removed low-quality reads, screened for chimeras using the Pintail algorithm and retained singletons as described by Huse et al. (2007). The priming sites and amplicon length used in this study were designed to improve the quality of the sequences by allowing us to detect both proximal and distal primers. The ability to sequence both primers with each read proved to be an important factor in assessing the quality of the sequence reads and our ability to remove low-quality sequences before analysis. We assigned operational taxonomic units (referred to here as ‘taxa') using the 2% single-linkage pre-clustering and pairwise alignment with average linkage clustering method (Huse et al., 2010). Three-percent cluster widths were used for all bacterial, archaeal and eukaryotic analyses. Taxonomic identifications for operational taxonomic units based on representative sequences were assigned using the Global Assignment of Sequence Taxonomy (Huse et al., 2008). Previous work by Huse et al. (2008) showed that taxonomy recovered from the V6 region in bacteria was equivalent to that recovered from the corresponding full-length molecule. To obtain a working classification for a given taxon, each sequence tag was mapped to the nearest sequence(s) in the SILVA-ARB database of over 1 million full-length 16S and 18S sequences (Pruesse et al., 2007), and assigned a taxonomic name based on the references. All sequences conform to the minimum information about a MARKer gene sequence (MIMARKS) standard (Yilmaz et al., 2011) and were deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA) under the accession number SRA049830.
Nonparametric Chao1, Shannon and Shannon Evenness alpha diversity estimates for Bacteria and Archaea were calculated with Mothur (v.1.17.1; http://www.mothur.org; Schloss et al., 2009). Parametric alpha diversity estimates were calculated using CatchAll v.1.0 (Bunge, 2011). We calculated eukaryotic richness estimates using the nonparametric Chao2 estimator (Amaral-Zettler et al., 2009) as implemented in the program SPADE (Chao and Shen, 2010) based on presence/absence matrices using separate paired replicates as input. We performed alpha diversity calculations for Bacteria and Archaea on pooled sequences from duplicate samples in order to increase the number of sequences per sample and reduce the bias in our estimates of diversity. For Bacteria and Archaea, we pooled sequences from lake epilimnion and hypolimnion for each season to better represent the total diversity in the lake for each season. We performed these calculations using full data sets and using reduced data sets in which the number of sequences per sample was made equal through random resampling (10 027 sequences per sample for Bacteria, 433 for Archaea and 2524 for Eukarya). Beta diversity estimates were determined for all samples (that is, no pooling). Beta diversity estimates for Bacteria and Archaea were calculated as the Morisita–Horn index (Cmh) in order to account for variation in sampling effort (Wolda, 1981; Magurran, 1988). Beta diversity for Eukarya was calculated as the Sørensen index (Qs) using presence/absence data. Multidimensional scaling was done using Primer-E (v.6) (Clarke, 1993). We also performed beta diversity calculations using reduced data sets in which the number of sequences per sample was made equal with random resampling (1770 sequences per sample for Bacteria, 108 for Archaea and 1206 for Eukarya).
To investigate the biogeography of rare bacterial taxa, we defined rare taxa as those that make up <0.1% of the pooled sequences from duplicate samples for each environment, and abundant taxa as those that make up >0.1% of the pooled sequences. We chose this level because it is generally considered the lower detection limit for PCR-based community fingerprinting techniques such as denaturing gradient gel electrophoresis (Kan et al., 2006). Rare lake taxa are those that are not abundant in any lake samples, and rare upslope taxa are those that are not abundant in soil water or headwater stream water.
Results
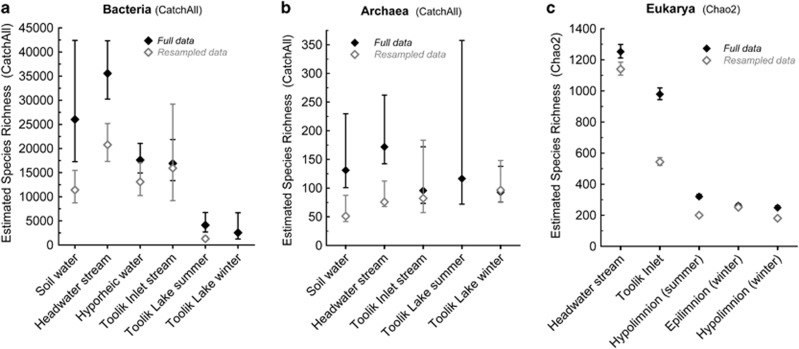
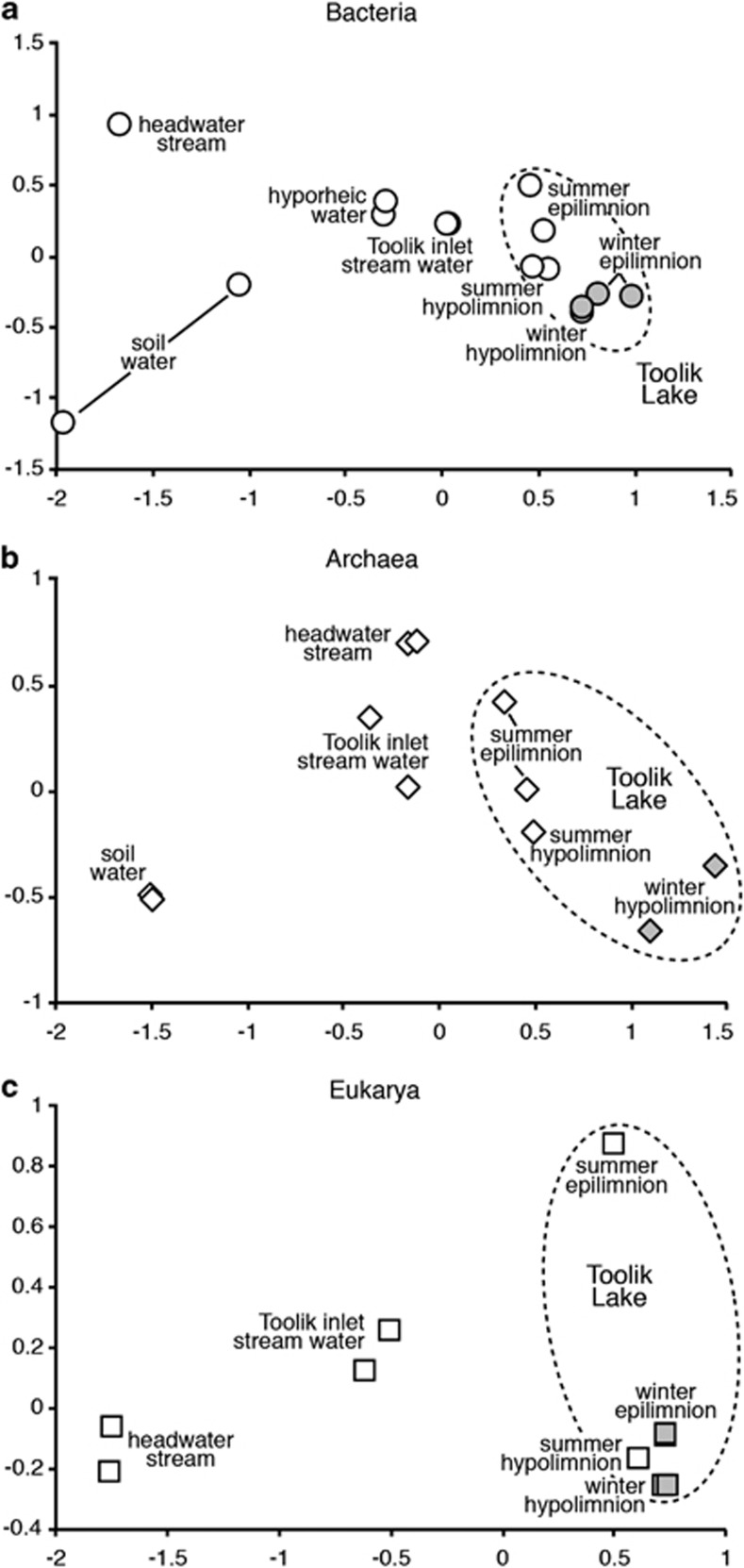
Alpha diversity (species richness) of bacterial and eukaryotic communities varied strongly across the landscape, and was relatively low in Toolik Lake and high in upslope environments (Figure 1, Supplementary Table S2). Alpha diversity of Archaea was considerably lower than for Bacteria and Eukarya, and did not vary systematically across the landscape (Figure 1). These patterns occurred for both full and resampled data sets. Beta diversity, visualized with multidimensional scaling diagrams (Figure 2), showed that microbial communities clustered by environment type (ANOSIM P<0.01 for lake vs other habitats for Bacteria, Archaea and Eukarya; ANOSIM P<0.05 for pairwise tests of bacteria samples grouped into winter lake, summer lake, inlet+hyporheic and headwater + soilwater). In Toolik lake, bacterial communities varied significantly by season (ANOSIM P<0.03) and eukaryotic communities varied significantly by depth (ANOSIM P<0.05). As shown by its central location on the MDS diagrams, communities in Toolik Inlet stream appeared to be a mixture of communities from soil water, headwater stream water and lake water, likely from lakes upstream of Toolik Inlet. Across all samples, the Cmh ranged from 0.01 to 0.99 for Bacteria and from 0.05 to 0.99 for Archaea. For Eukarya, the Qs ranged from 0.04 to 0.67. Cmh and Qs values changed only slightly when calculated with reduced data sets containing equal numbers of sequences per sample, and patterns in beta diversity were not significantly different based on ANOSIM analysis. On average, these values were higher for Bacteria by 0.003, lower for Archaea by 0.06 and lower for Eukarya by 0.01.
Figure 1.
Estimates of alpha diversity with Bonferroni-corrected confidence bounds calculated with CatchAll parametric models for pooled sequences from duplicate samples (when available, see Supplementary Table S1) for (a) Bacteria and (b) Archaea, and calculated as the Chao2 index with paired duplicate samples for (c) Eukarya. For Bacteria and Archaea, sequences from epilimnion and hypolimnion samples were pooled by season for Toolik Lake summer and Toolik Lake winter. Diversity estimates were calculated for all sequences (closed symbols) and for reduced sequence data sets that were randomly resampled to equal sample size (open symbols).
Figure 2.
Multidimensional scaling diagrams showing the degree of similarity among (a) Bacterial, (b) Archaeal and (c) Eukaryotic communities in duplicate samples (when available, see Supplementary Table S1). Bacterial and archaeal community similarity (Morisita–Horn) was calculated with relative abundance of sequences in taxa (97% sequence similarity). Eukaryotic similarity (Sørensen's Index) was calculated using the presence/absence of taxa. Winter samples are shaded for clarity. Bacteria from one headwater stream sample and Eukarya from soil water samples were omitted from this figure because of low sequence recovery.
Duplicate lake samples contained similar communities of Bacteria (Cmh averaged 0.96) and Eukarya (Qs averaged 0.66), but communities were different in duplicate samples from upslope environments (Figure 2). For example, the Cmh for duplicate soil bacteria communities was 0.43, and the Qs for headwater stream eukaryotic communities was 0.53. Archaeal communities showed the opposite pattern in which duplicate soil water and headwater stream samples contained very similar communities (Cmh averaged 0.99), and duplicate samples became increasingly different in downslope environments (Cmh averaged 0.75).
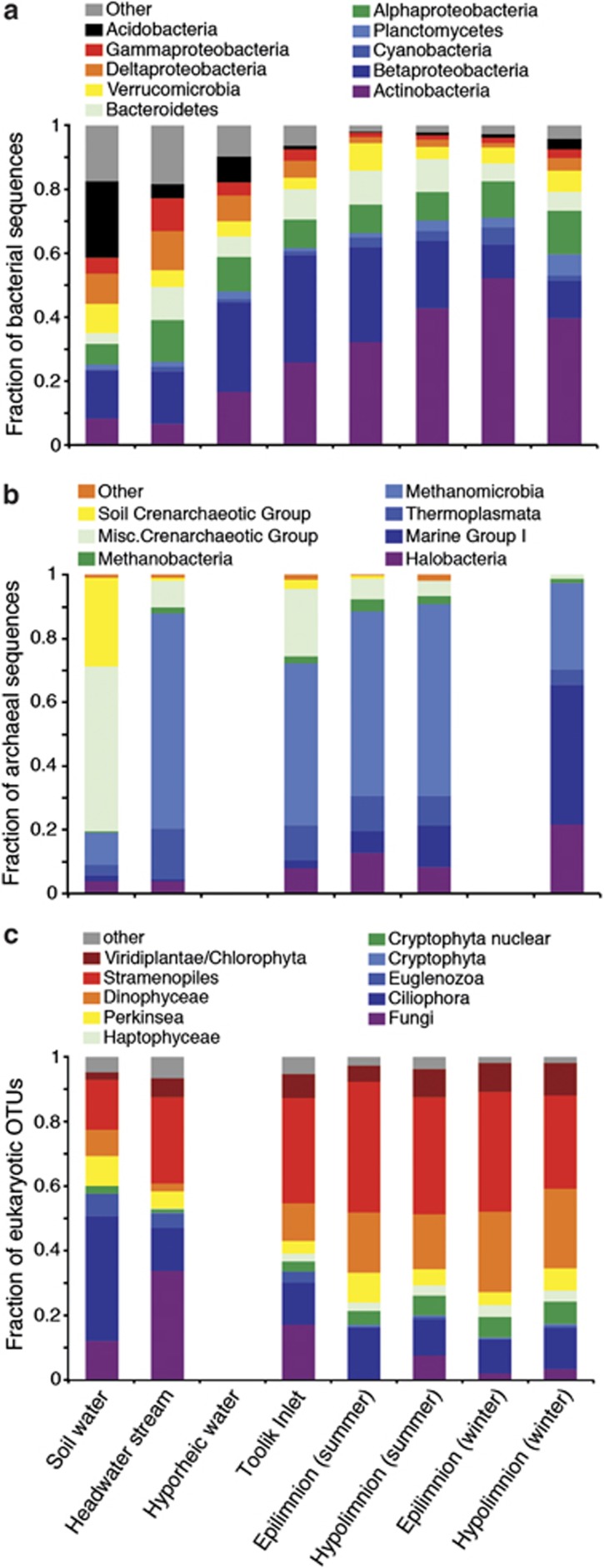
Bacterial communities shifted in composition along the landscape gradient from soil water and headwater stream communities of Acidobacteria, Gammaproteobacteria, Deltaproteobacteria, Verrucomicrobia and a diverse set of other phyla (‘Other' on Figure 3a) to lake communities dominated by Actinobacteria, Betaproteobacteria, Bacteroidetes and Alphaproteobacteria. Archaeal communities changed from soil water communities dominated by Crenarchaea to headwater stream communities dominated by Euryarchaea (Methanomicrobia and Thermoplasmata) to lake communities that include crenarchaeal Marine group I and Halobacteria (Figure 3b).
Figure 3.
Taxonomic information based on PCR-amplified small-subunit rRNA gene sequences and expressed as fraction of total sequences for (a) Bacteria and (b) Archaea, and as fraction of taxa (operational taxonomic units) for (c) Eukarya for each site using pooled sequences from duplicate samples (when available, see Supplementary Table S1).
Patterns in eukaryotic diversity were assessed using incidence rather than abundance-based approaches, because 18S rRNA gene copy number is highly variable (ranging from 1 to 10 000; Zhu et al., 2005; Auinger et al., 2008), potentially biasing diversity estimates based on sequence abundance. Eukaryotic taxa in soil water and the headwater stream included many fungal, ciliate and euglenozoan taxa, as well as stramenopiles, mainly in the phylum Bacillariophyta and the class Chrysophyceae (Figure 3c). Lake-influenced environments farther downstream contained more taxa related to the Dinophyceae, Haptophyceae and Cryptophyta. Note that taxa classified as metazoa, metaphyta (Viridiplantae/Streptophyta, Rhodophyta) and Unknown (‘Unknown Eukarya' and ‘environmental sample') were excluded from these analyses; they accounted for small percentages of eukaryotic taxa from soil water and headwater stream water (6% metazoa, 1% metaphyta, 5% unknown) and Toolik Lake (6%, 1%, 8%, respectively).
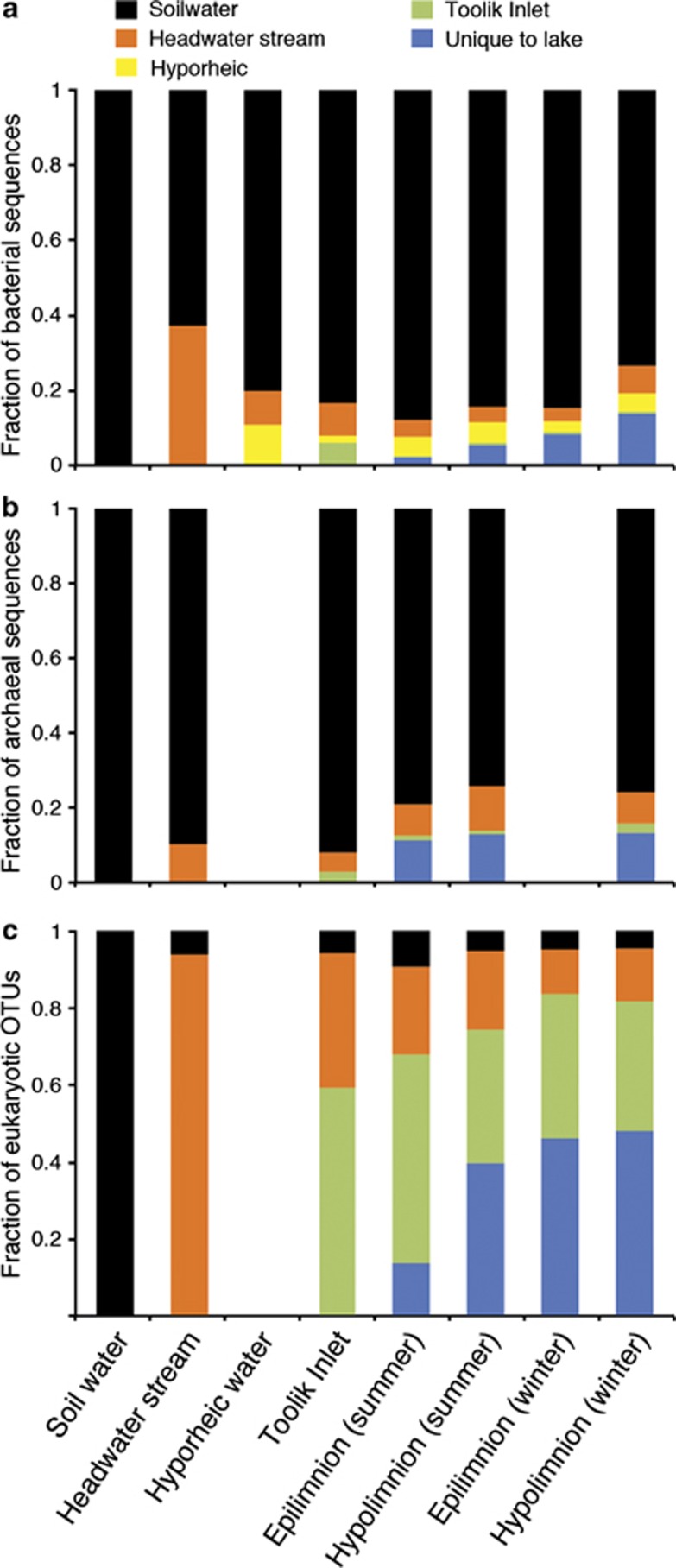
We assigned each taxon in our study to the farthest upslope environment where it first appeared, and found that for Bacteria and Archaea a substantial fraction of the sequences in Toolik Lake belonged to taxa that first appeared in soil water or the headwater stream (Figure 4). These taxa include the 39 most common bacterial taxa in Toolik Lake, and accounted for 89% of lake bacterial sequences and 85% of lake archaeal sequences. In contrast, few eukaryotic taxa in Toolik Lake (22%) belonged to taxa found in soil water or headwater stream water, and instead they first appeared in lake-influenced Toolik Inlet or were unique to the lake.
Figure 4.
DNA sequences from (a) Bacteria and (b) Archaea belonging to taxa (operational taxonomic units) categorized by the farthest upslope environment where they first appear, and expressed as a fraction of the total pooled sequences for each site (when available, see Supplementary Table S1). Relative number of taxa for Eukarya (c) categorized by the farthest upslope environment where they first appear.
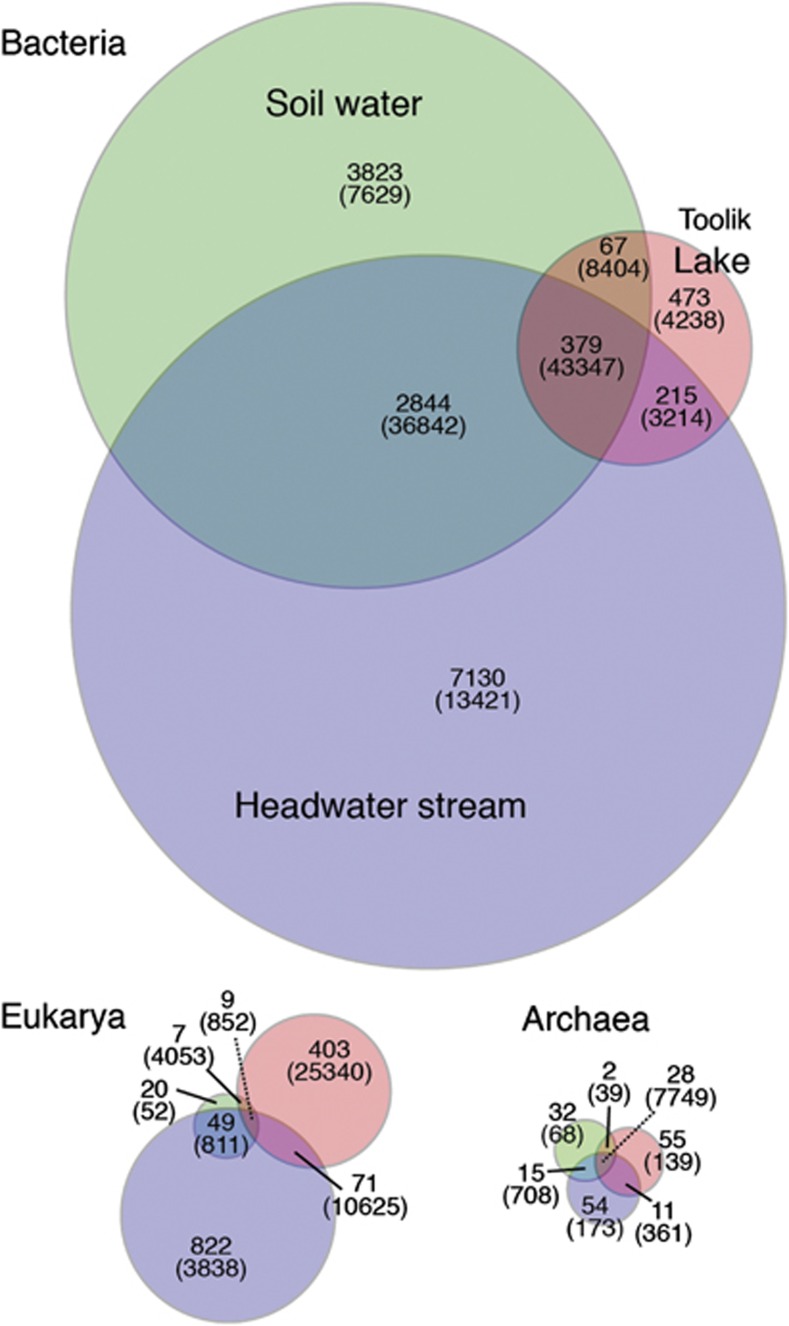
Venn diagrams demonstrate the strong overlap in bacterial and archaeal diversity between Toolik Lake and upslope environments (Figure 5). We found that 58% of bacterial taxa and 43% of archaeal taxa in Toolik Lake were also identified in soil water or headwater stream water. In contrast, only 18% of eukaryotic taxa in Toolik Lake were found in these upslope environments.
Figure 5.
Venn diagram showing the number of shared Bacterial, Archaeal and Eukaryotic taxa (in bold) among soil water, headwater stream and lake samples for pooled sequences from duplicate samples. The number of sequences associated with taxa is shown in parentheses. Taxa are defined by 97% sequence similarity. Circled areas are proportional to the number of taxa detected in each environment.
To investigate spatial patterns of the most abundant taxa in lake and upslope environments, we identified the 10 taxa in each environment with the largest average relative abundances. All 10 of the most abundant lake taxa were present in upslope environments as either abundant (6) or rare (4) taxa (Supplementary Figure S2a). Of the top 10 upslope taxa, only four appear in Toolik Lake, and all four would be considered abundant taxa in lake samples (Supplementary Figure S2b).
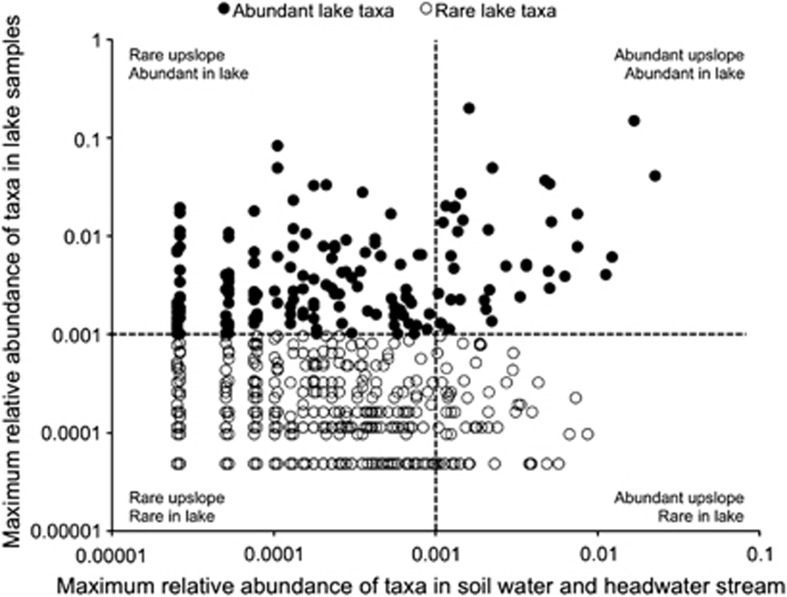
To compare the relative abundance of bacterial taxa in lake and upslope environments, we identified all taxa that appeared in both lake samples and upslope samples (661 taxa total, Figure 5), and plotted the maximum relative abundance of each taxon in lake samples against the maximum relative abundance in soil water and headwater stream samples (Figure 6). Most of the abundant lake taxa were rare in upslope environments (that is, <0.1% of the total sequences), but 23% of abundant lake taxa were also abundant in upslope environments (taxa in upper right quadrant of Figure 6). This pattern in which most of the abundant lake taxa were ‘rare' in upslope environments remains robust even if cutoffs other than 0.1% are used to define rare taxa (Figure 6).
Figure 6.
Maximum relative abundance of sequences for bacterial taxa that appear in at least one lake sample (epilimnion summer, hypolimnion summer, epilimnion winter, hypolimnion winter) and at least one upslope sample (soil water, headwater stream). Relative abundances were calculated for pooled duplicate samples from each location and date, and the maximum relative abundance was selected for this figure. Filled circles indicate taxa that were classified as abundant (>0.1%) in lake samples.
Discussion
Recent assessments of microbial diversity have identified an overlap between soil and freshwater microbial communities despite very large differences in habitat characteristics (Lozupone and Knight, 2007; Tamames et al., 2010). Lakes and larger rivers are planktonic environments containing free-living microbes and active microbial food webs that maintain a low and fairly constant abundance of microbial cells. In contrast, soils are considerably more complex environments dominated by surface-attached microbial communities that support much higher cell concentrations. For metazoans, this and other habitat differences lead to there being very little overlap in species diversity between soils and freshwater ecosystems. However, at the microscopic scale, soils contain microniches (for example, interstitial water, Fenchel, 1994) that share similarities with freshwater planktonic environments and may support ‘planktonic' microbes. Our results support this model, particularly in the wet or flooded soils overlying permafrost in large parts of the Arctic. Furthermore, the results suggest that, in contrast to metazoans, a substantial portion of bacterial and archaeal diversity found within surface freshwaters may originate in complex soil environments.
In support of the idea that surface water microbial diversity may originate in soils, we documented trends of decreasing alpha and beta diversity for bacterial and eukaryotic microbial communities across a connected landscape gradient from soil water and a headwater stream to a third-order stream and a terminal lake (Figure 1, Supplementary Table S2). For Bacteria, these observations are consistent with earlier observations of bacterial diversity within different ecosystems including soil waters (Judd et al., 2006, 2007), and streams and lakes (Crump et al., 2003, 2007). Bacterial alpha diversity in soil water and a headwater stream was on the same scale as previous pyrosequence-based estimates of bacterial diversity in soils, which range from thousands to tens of thousands of taxa based on 97% DNA sequence similarity of 16S rRNA genes (Roesch et al., 2007; Uroz et al., 2010). Bacterial alpha diversity in Toolik Lake was much lower and was on the same scale as in other lakes and the surface ocean (Shaw et al., 2008; Kirchman et al., 2010; Logue, 2010). This 10-fold decrease in diversity resulted from the loss of many taxa introduced from upslope environments combined with the increase in relative abundance of a handful of taxa that presumably are best adapted to lake conditions. Eukaryotic alpha diversity was highest in the headwater stream and decreased downslope approximately fivefold. For each major taxonomic group, the diversity peaked in the headwater stream, suggesting that small streams are the initial mixing zones for communities from various upslope terrestrial environments. Archaeal alpha diversity was nearly two orders of magnitude lower than bacterial diversity, as observed in many environments (Roesch et al., 2007; Aller and Kemp, 2008), and, unlike bacterial and eukaryotic alpha diversity, did not show systematic variation across the landscape.
The taxonomic composition of microbial communities shifted across the landscape, but the largest shifts were due to changes in only a few major taxonomic groups (Figure 3). Bacterial communities shifted from diverse soil water and headwater stream communities to lake communities dominated by Actinobacteria and Betaproteobacteria. Archaeal communities showed a similar shift moving downslope, punctuated by increases in Methanomicrobia in the headwater stream and crenarchaeal Marine group I in the lake. Shifts in eukaryotic communities reflected the environments through which water flowed across the landscape, starting with a typical soil community of fungi, ciliates and euglenozoa, and adding stramenopile taxa in shallow streams that support epilithic diatom communities. Lake-influenced environments contained more typical planktonic eukaryotic taxa related to the Dinophyceae, Haptophyta and Cryptophyta. In addition, consistent with prior results from Toolik Lake (Crump et al., 2003), the difference between the summer surface-water community and the winter and summer deep-water communities appears to be strongly related to the inoculation of taxa from Toolik Inlet stream into the upper layers of the lake (Figure 2).
The diversity and dynamics of rare taxa in microbial communities were virtually unexplored before next-generation DNA sequencing, and the ecological role of these organisms is still under debate (Pedros-Alio, 2006; Sogin et al., 2006; Galand et al., 2009). Nearly all taxa in the soil and headwater stream were defined as rare (98% and 99%, respectively), and these taxa comprised a large percentage of the total DNA sequences from these habitats (56% and 75%, respectively). A smaller fraction of the taxa in lake samples were defined as rare (average 74%), and, in contrast to upslope habitats, the rare lake taxa comprised only a small percentage of the total DNA sequences (average 9%). This pattern in the numerical dominance of rare taxa is reflected in rank abundance curves for bacteria in which the slopes for soil water and headwater stream communities are shallower than for lake communities (Supplementary Figure S3). This pattern is also demonstrated by calculations of Shannon evenness, which show that soil water bacterial community diversity was more even than diversity in lake communities (Supplementary Table S2). Evenness is rarely reported for microbial communities, but our results agree with the general observations that soil microbial communities are more diverse than aquatic communities (Tringe et al., 2005; Tamames et al., 2010). This pattern in the numerical dominance of rare taxa appears to be a fundamental difference between terrestrial and aquatic bacterial communities, and may apply to locations other than the arctic tundra.
To test the idea that patterns of microbial diversity on a landscape are controlled by hydrological connections between upslope and downslope habitats, each taxon in our study was assigned to the farthest upslope environment where it first appeared. In Toolik Lake, for Bacteria and Archaea a substantial fraction of the sequences (58% and 43%, respectively) belonged to taxa that first appeared in soil water or the headwater stream (Figure 4). These taxa include the 39 most common bacterial taxa in Toolik Lake, and accounted for 89% of lake bacterial sequences and 85% of lake archaeal sequences (Figures 3 and 4). In contrast, only 18% of eukaryotic taxa in Toolik Lake originated in upslope habitats, and instead they first appeared in lake-influenced Toolik Inlet stream or were unique to the lake. This is perhaps not surprising because the eukaryotic community in the lake includes a diverse assemblage of phytoplankton that are unlikely to grow in soil water, but this pattern also held for heterotrophic taxa where, for example, only 18% of the ciliate taxa were also detected upstream. This suggests that eukaryotic diversity in lakes is less dependent on dispersal from terrestrial environments than is bacterial and archaeal diversity.
Rare taxa in aquatic environments may be capable of filling new niches created by temporal and spatial changes in environmental conditions (Sogin et al., 2006; Jones and Lennon, 2010). Our results support this idea for bacteria because most of the abundant lake taxa that first appeared in upslope environments (128 of 166 operational taxonomic units) were considered rare in the soil and headwater stream (Figure 6, upper left quadrant). However, many of these abundant lake taxa (23%) were also abundant in upslope environments. These patterns in species abundance distributions are robust even if we define rare taxa at levels of relative abundance other than 0.1% (Figure 6). For example, if we define rare taxa as those with a relative abundance of 0.03%, then 63% (172 of 274) of the abundant lake taxa that appear in upslope environments would be considered rare in soil and the headwater stream. Given these results it appears that although the formation of landscape-level patterns in microbial communities is complicated, within tundra ecosystems the process involves both the generalist taxa that are abundant everywhere and the specialist taxa that are rare in some environments but when introduced can become abundant in others.
The mechanisms forming biogeographic patterns on the landscape can be set in a metacommunity perspective, where local communities are linked by dispersal and influenced by extinction and interactions among species (Wilson, 1992). For dispersal, there is an ongoing discussion concerning the dispersal capabilities of microorganisms and the temporal and spatial scales over which dispersal can influence microbial biogeography (Martiny et al., 2006; Telford et al., 2006). We found that bacterial communities in a headwater stream are similar to soil water communities (Figures 2, 3, 4, 5), suggesting that immigration (or perhaps more properly, advection) of soil bacteria strongly influences the first receiving water body because the ‘mass effect' of dispersing organisms exceeds the rate of local extinction (for example, Leibold et al., 2004). In other words, microbial diversity in soil water and headwater streams is similar because the time bacteria spend in the stream (their ‘residence time') is too short to allow communities to change before they enter a lake environment (Crump et al., 2007). This is perhaps not surprising, but it calls into question the role of dispersal into environments such as lakes where dilution of dispersed species is high and residence time is long enough to allow extinction of dispersed microbes. One study (Fierer et al., 2010) suggests that dispersal may be particularly important during initial colonization of previously sterile habitats (for example, metal and concrete pipes, decomposing wood). Our results suggest that dispersal mass effects might continue to be important in aquatic habitats with longer residence times that support established microbial communities, although it is still unclear which dispersed taxa are viable in their new environments.
Similarly, upslope seeding of abundant lake taxa via wind probably occurs in this system when periodic storm events cause aerosol formation in lakes (Grammatika and Zimmerman, 2001). Over very long time scales, this feedback process may help seed populations of lake organisms into soil waters in the region, and thus homogenize the regional metacommunity, although the patterns we observed in diversity still indicate quite heterogeneous distributions of taxa. However, the concentration of bacterial cells in aerosols is extremely low (Aller et al., 2005), and the magnitude of upslope dispersal via wind is probably very much lower than downslope dispersal via water flow. In addition, the high concentration of cells in soils makes it unlikely that recently dispersed wind-blown organisms could be detected in the highly concentrated soil environment using our present techniques.
A second major process controlling patterns of diversity is the ‘species sorting' that takes place during ecological interactions (for example, predation and competition), and in response to physical and chemical limits that exclude some species from particular environments. Several studies on lakes demonstrate that species sorting is the dominant mechanism for shifting diversity through time, whereas mass effects of dispersal are only important in systems with short residence times (Lindström et al., 2006; Shade et al., 2007; Nelson, 2009). We propose that dispersal of upslope taxa may influence downslope biodiversity and even guide seasonal succession by seeding freshwater communities with rare organisms that become dominant during species sorting. For example, the fact that 4 of the 10 most abundant bacterial taxa in Toolik Lake were found as rare taxa upslope is most likely explained by dispersal followed by species sorting within the lake, especially given the assumption that lakes are unimportant in reverse transport of bacteria back into soils (Supplementary Figure S2). Although it is unclear whether preferential growth or grazing is the dominant process at work, what is clear is that both rare and common taxa stored in terrestrial environments may serve as reservoirs of organisms that determine downslope community composition, or that are responsible for reassembly of nearly identical communities year after year in many planktonic ecosystems (for example, Crump et al., 2003, 2009; Crump and Hobbie, 2005; Shade et al., 2007; Nelson, 2009).
This ‘landscape reservoir'concept also provides a spatial dimension to a theoretical model in which microbial dormancy and reduced mortality maintain microbial diversity and prevent the extinction of rare taxa, which can then reemerge as abundant taxa when conditions are favorable (Jones and Lennon, 2010). Soils, hyporheic zones, sediments and other environments dominated by biofilms are well suited for maintaining dormant microbial populations because of the protection from grazing mortality within biofilms (for example, Clarholm, 1981). In contrast, open water provides few refugia for microbes and subjects them to intense grazing pressure (Jurgens and Matz, 2002; Gobler et al., 2008). Advection of taxa from refugia in upslope environments may enhance the source of rare taxa in planktonic environments, and prevent stochastic emergence of functionally identical communities (Sloan et al., 2006) following seasonal shifts in environmental conditions. Our finding that 56% of abundant bacterial taxa in Toolik Lake were rare taxa in upslope environments suggests that many rare taxa are quite ‘adaptable' to different niches along the hydrological continuum. In turn, we suggest that many rare taxa are viable yet relatively inactive (Jones and Lennon, 2010) during transport, until conditions change and their populations can grow to dominate in a downstream community, in much the same way as rare taxa in the coastal ocean can become dominant when seasonal environmental conditions become favorable (Campbell et al., 2011).
Overall, this study showed that species sorting of both rare and common taxa has a fundamental role in determining community composition as taxa from different habitats are mixed and transported across hydrologically linked ecosystems. In addition, several lines of evidence indicate, for the first time to our knowledge, that dispersal from terrestrial and upslope habitats can be quantitatively important in controlling the patterns of diversity of downslope surface waters, even in ecosystems such as arctic tundra where the bulk of spring snow melt flows over the still frozen soils. This model of landscape biogeography may also apply to regions with more complex flowpaths that include multiple soil layers and deep groundwater environments, such as mid-latitude regions in which spring snow melt flows through thawed soils and provides more extensive contact and presumably greater transfer of soil microbes into aquatic communities. Although we demonstrated that both species sorting and mass effects through dispersal are prominent controls on microbial community structure, it remains to be seen how these controls vary over time or differentially affect rare versus dominant taxa. It is clear, however, that the patterns of change in diversity over space and time in arctic lakes and streams would be difficult to understand without considering upslope sources of diversity, and we anticipate that future changes in soil environments will be reflected in surface-water microbial diversity.
Acknowledgments
Funding was provided by NSF DEB-0717390 to LAZ (MIRADA-LTERS); DEB-0639790 to BCC and DEB-0639805 to GWK (Arctic LTREB) and DEB-1026843 to GWK (Arctic LTER). We thank Joanna Green and Cody Johnson for sample collection, and Elizabeth McCliment for help with pyrosequencing.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aller JY, Kemp PF. Are Archaea inherently less diverse than Bacteria in the same environments. FEMS Microbiol Ecol. 2008;65:74–87. doi: 10.1111/j.1574-6941.2008.00498.x. [DOI] [PubMed] [Google Scholar]
- Aller JY, Kuznetsova MR, Jahns CJ, Kemp PF. The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. J Aerosol Sci. 2005;36:801–812. [Google Scholar]
- Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One. 2009;4:e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auinger BM, Pfandl K, Boenigk J. Improved methodology for identification of protists and microalgae from plankton samples preserved in Lugol's iodine solution: Combining microscopic analysis with single-cell PCR. Appl Environ Microbiol. 2008;74:2505–2510. doi: 10.1128/AEM.01803-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström AK, Jansson M. Bacterioplankton production in humic Lake Örträsket in relation to input of bacterial cells and input of allochthonous organic carbon. Microb Ecol. 2000;39:101–115. doi: 10.1007/s002480000007. [DOI] [PubMed] [Google Scholar]
- Bunge J. Estimating the number of species with CatchAll. Proc Pac Symp Biocomput. 2011;16:121–130. doi: 10.1142/9789814335058_0014. [DOI] [PubMed] [Google Scholar]
- Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA. 2011;108:12776–12781. doi: 10.1073/pnas.1101405108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Shen T-J.2010Program SPADE (Species Prediction And Diversity Estimation)Available at: : http://chao.stat.nthu.edu.tw/ .
- Clarholm M. Protozoan grazing of bacteria in soil—impact and importance. Microb Ecol. 1981;7:343–350. doi: 10.1007/BF02341429. [DOI] [PubMed] [Google Scholar]
- Clarke KR. Nonparametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- Crump BC, Adams HE, Hobbie JE, Kling GW. Biogeography of bacterioplankton in lakes and streams of an arctic tundra catchment. Ecology. 2007;88:1365–1378. doi: 10.1890/06-0387. [DOI] [PubMed] [Google Scholar]
- Crump BC, Hobbie JE. Synchrony and seasonality of bacterioplankton communities in two temperate rivers. Limnol Oceanogr. 2005;50:1718–1729. [Google Scholar]
- Crump BC, Kling GW, Bahr M, Hobbie JE. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl Environ Microbiol. 2003;69:2253–2268. doi: 10.1128/AEM.69.4.2253-2268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Peterson BJ, Raymond PA, Amon RMW, Rinehart A, McClelland JW, et al. Circumpolar synchrony in big river bacterioplankton. Proc Natl Acad Sci USA. 2009;106:21208–21212. doi: 10.1073/pnas.0906149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T. Microbial ecology on land and sea. Philos Trans R Soc Lond Ser B-Biol Sci. 1994;343:51–56. [Google Scholar]
- Fierer N, Morse JL, Berthrong ST, Bernhardt ES, Jackson RB. Environmental controls on the landscape-scale biogeography of stream bacterial communities. Ecology. 2007;88:2162–2173. doi: 10.1890/06-1746.1. [DOI] [PubMed] [Google Scholar]
- Fierer N, Nemergut D, Knight R, Craine JM. Changes through time: integrating microorganisms into the study of succession. Res Microbiol. 2010;161:635–642. doi: 10.1016/j.resmic.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci USA. 2009;106:22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobler CJ, Davis TW, Deonarine SN, Saxton MA, Lavrentyev PJ, Jochem FJ, et al. Grazing and virus-induced mortality of microbial populations before and during the onset of annual hypoxia in Lake Erie. Aquat Microb Ecol. 2008;51:117–128. [Google Scholar]
- Grammatika M, Zimmerman WB. Microhydrodynamics of flotation processes in the sea surface layer. Dyn Atmos Oceans. 2001;34:327–348. [Google Scholar]
- Hobbie JE, Corliss TL, Peterson BJ. Seasonal patterns of bacterial abundance in an arctic lake. Arctic Alpine Res. 1983;15:253–259. [Google Scholar]
- Huber JA, Welch DBM, Morrison HG, Huse SM, Neal PR, Butterfield DA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Huse S, Huber J, Morrison H, Sogin M, Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genetics. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, McMahon KD. Species-sorting may explain an apparent minimal effect of immigration on freshwater bacterial community dynamics. Environ Microbiol. 2009;11:905–913. doi: 10.1111/j.1462-2920.2008.01814.x. [DOI] [PubMed] [Google Scholar]
- Judd KE, Crump BC, Kling GW. Variation in dissolved organic matter controls bacterial production and community composition. Ecology. 2006;87:2068–2079. doi: 10.1890/0012-9658(2006)87[2068:vidomc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Judd KE, Crump BC, Kling GW. Bacterial responses in activity and community composition to photo-oxidation of dissolved organic matter from soil and surface waters. Aquat Sci. 2007;69:96–107. [Google Scholar]
- Jurgens K, Matz C. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek. 2002;81:413–434. doi: 10.1023/a:1020505204959. [DOI] [PubMed] [Google Scholar]
- Kan JJ, Wang K, Chen F. Temporal variation and detection limit of an estuarine bacterioplankton community analyzed by denaturing gradient gel electrophoresis (DGGE) Aquat Microb Ecol. 2006;42:7–18. [Google Scholar]
- Kirchman DL, Cottrell MT, Lovejoy C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol. 2010;12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- Kling GW, Kipphut GW, Miller MC, O'Brien WJ. Integration of lakes and streams in a landscape perspective: the importance of material processing on spatial patterns and temporal coherence. Freshw Biol. 2000;43:477–497. [Google Scholar]
- Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, et al. The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett. 2004;7:601–613. [Google Scholar]
- Lindström ES, Forslund M, Algesten G, Bergstrom AK. External control of bacterial community structure in lakes. Limnol Oceanogr. 2006;51:339–342. [Google Scholar]
- Logue JB.2010Factors influencing the biogeography of bacteria in fresh waters—a metacommunity approachPh.D. thesis,Uppsala University, Uppsala, Sweden [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magurran AE.1988Ecological Diversity and Its Measurementvol. 179.Princeton University Press: Princeton, NJ [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- McDowell-Boyer LM, Hunt JR, Sitar N. Particle transport through porous media. Water Resour Res. 1986;22:1901–1921. [Google Scholar]
- Nelson CE. Phenology of high-elevation pelagic bacteria: the roles of meteorologic variability, catchment inputs and thermal stratification in structuring communities. ISME J. 2009;3:13–30. doi: 10.1038/ismej.2008.81. [DOI] [PubMed] [Google Scholar]
- O'Brien WJ, Bahr M, Hershey AE, Hobbie JE, Kipphut GW, Kling GW, et al. 1997The limnology of Toolik LakeIn: Milner A, Oswood MW (eds).Freshwaters of Alaska: Ecological Syntheses (Ecological Studies, Vol 119) Springer-Verlag: New York; 61–106. [Google Scholar]
- Pedros-Alio C. Marine microbial diversity: can it be determined. Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A, Tiedje JM. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc Natl Acad Sci USA. 2007;104:2761–2766. doi: 10.1073/pnas.0610671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-Source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Kent AD, Jones SE, Newton RJ, Triplett EW, McMahon KD. Interannual dynamics and phenology of bacterial communities in a eutrophic lake. Limnol Oceanogr. 2007;52:487–494. [Google Scholar]
- Shaw AK, Halpern AL, Beeson K, Tran B, Venter JC, Martiny JBH. It's all relative: ranking the diversity of aquatic bacterial communities. Environ Microbiol. 2008;10:2200–2210. doi: 10.1111/j.1462-2920.2008.01626.x. [DOI] [PubMed] [Google Scholar]
- Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored ‘rare biosphere. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamames J, Abellan JJ, Pignatelli M, Camacho A, Moya A. Environmental distribution of prokaryotic taxa. BMC Microbiol. 2010;10 doi: 10.1186/1471-2180-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford RJ, Vandvik V, Birks HJB. Dispersal limitations matter for microbial morphospecies. Science. 2006;312:1015–1015. doi: 10.1126/science.1125669. [DOI] [PubMed] [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Uroz S, Buee M, Murat C, Frey-Klett P, Martin F. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep. 2010;2:281–288. doi: 10.1111/j.1758-2229.2009.00117.x. [DOI] [PubMed] [Google Scholar]
- Whalen SC, Cornwell JC. Nitrogen, phosphorus and organic carbon cycling in an arctic lake. Can J Fish Aquat Sci. 1985;42:797–808. [Google Scholar]
- Wilson DS. Complex interactions in metacommunities, with implications for biodiversity and higher levels of selection. Ecology. 1992;73:1984–2000. [Google Scholar]
- Wolda H. Similarity indices, sample size and diversity. Oecologia. 1981;50:296–302. doi: 10.1007/BF00344966. [DOI] [PubMed] [Google Scholar]
- Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotech. 2011;29:415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Massana R, Not F, Marie D, Vaulot D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol. 2005;52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.