Abstract
The Deepwater Horizon oil spill in the Gulf of Mexico resulted in a deep-sea hydrocarbon plume that caused a shift in the indigenous microbial community composition with unknown ecological consequences. Early in the spill history, a bloom of uncultured, thus uncharacterized, members of the Oceanospirillales was previously detected, but their role in oil disposition was unknown. Here our aim was to determine the functional role of the Oceanospirillales and other active members of the indigenous microbial community using deep sequencing of community DNA and RNA, as well as single-cell genomics. Shotgun metagenomic and metatranscriptomic sequencing revealed that genes for motility, chemotaxis and aliphatic hydrocarbon degradation were significantly enriched and expressed in the hydrocarbon plume samples compared with uncontaminated seawater collected from plume depth. In contrast, although genes coding for degradation of more recalcitrant compounds, such as benzene, toluene, ethylbenzene, total xylenes and polycyclic aromatic hydrocarbons, were identified in the metagenomes, they were expressed at low levels, or not at all based on analysis of the metatranscriptomes. Isolation and sequencing of two Oceanospirillales single cells revealed that both cells possessed genes coding for n-alkane and cycloalkane degradation. Specifically, the near-complete pathway for cyclohexane oxidation in the Oceanospirillales single cells was elucidated and supported by both metagenome and metatranscriptome data. The draft genome also included genes for chemotaxis, motility and nutrient acquisition strategies that were also identified in the metagenomes and metatranscriptomes. These data point towards a rapid response of members of the Oceanospirillales to aliphatic hydrocarbons in the deep sea.
Keywords: Gulf oil spill, Deepwater Horizon, Oceanospirillales, single-cell genomics, metagenomics, metatranscriptomics
Introduction
On 20 April 2010, the Deepwater Horizon oil rig exploded and sank, resulting in an unremitting flow of oil from April 2010 to July 2010 into the Gulf of Mexico, for a total of approximately 4.9 million barrels (779 million liters, ±10%) (Federal Interagency Solutions Group, 2010). The MC252 oil fraction was comprised of a complex mixture of hydrocarbons including saturated hydrocarbons (74%), aromatic hydrocarbons (including polycyclic hydrocarbons (PAHs)) which reached maximal concentrations of 1200 mg l−1 at the surface (Hazen et al., 2010) (16%), and polar compounds (10%) (Reddy et al., 2012). During the spill, an oil plume was detected at depths of approximately 1000–1300 m (Camilli et al., 2010; Hazen et al., 2010). The deep-sea oil plume was reported to contain gaseous components (Valentine et al., 2010; Kessler et al., 2011), as well as non-gaseous, more recalcitrant compounds such as benzene, toluene, ethylbenzene and total xylenes (BTEX) at concentrations ranging 50–150 μg l−1 (Camilli et al., 2010; Hazen et al., 2010). This influx of hydrocarbons had a significant impact on the indigenous microbial community structure (Hazen et al., 2010; Valentine et al., 2010; Kessler et al., 2011; Redmond and Valentine, 2011), including enrichment of uncultivated members of the Oceanospirillales early in the spill history (Hazen et al., 2010; Redmond and Valentine, 2011). The lack of a cultivated isolate of the Oceanospirillales from the plume precluded a clear understanding of the direct physiological and ecological consequences of the hydrocarbons on this group of microorganisms.
The documented shifts in the microbial community structure over time in response to the deep-sea plume of hydrocarbons have been shown by DNA-based methods such as cloning and sequencing of 16S rRNA genes (Hazen et al., 2010; Valentine et al., 2010; Kessler et al., 2011; Redmond and Valentine, 2011) and microarray analysis of functional genes (Lu et al., 2011). Cloning and sequencing revealed a clear temporal succession of bacteria in the deep-sea hydrocarbon plume from a community dominated by Oceanospirillales (Hazen et al., 2010; Redmond and Valentine, 2011) to Colwellia and Cycloclasticus (Valentine et al., 2010; Redmond and Valentine, 2011) and finally to methylotrophic bacteria (Kessler et al., 2011). To date, however, no deep-sequencing approach has been used to analyze the microbial community structure, including rare members of the community, and their function. In addition, there is no information about what microorganisms were active or which functional genes were actually expressed in response to the oil spill.
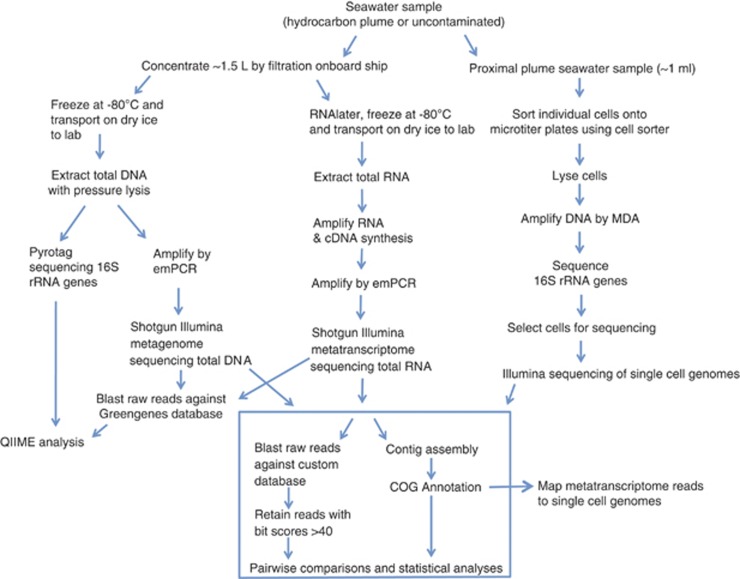
Here we aimed to determine the specific roles of the Oceanospirillales that were enriched in the plume early in the spill history. In addition, we aimed to determine which functional genes and pathways were expressed in the deep-sea plume. To address these aims, we not only analyzed the functional gene repertoire in total DNA extracted from metagenomic samples but also extracted and sequenced total RNA metatranscriptomes to determine which genes were highly expressed and representative of active members of the community. In addition, to specifically characterize the functional roles of the dominant Oceanospirillales, we isolated and sequenced single-representative cells. For all of these analyses, we used the Illumina sequencing platform (Illumina, San Diego, CA, USA), which resulted in over 60 Gb of data. To analyze and integrate these large 'omics' data sets, including raw, unassembled reads, we used several novel bioinformatics approaches, which are outlined in Figure 1. For this study, we focused on samples that were collected during the oil spill between 27 and 31 May 2010 (Hazen et al., 2010) for in-depth phylogenetic and functional analyses: two plume samples, one proximal (1.5 km from the wellhead) and one distal (11 km from the wellhead), and one uncontaminated sample collected at plume depth (40 km from the wellhead) (Supplementary Figure S1).
Figure 1.
Methods schematic. Each type of molecular approach—metagenomics, metatranscriptomics and single cell genomics—is shown, as are the subsequent, novel bioinformatics approaches that were used to analyze the various data sets.
Materials and methods
Sample collection
From each station, 1–5 l of seawater were filtered through a 0.2-μm diameter filter from the Gulf of Mexico during two monitoring cruises from 27 May 2010 to 2 June 2010 on the R/V Ocean Veritas and R/V Brooks McCall. Detailed information regarding sample collection can be found in Hazen et al. (2010).
DNA extraction
DNA was extracted from microbial cells collected onto filters using a modified Miller method (Miller et al., 1999), with the addition of a pressure lysis step to increase cell-lysis efficiency. One-half of each filter was placed into a Pressure Biosciences FT500 Pulse Tube (Pressure Biosciences, Easton, MA, USA). A total of 300 μl of Miller phosphate buffer and 300 μl of Miller SDS lysis buffer were added and mixed. A solution of 600 μl phenol:chloroform:isoamyl alcohol (25:24:1) was then added. The samples were subjected to pressure cycling at 35 000 psi for 20 s and 0 psi for 10 s for a total of 20 cycles using the Barocycler NEP3229 (Pressure Biosciences). After pressure cycling, the sample material was transferred to a Lysing Matrix E tube (MP Biomedicals, Solon, OH, USA) and the samples were subjected to bead beating at 5.5 m s−1 for 45 s in a FastPrep instrument (MP Biomedicals). The tubes were centrifuged at 16 000 g for 5 min at 4 °C, 540 μl of supernatant was transferred to a 2-ml tube and an equal volume of chloroform was added. The individual samples were mixed by inversion and then centrifuged at 10 000 g for 5 min. A total of 400 μl of the aqueous phase was transferred to another tube and two volumes of Solution S3 (MoBio, Carlsbad, CA, USA) were added and mixed by inversion. The rest of the clean-up procedures followed the instructions in the MoBio Soil DNA extraction kit. Samples were recovered in 60 μl Solution S5 and stored at −20 °C.
16S rRNA gene sequencing and analysis
16S rRNA gene sequences were amplified from the DNA extracts using the primer pair 926wF (5′-AAACTYAAAKGAATTGRCGG-3′) and 1392R (Lane, 1991) as previously described (Kunin et al., 2010). The reverse primer included a 5-bp barcode for multiplexing of samples during sequencing. Emulsion PCR and sequencing of the PCR amplicons was performed at DOE's Joint Genome Institute following the manufacturer's instructions for the Roche (Branford, CT, USA) 454 GS Titanium technology (Allgaier et al., 2010). A total of 87 000 pyrotag sequences were obtained and analyzed using QIIME (Caporaso et al., 2010a). Briefly, 16S rRNA gene sequences were clustered with uclust (Edgar, 2010) and assigned to operational taxonomic units (OTUs) with 97% similarity. Representative sequences from each OTU were aligned with Pynast (Caporaso et al., 2010b) using the Greengenes (DeSantis et al., 2006) core set. Taxonomy was assigned using the Greengenes 16S rRNA gene database (version 6 October 2010). As the number of sequence reads in each sample varied, the data set was rarified prior to alpha diversity calculations.
RNA extraction and amplification
Immediately following sampling and filtration at the proximal sampling station, samples intended for RNA extractions were placed in RNAlater (Ambion, Foster City, CA, USA) to prevent RNA degradation. Samples were stored according to the manufacturer's protocol (in RNAlater at −80 °C) until the time of extraction. Total RNA was extracted from the proximal and distal plume stations, as well as from the uncontaminated sample from plume depth, as previously described (DeAngelis et al., 2010). The quantity and quality of extracted RNA was checked using a Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Specifically, the RNA integrity was verified by determining the RNA integrity number. For our samples, the RNA integrity number was ∼9 on a scale of 1–10, with 10 indicating that no degradation had occurred. Insufficient RNA was obtained from the uncontaminated sample for downstream processing. Total RNA from the proximal and distal plume stations was amplified using the Message Amp II-Bacteria Kit (Ambion) following the manufacturer's instructions. First-strand synthesis of cDNA from the resulting antisense RNA was carried out with the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). The SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) was used to synthesize double-stranded cDNA. cDNA was purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA, USA). Poly(A) tails were removed by digesting purified DNA with BpmI for 3 h at 37 °C. Digested cDNA was purified with QIAquick PCR purification kit (Qiagen).
Emulsion PCR
To increase yields required for sequencing, DNA and cDNA were amplified by emulsion PCR. A detailed description of this method can be found in Blow et al. (2008). Briefly, DNA for metagenomic samples was sheared (cDNA was not sheared) using the Covaris S-Series instrument (Covaris, Woburn, MA, USA). DNA and cDNA were end-repaired using the End-It DNA End-Repair Kit (Epicentre Biotechnologies, Madison, WI, USA). End-repaired DNA and cDNA were then ligated with Illumina Paired End Adapters 1 and 2. For each sample, 10 ng was used for emulsion PCR. Emulsion PCR reagents and thermal cycler protocols were as previously described (Blow et al., 2008). Amplified products were cleaned with a PCR mini-elute column (Qiagen), visualized and ∼300 bp fragments were excised from a 2% agarose gel.
Sequencing
Metagenomic shotgun sequencing libraries of the samples were sequenced using the Illumina GAIIx 2 × 114 bp pair-end technology. The Illumina sequencing platform was used to generate 14–17 Gb of sequence data per sample.
cDNA was sequenced using the Illumina GAIIx sequencing platform. cDNA was quantified and clustered accordingly onto one lane of a flow cell on Illumina's cBot Cluster Generation System. After cluster generation, the flow cell was transferred to a GAIIx and was sequenced for 100 cycles for read 1. Then, turnaround chemistry was performed by the paired-end module, which prepared the flow cell for read 2 sequencing. Another 100 cycles of sequencing followed, resulting in 100 bp paired-end reads.
Sequence assembly and analysis
Raw Illumina metagenomic reads (∼113 bp in length) were trimmed using a minimum quality cutoff of 3. Both trimmed and untrimmed reads were kept for further assembly. Paired-end Illumina reads were assembled using SOAPdenovo (http://soap.genomics.org.cn/soapdenovo.html) at a range of Kmers (21, 23, 25, 27, 29 and 31) for both trimmed and untrimmed reads. Default settings for all SOAPdenovo assemblies were used (flags: –d 1 and –R). Contigs generated by each assembly (12 total contig sets) were merged using a combination of in-house Perl script. Contigs were then sorted into two pools based on length. Contigs <1800 bp were assembled using Newbler (Life Technologies, Carlsbad, CA, USA) in an attempt to generate larger contigs (flags: −tr, −rip, −mi 98 and −ml 60). All assembled contigs >1800 bp, as well as the contigs generated from the final Newbler run, were combined using minimus 2 (AMOS, http://sourceforge.net/projects/amos) and the default parameters for joining. Minimus2 is an overlap-based assembly tool that is useful for combining low numbers of longer sequences, as are found in assembled contigs. Assembly of the total of 368 million paired-end quality filtered metagenome sequence reads that averaged 113 bp in length (45 Gb) resulted in 1.1 million contigs. These contigs had an average N50 length of 382 bp (N50 is the length of the smallest contig in the set of largest contigs that have a combined length that represents at least 50% of the assembly (Miller et al., 2010)). Assembled data was annotated in IMG (Markowitz et al., 2008). Cluster of Orthologous (COG) annotations for both plume samples and the uncontaminated sample, including average fold, were exported. A pairwise statistical comparison of COGs in each of the three samples was carried out using STAMP (Parks and Beiko, 2010). Raw Illumina metatranscriptomic reads (∼100 bp in length) were assembled using the CLC Genomics Workbench (version 4.0.3; CLC Bio, Cambridge, MA, USA). Paired-end reads were assembled using the following parameters: mismatch cost 2, insertions cost 3, deletion cost 3, length fraction 0.5 and similarity 0.8. The minimum contig length was set to 200 bp. Assembled metatranscriptomic data was annotated using CAMERA (v2.0.6.2) (Seshadri et al., 2007).
blastn
Single reads from each metagenomic and metatranscriptomic sample was searched against the Greengenes (DeSantis et al., 2006) database of 16S rRNA genes using blastn with a bit score cutoff of >100. For each sequence, the blast result with the highest bit score was selected.
tblastn
Raw metagenomic, metatranscriptomic and single-cell reads were searched against a subset of proteins (∼12 000 archaeal and bacterial proteins) involved in hydrocarbon degradation from the GeoChip (He et al., 2010) database. This database was selected because, to our knowledge, this is the only curated database of the nearly complete pathways for hydrocarbon degradation. Paracel blast was used with the tblastn algorithm, allowing all possible hits and using a bit score cutoff of >40. For each sequence, the blast result with the highest bit score was selected. Although putative and potential proteins were part of the overall database searched, only characterized proteins were included in the final data analysis and presentation. A pairwise statistical comparison of the results of the metagenomic and metatranscriptomic blast analyses was carried out using STAMP (Parks and Beiko, 2010), using the a two-sided Chi-square test (with Yates) statistic with the DP: asymptotic-CC confidence interval method and the Bonferroni multiple test correction. A P-value of >0.05 was used with a double-effect size filter (difference between proportions effect size <1.00 and a ratio of proportions effect size <2.00).
Single-cell sorting, whole-genome amplification and screening
Cells were collected following the clean sorting procedures detailed by Rodrigue et al. (2009). Briefly, single cells from the proximal plume water sample were sorted by the Cytopeia Influx Cell Sorter (BD Biosciences, Franklin Lakes, NJ, USA) into three 96-well plates containing 3 μl of ultraviolet-treated TE. The cells were stained with SYBR Green I (Invitrogen) and illuminated by a 488-nm laser (Coherent Inc., Santa Clara, CA, USA). The sorting window was based on the size determined by side scatter and green fluorescence (531/40 bp filter). Single cells were lysed for 20 min at room temperature using alkaline solution from the Repli-G UltraFast Mini Kit (Qiagen) according to the manufacturer's instructions. After neutralization, the samples were amplified using the RepliPHI Phi29 reagents (Epicentre Biotechnologies). Each 50-μl reaction contained Phi29 Reaction Buffer (1 × final concentration), 50 μℳ random hexamers with the phosphorothioate bonds between the last two nucleotides at the 3′ end d (IDT), 0.4 mℳ dNTP, 5% DMSO (Sigma, St Louis, MO, USA), 10 mℳ DTT (Sigma), 100 U Phi29 and 0.5 mℳ Syto 13 (Invitrogen). A mastermix of multiple displacement amplification (MDA) reagents minus the Syto 13 sufficient for a 96-well plate was ultraviolet-treated for 60 min for decontamination. Syto 13 was then added to the mastermix, which was added to the single cells for real-time MDA on the Roche LightCycler 480 for 17 h at 30 °C. All steps of single-cell handling and amplification were performed under most stringent conditions to reduce the introduction of contamination. Single-cell MDA products were screened using Sanger sequencing of 16S rRNA gene amplicons derived from each MDA product. A total of 16 Oceanospirillales cells were obtained. Three single-amplified genomes were identified as being 95% similar to the dominant Oceanospirillales OTU, and of high sequence quality (16S rRNA gene) and pursued for whole-genome sequencing.
Single-cell Illumina sequencing, quality control and assembly
Single-cell amplified DNA of three Oceanospirillales cells was used to generate normalized, indexed Illumina libraries. Briefly, 3 μg of MDA product was sheared in 100 μl using the Covaris E210 (Covaris) with the setting of 10% duty cycle, intensity 5 and 200 cycle per burst for 6 min per sample and the fragmented DNA purified using QIAquick columns (Qiagen) according to the manufacturer's instructions. The sheared DNA was end-repaired, A-tailed and ligated to the Illumina adaptors according to the Illumina standard paired-end protocol. The ligation product was purified using AMPure SPRI beads, then underwent normalization using the Duplex-Specific Nuclease Kit (Axxora, San Diego, CA, USA). The normalized libraries were then amplified by PCR for 12 cycles using a set of two indexed primers and the library pool was sequenced using an Illumina GAIIx sequencer according to the manufacturer's protocols (run mode 2 × 150 bp). Approximately 2.5 Gb (16 797 846 reads) of sequence data was collected from the Oceanospirillales single-cell genomes. The Illumina single-amplified genome data was quality controlled using GC content and blast analysis and no contamination was detectable in two of the single-amplified genomes, whereas the third single-amplified genome was excluded from the analysis due to the presence of contaminating sequences. Reads from these two single cells were assembled using Velvet (Zerbino and Birney, 2008). To estimate genome-sequence completeness, the annotated, assembled draft genome data was compared with core COGs for Proteobacteria and Gammaproteobacteria (number of identified core COGs/number of expected core COGs).
Mapping and analysis
Unassembled metatranscriptomic reads were mapped to the Oceanospirillales single-cell draft genome using the CLC Genomics Workbench (CLC bio), using the following parameters: mismatch cost 2, insertions cost 3, deletion cost 3, length fraction 0.5 and similarity 0.8. Assembled single-cell data was annotated using CAMERA (v2.0.6.2) (Seshadri et al., 2007). The Interactive Pathways Explorer v2 (Letunic et al., 2008) was used to map the assembled, annotated metatranscriptome with an assembled, annotated Oceanospirillales single-cell draft genome. Clustered regularly interspaced short palindromic repeat regions were identified in the draft genome using CRISPRFinder (Grissa et al., 2007).
Cell counts
Cell counts were carried out as described in Hazen et al. (2010). Briefly, samples were preserved in 4% formaldehyde and stored at 4 °C until the time of analysis. Filtered cells were stained with Acridine Orange and imaged with a Zeiss Axioskop (Carl Zeiss, Inc., Oberkochen, Germany) microscope.
Infrared spectromicroscopy and data processing
Synchrotron radiation-based Fourier-transform infrared measurements and analyses were conducted at the infrared beamline of the Advanced Light Source (http://infrared.als.lbl.gov/) on thin layers of fresh samples. Samples consisted of 25 ml of seawater from the proximal plume station. A total of 10 subsamples were randomly collected using a glass pipette. Samples were placed between a gold-coated Si wafer and a SiNx window. Photons emitted over a mid-infrared wavenumber range of 4000 to 650 cm−1 were focused through the samples by the Nicolet Nic-Plan IR microscope (with a numerical aperture objective of 0.65), which was coupled to a Nicolet Magna 760 FTIR bench (Thermo Scientific Inc., Waltham, MA, USA). The entire view-field was 200 × 150 μm2, which was typically divided into equal-sized 2 × 2 μm2 squares before raster scanning. The synchrotron radiation-based Fourier-transform infrared transflectance spectra at each position were collected using a single-element mercury cadmium telluride detector at a spectral resolution of 4 cm-1 with 32 co-added scans and a peak position accuracy of 1/100 cm−1. In transflectance, the synchrotron infrared beam transmitted through the cells, reflected off the gold-coated surface and then transmitted through the sample a second time before reaching the detector. Background spectra were acquired from neighboring locations without any cells and used as reference spectra for both samples and standards to remove background H2O and CO2 absorptions. All synchrotron radiation-based Fourier-transform infrared transflectance spectra were subjected to an array of data preprocessing and processing calculations using Thermo Electron's Omnic version 7.3. The processing includes the computation conversion of transflectance to absorbance, spectrum baseline removal and univariate analysis. In the univariate analysis, the calculated infrared absorbance at each wavenumber in the mid-infrared region can also be related to the relative concentration of a particular chemical component through the Beer–Lambert Law. Because analysis of each spectral absorption band provides a single absorption value (representing the relative abundance of a chemical component), we also constructed two-dimensional images to visualize the relative abundance of petroleum products and microbial biomolecules.
Hydrocarbon analysis
The profile of Macondo crude oil (collected on 22 May 2010 directly from the Discovery Enterprise drill ship located above the wellhead) was determined by gas chromatography–mass spectrometry using an Agilent 6890N (Agilent Technologies). Triplicate samples of 0.2 μl of raw oil were directly injected to the column with no sample cleanup. This method was used to enable detection of low-molecular weight compounds that would be lost during sample processing or masked due to interference from solvent peaks. The Agilent 6890N was equipped with a 5972 mass selective detector and operated in SIM/SCAN mode. The injection temperature was 250 °C, detector temperature was 300 °C and column used was 60 m Agilent HP-1MS with a flow rate of 2 ml min−1. The oven temperature program included a 50°C hold for 3 min ramped to 300 °C at 4 °C min−1 with a final 10-min hold at 300 °C. Compound identification was determined from selective ion monitoring coupled with comparison with the known standards and compound spectra in the NIST 08MS library. Compounds were reported as fractions of total oil in Supplementary Figure S2 from averages of triplicate injections, the error bars indicating s.d.
Hydrocarbon concentrations in all samples (Supplementary Table S1) were determined from water samples that were collected in the field and directly filtered through Sterivex filters (0.22 μm; Millipore, Billerica, MA, USA) as described previously (Hazen et al., 2010). Oil biomarkers from the plume samples matched to those observed from the Macondo well.
Volatile aromatic hydrocarbons were measured using USEPA (US Environmental Protection Agency) methods 5030/8260b on an Agilent 6890 GC with a 5973 mass spectrometer detector. Initial oven temperature 10 °C, initial time 3 min, ramp 8–188 °C min−1, then 16–220 °C min−1 and hold for 9 min. Split ratio 25:1. Restek Rtx-VMS capillary column, 60 m length by 250 μm diameter and 1.40 μm film. Scan 50–550 m z−1.
Results and discussion
Throughout our analyses, we found differences in the microbial community structures of the samples collected from the two plume sites due to the differences in the amount of time the respective indigenous deep-sea microbes were exposed to hydrocarbons. Our samples were collected during the Deepwater Horizon spill within 24 h following the failed top kill effort (29 May 2010; proximal station). This effort resulted in a large influx of hydrocarbons into the deep sea on the dates that we sampled. Because of the movement of water in marine currents, we took the current velocity into account (6.7 km per day; Camilli et al., 2010; Hazen et al., 2010) when calculating the length of time that microbes in our samples had been exposed to hydrocarbons from the oil spill. Based on these calculations, the microbial communities would have been exposed to hydrocarbons for approximately 6 h by the time the plume reached the proximal station, whereas by the time the plume reached the distal station, the microbes would have been exposed to hydrocarbons for approximately 39 h. Hydrocarbons were not detected in the uncontaminated sample collected from plume depth.
Analysis of our combined DNA sequence data (16S rRNA gene sequences from 454 ‘pyrotag sequencing' and ‘total metagenomic DNA') revealed that the plume samples had a lower microbial diversity than samples outside the plume (Supplementary Figure S3 and Table 1), with an enrichment of Oceanospirillales (Figure 2 and Supplementary Tables S2 and S3), as previously reported (Hazen et al., 2010; Redmond and Valentine, 2011). In the pyrotag data, one Oceanospirillales OTU comprised up to 80–90% of the proximal and distal plume communities, respectively, whereas it comprised only 3% of the total community in the uncontaminated sample (Figure 2 and Supplementary Table S2). Similarly, in the metagenome data, the Oceanospirillales comprised >60% of both plume samples, compared with 5% in the uncontaminated sample in the metagenome data (Figure 2 and Supplementary Table S3). This observed bloom of Oceanospirillales corresponded with an increase in bacterial cell densities in the plume from 5.47±2.68 × 103 cells per ml in the uncontaminated sample to 1.44±0.47 × 105 cells per ml in the proximal plume and 2.68±0.48 × 105 cells per ml in the distal plume (see Hazen et al., 2010 (Figure 4a)).
Table 1. Diversity metrics of rarified bacterial and archaeal 16S rRNA 454-pyrotag sequences.
| Sample | Chao1a | Chao1 (lower bound)b | Chao1 (upper bound)b | ACEc | Simpsond | Singletonse | Doubletonse |
|---|---|---|---|---|---|---|---|
| Distal plume | 394.53 | 273.32 | 628.27 | 443.80 | 0.58 | 91.78 | 15.96 |
| Proximal plume | 806.71 | 626.34 | 1093.50 | 911.07 | 0.57 | 198.93 | 38.47 |
| Uncontaminated | 1722.58 | 1507.22 | 2007.66 | 1849.08 | 0.96 | 481.80 | 126.04 |
Species richness (Chao, 1984).
Confidence intervals (Chao et al., 1992).
Species richness (Chao et al., 2000).
Species diversity (Simpson, 1949).
Singeltons are species with only one individual. Doubletons are species with only two individuals (Colwell and Coddington, 1994).
Figure 2.
Relative abundance of bacteria and archaea in the proximal and distal plume samples and in the uncontaminated sample collected from plume depth. (a) Relative OTU abundance of rarified 16S rRNA gene 454-pyrotag data. Universal primers for archaea and bacteria were used. Taxonomy was assigned using the Greengenes (DeSantis et al., 2006) 16S rRNA gene database. (b) Raw, unassembled metagenomic and metatranscriptomic reads were compared with the Greengenes (DeSantis et al., 2006) database. Less-abundant bacteria and archaea are grouped under the category ‘other.' The complete list of bacteria and archaea observed in these analyses are presented in Supplementary Tables S2, S3 and S4.
Recently, we used a GeoChip (He et al., 2010) functional gene microarray to determine which functional genes were prevalent in the plume and found several hydrocarbon degradation genes having a higher relative abundance in the plume (Lu et al., 2011). However, those data were not sufficient for determination of the biodegradation pathways or whether such pathways were actually expressed or attributed to a particular microorganism in the plume. Here we examined deep metagenome sequence data for genes and the pathways involved in hydrocarbon degradation. We found that the entire pathway for degradation of n-alkanes was represented and abundant in the metagenome data from the plume samples (Figure 3). Alkane oxidation is initiated by monooxygenases, yielding alcohols as intermediates, which are converted to aldehydes and fatty acids by alcohol and aldehyde dehydrogenases (Sabirova et al., 2006). In our study, we observed genes corresponding to alkane monooxygenases, a group of enzymes with broad substrate specificity. In addition, the nearly complete pathway for cyclohexane degradation (alkane monooxygenase→cyclohexanol dehydrogenase→cyclohexanone monooxygenase→→beta oxidation) (Sabirova et al., 2006) was observed and abundant in the metagenomes (Figure 3). We also found a specific alkane gene (alkane-1 monooxygenase), as also reported by Lu et al. (2011), that was more abundant in the plume than outside of the plume. However, in contrast to Lu et al. (2011), we found that genes involved in degradation of aromatic compounds were less abundant than those involved in alkane degradation (Figure 3; see Supplementary Figure S2 for Macondo crude oil constituents and Supplementary Table S1 for n-alkane, cyclohexane, methylcyclohexane, BTEX and PAH concentrations in the plume samples). For example, genes coding for ethylbenzene, toluene and PAH degradation were significantly (P<0.05) less abundant in both plume samples compared with the uncontaminated sample. The abundance of genes involved in alkane degradation compared with those involved in degradation of aromatic compounds in our data set is consistent with the ease of degradation of the respective hydrocarbons (Das and Chandran, 2011) and suggested that the plume was enriched with populations having the capacity for degradation of alkanes. Additional evidence for biodegradation of alkanes in the plume samples was presented in our previous study (Hazen et al., 2010) that reported oil half-lives in the plume of 1.2–6.1 days for C13–C26 n-alkanes. It should be noted that biodegradation of hydrocarbons in the plume was carried out without significant oxygen depletion (oxygen saturation averaged 59–67% inside and outside the plume, respectively) (Hazen et al., 2010).
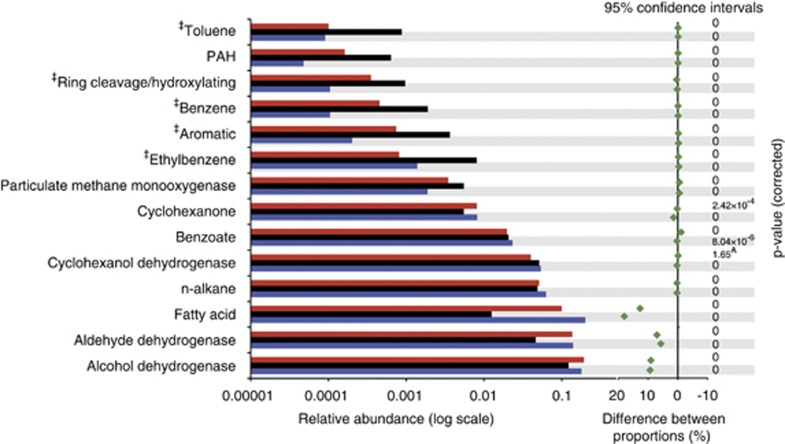
Figure 3.
Analysis of genes involved in hydrocarbon degradation in the metagenome data. Blue bars denote the distal station metagenome, black bars denote the uncontaminated sample metagenome and red bars denote the proximal station metagenome. Raw, unassembled metagenomic reads were compared with proteins involved in hydrocarbon degradation, using a custom database using the tblastn algorithm. A bit score cutoff of ⩾40 was used. Genes were grouped according to function. AIndicates that a corrected P-value was not significant. Gene categories denoted with an ‘‡' indicate a similar substrate degradation pathway in which the different substrates are degraded by the same enzyme (simple ring oxygenases). A complete list of all gene categories is provided in Supplementary Table S6.
To determine the active microbial community composition and expressed functions in the plume interval, we extracted high quality total RNA from the proximal and distal plume stations and sequenced the samples using the Illumina platform, resulting in a total of 140 million paired-end reads (15 Gb). To assign microbial identities, the unassembled metatranscriptome data (70 million single reads) was compared with a Greengenes (DeSantis et al., 2006) database using blastn. We found that Oceanospirillales was not only the most abundant member of the community but also was active with a relative abundance of transcripts of 46% in the proximal plume station sample and 69% in the distal plume station sample (Figure 2 and Supplementary Table S4). Other members of the community that were active included Alteromonadales (11% relative abundance proximal plume/9% relative abundance distal plume), Deltaproteobacteria (10%/1%), Pseudomonadales (6%/4%) and SAR86 (3%/1%) (Figure 2 and Supplementary Table S4). These community members were also relatively abundant in our metagenome data (Figure 2 and Supplementary Table S3). Therefore, the dominant members of the community that were enriched by the deep-sea plume were also active in the plume.
Previous analysis of samples from the deep-sea plume using DNA–based analyses reported other microbial clades that were more or less abundant at different sampling times. For example, members of the Colwelliaceae were detected as dominant community members in the deep-sea plume in samples collected in mid-June 2010 (Valentine et al., 2010). In addition, microcosm experiments with labeled ethane and propane were dominated by Colwellia, with some Oceanospirillales increasing in abundance (Redmond and Valentine, 2011). Thus, these authors suggested that Colwellia was primarily responsible for in situ ethane and propane oxidation, with perhaps, Oceanospirillales also having a role (Redmond and Valentine, 2011). However, cross-feeding could not be excluded (Redmond and Valentine, 2011). Although the Colwelliaceae were not abundant at <1% relative abundance in our samples collected in late May, we found that they were represented in the active microbial community in both of our plume samples (Figure 2 and Supplementary Table S4). However, other members of the community that were previously reported to be abundant (Valentine et al., 2010), such as Cycloclasticus, which has members that are able to degrade simple and PAH aromatics (Dyksterhouse et al., 1995), although present in the pyrotag data at low abundances (Supplementary Table S2), were not represented in our metagenome or metatranscriptome data (Supplementary Tables S3 and S4). In addition, the methylotrophs (Methylococcales and Methylophaga), although rare, at <1% relative abundance in the plume samples, were active (Supplementary Table S4). Further, the type II methanotrophs Methylosinus, Methylocystis and Methylocella were observed in both plume samples, although at very low levels (<0.01% relative abundance). The metatranscriptome data thus revealed for the first time that Oceanospirillales was the dominant active member of the microbial community in the deep-sea plume samples, which we collected in late May, in addition to some other members of the community, including some rare members.
We next determined what functions were expressed in the active microbial community enriched in the plume, with a focus on hydrocarbon degradation genes. A total of 70 million single, unassembled reads resulting from the metatranscriptome sequencing were compared with a hydrocarbon-degradation gene database. Differences in relative abundances of active degradation genes (RNA transcripts) in the plume samples were more pronounced compared with the DNA analyses. The metatranscriptome data largely supported our metagenome data; for example, finding that alkane monooxygenases were highly expressed, with the same pathways for alkane, and specifically for cyclohexane degradation present and abundant (Figure 4). This finding suggests that alkane degradation was the dominant hydrocarbon degradation pathway expressed in the plume at the time interval we sampled. Genes coding for degradation of simple and PAH aromatics were either expressed at low levels or not at all (Figure 4). Reddy et al. (2012) determined the composition of oil and gas that was emitted from the Macondo well and reported that BTEX compounds were the most abundant hydrocarbons larger than C1–C5 in the plume. However, our findings indicate that of the BTEX compounds, only those genes coding for ethylbenzene degradation were expressed, and only in the proximal plume sample (Figure 4). This finding suggests that the more recalcitrant compounds were not being actively degraded at the time when we sampled. Although the samples analyzed by Reddy et al. (2012) were collected at later time points than ours (mid to late June), their findings of negligible biodegradation of BTEX compounds over 4 days in the deep-sea plume are consistent with our findings.
Figure 4.
Analysis of genes involved in hydrocarbon degradation in the metatranscriptome data. Blue bars denote the distal station metatranscriptome and red bars denote the proximal station metatranscriptome. Raw, unassembled metatranscriptome reads were compared with proteins involved in hydrocarbon degradation, using a custom database using the tblastn algorithm. A bit score cutoff of ⩾40 was used. Genes were grouped according to function. An asterisk indicates that the difference in relative abundance of a particular gene group in the proximal station metatranscriptome compared with the distal station metatranscriptome was statistically significant. Gene categories denoted with an ‘‡' indicate a similar substrate degradation pathway in which the different substrates are degraded by the same enzyme (simple ring oxygenases). Within this category, ring cleavage/hydroxylating enzymes were observed at very low abundance and only in the proximal plume station. Simple ring oxygenases that are involved in the degradation of benzene, toluene and PAHs were not observed in the metatranscriptome data. A complete list of all gene categories is provided in Supplementary Table S6.
Our study also revealed that a diversity of particulate methane monooxygenase (Pmo) genes, but no detectable soluble methane monooxygenases, were expressed in the plume and at higher levels with distance from the wellhead and over time (that is, 1.5–3 days to reach the distal station). Although pmo genes were expressed in the oil plume, their relative levels were still less than those for genes coding for alkane degradation (Figure 4). These results were surprising, given that methane was the most abundant hydrocarbon released during the spill (Kessler et al., 2011) with concentrations ranging 20–50-fold higher than background levels (Valentine et al., 2010 and references therein). Our data, as well as those of Valentine et al. (2010) and Kessler et al. (2011), suggested a lag time in the response of methanotrophs to the plume, relative to the initial bloom of Oceanospirillales capable of oxidation of alkanes. However, our findings suggest that methane oxidation was actively occurring in the plume samples presented here, which is earlier in the spill history than has previously been suggested (Valentine et al., 2010; Kessler et al., 2011).
Because of the dominance of members of the Oceanospirillales in the plume samples and the recalcitrant nature of members of this order to cultivation, we specifically targeted this group for single-cell genome sequencing. We sorted water collected from the proximal plume station by fluorescence-activated cell sorting. The single cells were lysed and genomic DNA was amplified using MDA. Subsequently, the single cells were screened on the basis of their 16S rRNA gene sequences for those with high sequence quality and that were >95% similar to the dominant Oceanospirillales OTU. After sequencing on the Illumina platform, two of these cells yielded high-quality sequences, which were concatenated and assembled, resulting in a single-draft genome. The single cells were most closely related (partial 16S rRNA gene) to an uncultured Oceanospirillales (99% similar) from the oil spill (Redmond and Valentine, 2011). Closest cultured representatives were Oleispira antarctica (Yakimov et al., 2003) (97% similar) and Thalassolituus oleivorans (97% similar), both of which degrade aliphatic hydrocarbons (C10–C18 and C7–C20, respectively) (Yakimov et al., 2003; Yakimov et al., 2004). However, genome sequences are not available for either of these isolates. There are 10 Oceanospirillales genome sequences available in IMG (Markowitz et al., 2008), the most well characterized being Alcanivorax borkumensis (Schneiker et al., 2006). As a rough estimate, the assembled single-cell Oceanospirillales draft genome (1.9 Mb genome with 876 contigs, N50 of 5030 bp and longest contig 25 481 bp) represented more than half a complete genome based on comparisons to the 3.1-Mb genome of A. borkumensis. A. borkumensis is typically found at low abundance in unpolluted marine environments (Schneiker et al., 2006), but can represent as much as 90% of petroleum-degrading microbial communities (Harayama et al., 1999). The 16S rRNA gene sequences for our single cells were <88% similar to A. borkumensis, and thus represent a different genus within the Oceanospirillales. Additionally, by comparison of the annotated COGs from the draft genome assembly with those within the Gammaproteobacteria, the draft genome was 53% complete at the phylum level and 52% complete at the sub-phylum level. We also examined all of the raw, unassembled reads for each single-cell genome to ensure that all of the sequence data were analyzed.
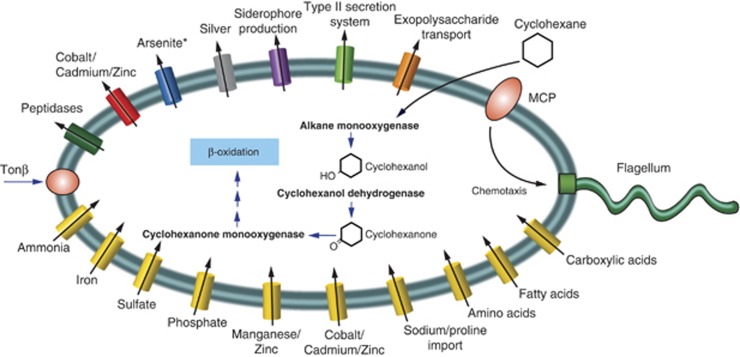
Within the draft genome, we used CAMERA (Seshadri et al., 2007) to obtain gene annotations in the assembled contigs. The annotations included putative genes encoding methyl-accepting chemotaxis proteins, flagella, pili and signal transduction mechanisms, all of which were present in the metagenomes and expressed in the plume interval (Figure 5, Supplementary Figures S4 and S5). Physical evidence of microbial cell attraction to oil in the proximal plume sample was also provided by synchrotron radiation-based Fourier-transform infrared spectromicroscopy that revealed sharp absorptions at ∼1640 and ∼1548 cm−1 in the fingerprint region (between 1800 and 900 cm−1) that are interpreted as Macondo oil droplets surrounded by microorganisms (Supplementary Figure S6). Together, the physical and molecular evidence suggest that bacterial cells were actively attracted to and interacted with oil in the hydrocarbon plume.
Figure 5.
Oceanospirillales single-cell metabolic reconstruction using COG annotations of assembled sequence data and the blast comparison of unassembled single-cell reads to genes involved in hydrocarbon degradation. All genes in the single-cell metabolic reconstruction were present in the metagenomes and most were expressed in the metatranscriptome, except for those with an asterisk following the gene name.
Several key functions were recently identified as important for several low-abundance marine surface bacteria to rapidly respond and bloom when conditions become more energy-rich (Yooseph et al., 2010). These included the capacity for chemotaxis and motility, which we found in the draft genome, the metagenomes and metatranscriptome. Clustered regularly interspaced short palindromic repeat regions to protect from phage predation (Yooseph et al., 2010) were also identified in the Oceanospirillales draft genome, suggesting a mechanism for avoiding phage predation.
Closer investigation of the draft genome revealed genes for uptake of a suite of nutrients (Figure 5), all of which were also found in the metagenomes and expressed in the plume metatranscriptome. For example, COGs involved in uptake of nitrogen (ammonia permease), phosphate (ABC-type phosphate/phosphonate transport system, permease component), iron (ABC-type Fe3+ siderophore transport system, permease component, siderophore interacting protein and Fe2+ transport system proteins), sulfur (sulfate permease and related transporters), Cobalt, Cadmium and Zinc (transporters) were detected in all three data sets (see Supplementary Table S5).
We also analyzed the unassembled Oceanospirillales single-cell reads for genes involved in hydrocarbon degradation and searched for genes with closest similarities to previously characterized genes based on bit scores ⩾40. Consistent with what we observed in the metagenomic and metatranscriptomic data, the Oceanospirillales draft genome had genes with closest similarities to those coding for the cyclohexane degradation pathway (Figure 5). This aliphatic degradation pathway is similar to what was proposed for A. borkumensis (Schneiker et al., 2006). We did not find evidence in the draft genome for ethane or propane oxidation, which Redmond and Valentine (2011) suggested as a potential metabolic role for the Oceanospirillales observed in their SIP experiments.
Conclusion
In this study, we determined that the dominant and active, yet uncultured, Oceanospirillales possessed genes that encode the nearly complete pathway for cyclohexane degradation. This pathway was present in the single cells, the metagenomes and expressed in the plume metatranscriptomes. The capacity of the Oceanospirillales representatives for chemotaxis, motility, and for degradation of alkanes, may have enabled these cells to actively aggregate and increase in numbers in the plume and to scavenge nutrients using a suite of transporters and siderophores. In addition, by using a shotgun metatranscriptome approach, for the first time, we were able to determine which hydrocarbon degradation pathways and other functions were actively expressed in the deep-sea at the time we sampled, to ascribe these pathways to particular groups of microorganisms and to elucidate how these active processes shifted in response to the hydrocarbon plume. Given that the Gulf of Mexico experiences frequent, natural oil spills, elucidating the role of Oceanospirillales in oil disposition provides critical data in understanding how members of the deep-sea microbial community can rapidly respond and become enriched in the presence of hydrocarbons.
Acknowledgments
This work was supported by a subcontract from the University of California at Berkeley, Energy Biosciences Institute to Lawrence Berkeley National Laboratory under its US Department of Energy contract DE-AC02-05CH11231. The SR-FTIR work was conducted at the infrared beamline at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy. We are extremely grateful to Michael D Curtin for his assistance with Paracel BLAST. We are thankful for the help of Theresa Pollard with handling shipping, ordering and transportation of supplies and people to and from the field. We also thank the captain, crew and science teams aboard the R/V Ocean Veritas and the R/V Brooks McCall. Lastly, we thank Damon Tighe and Janey Lee at Joint Genome Institute for single-cell sorting and amplification and Chien-Chi and Matthew Scholz for metagenome assemblies.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allgaier M, Reddy A, Park JI, Ivanova N, D'Haeseleer P, Lowry S, et al. Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS One. 2010;5:e8812. doi: 10.1371/journal.pone.0008812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Zhang T, Woyke T, Speller CF, Krivoshapkin A, Yang DY, et al. Identification of ancient remains through genomic sequencing. Genome Res. 2008;18:1347–1353. doi: 10.1101/gr.076091.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli R, Reddy CM, Yoerger DR, Van Mooy BA, Jakuba MV, Kinsey JC, et al. Tracking hydrocarbon plume transport and biodegradation at Deepwater Horizon. Science. 2010;330:201–204. doi: 10.1126/science.1195223. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010a;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010b;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Non-parametric estimation of the number of classes in a population. Scan J Stat. 1984;11:265–270. [Google Scholar]
- Chao A, Lee SM, Jeng SL. Estimating population size for capture-recapture data when capture probabilities vary by time and individual animal. Biometrics. 1992;48:201–216. [PubMed] [Google Scholar]
- Chao A, Hwang WH, Chen YC, Kuo CY. Estimating the number of shared species in two communities. Stat Sinica. 2000;10:227–246. [Google Scholar]
- Colwell RK, Coddington JA. Estimating terrestrial biodiversity through extrapolation. Philos T Roy Soc B. 1994;345:101–118. doi: 10.1098/rstb.1994.0091. [DOI] [PubMed] [Google Scholar]
- Das N, Chandran P. Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnol Res Int. 2011;2011:1–13. doi: 10.4061/2011/941810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis KM, Silver WL, Thompson AW, Firestone MK. Microbial communities acclimate to recurring changes in soil redox potential status. Environ Microbiol. 2010;12:3137–3149. doi: 10.1111/j.1462-2920.2010.02286.x. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a Chimera-Checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyksterhouse S, Gray J, Herwig R, Lara J, Staley J. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int J Syst Bacteriol. 1995;45:116–123. doi: 10.1099/00207713-45-1-116. [DOI] [PubMed] [Google Scholar]
- Federal Interagency Solutions Group 2010. Oil Budget Calculator Science and Engineering Team. Oil Budget Calculator Technical Documentation.
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S, Kishira H, Kasai Y, Shutsubo K. Petroleum biodegradation in marine environments. J Mol Microbiol Biotechnol. 1999;1:63–70. [PubMed] [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL, et al. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010;4:1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- Kessler JD, Valentine DL, Redmond MC, Du M, Chan EW, Mendes SD, et al. A persistent oxygen anomaly reveals the fate of spilled methane in the deep gulf of Mexico. Science. 2011;331:312–315. doi: 10.1126/science.1199697. [DOI] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackenbrandt E, Goodfellow M (eds)Nucleic acid techniques in bacterial systematics John Wiley: New York; 115–148. [Google Scholar]
- Letunic I, Yamada T, Kanehisa M, Bork P. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem Sci. 2008;33:101–103. doi: 10.1016/j.tibs.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Lu Z, Deng Y, Van Nostrand JD, He Z, Voordeckers J, Zhou A, et al. Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J. 2011;6:451–460. doi: 10.1038/ismej.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Ivanova NN, Szeto E, Palaniappan K, Chu K, Dalevi D, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DN, Bryant JE, Madsen EL, Ghiorse WC. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol. 1999;65:4715–4724. doi: 10.1128/aem.65.11.4715-4724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- Reddy CM, Arey JS, Seewald JS, Sylva SP, Lemkau KL, Nelson RK, et al. 2012Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill Proc Natl Acad Sci USAe-pub ahead of print 3 February 2012;doi: 10.1073/pnas.1101242108 [DOI] [PMC free article] [PubMed]
- Redmond MC, Valentine DL.2011Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill Proc Natl Acad Sci USAe-pub ahead of print 3 October 2011;doi: 10.1073/pnas.1108756108 [DOI] [PMC free article] [PubMed]
- Rodrigue S, Malmstrom RR, Berlin AM, Birren BW, Henn MR, Chisholm SW. Whole genome amplification and de novo assembly of single bacterial cells. PLoS One. 2009;4:e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirova JS, Ferrer M, Regenhardt D, Timmis KN, Golyshin PN. Proteomic insights into metabolic adaptations in Alcanivorax borkumensis induced by alkane utilization. J Bacteriol. 2006;188:3763–3773. doi: 10.1128/JB.00072-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiker S, dos Santos VAPM, Bartels D, Bekel T, Brecht M, Buhrmester J, et al. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat Biotech. 2006;24:997–1004. doi: 10.1038/nbt1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M. CAMERA: A Community Resource for Metagenomics. PLoS Biol. 2007;5:e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson EH. Measurement of Diversity. Nature. 1949;163:688–688. [Google Scholar]
- Valentine DL, Kessler JD, Redmond MC, Mendes SD, Heintz MB, Farwell C, et al. Propane respiration jump-starts microbial response to a deep oil spill. Science. 2010;330:208–211. doi: 10.1126/science.1196830. [DOI] [PubMed] [Google Scholar]
- Yakimov MM, Giuliano L, Gentile G, Crisafi E, Chernikova TN, Abraham W-R, et al. Oleispira antarctica gen. nov., sp. nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int J Syst Evol Microbiol. 2003;53:779–785. doi: 10.1099/ijs.0.02366-0. [DOI] [PubMed] [Google Scholar]
- Yakimov MM, Giuliano L, Denaro R, Crisafi E, Chernikova TN, Abraham WR, et al. Thalassolituus oleivorans gen. nov., sp. nov., a novel marine bacterium that obligately utilizes hydrocarbons. Int J Syst Evol Microbiol. 2004;54:141–148. doi: 10.1099/ijs.0.02424-0. [DOI] [PubMed] [Google Scholar]
- Yooseph S, Nealson KH, Rusch DB, McCrow JP, Dupont CL, Kim M, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468:60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- Zerbino D, Birney E. Velvet: algorithms for de novo short read assembly using De Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.