Abstract
Pyrosequencing analysis of 16S rRNA genes was used to study temporal dynamics of groundwater bacteria and archaea over 10 months within three well clusters separated by ∼30 m and located 250 m from the Columbia River on the Hanford Site, WA. Each cluster contained three wells screened at different depths ranging from 10 to 17 m that differed in hydraulic conductivities. Representative samples were selected for analyses of prokaryotic 16S and eukaryotic 18S rRNA gene copy numbers. Temporal changes in community composition occurred in all nine wells over the 10-month sampling period. However, there were particularly strong effects near the top of the water table when the seasonal rise in the Columbia River caused river water intrusion at the top of the aquifer. The occurrence and disappearance of some microbial assemblages (such as Actinobacteria ACK-M1) were correlated with river water intrusion. This seasonal impact on microbial community structure was greater in the shallow saturated zone than deeper zone in the aquifer. Spatial and temporal patterns for several 16S rRNA gene operational taxonomic units associated with particular physiological functions (for example, methane oxidizers and metal reducers) suggests dynamic changes in fluxes of electron donors and acceptors over an annual cycle. In addition, temporal dynamics in eukaryotic 18S rRNA gene copies and the dominance of protozoa in 18S clone libraries suggest that bacterial community dynamics could be affected not only by the physical and chemical environment but also by top-down biological control.
Keywords: community structure, Hanford Site, unconfined aquifer, spatiotemporal dynamics, hyporheic zone
Introduction
Shallow aquifers comprise a critical worldwide water resource for domestic, industrial and agricultural use. These aquifers are now known to contain metabolically diverse and active populations (Balkwill and Ghiorse, 1985). These microbes perform critical functions in biogeochemical cycles and attenuation of contaminants (Yagi et al., 2010) and are linked to the emission of greenhouse gases (Minamikawa et al., 2010; Elberling et al., 2011). Yet, because it is difficult to comprehensively sample subsurface environments, the diversity and dynamics of microbes in groundwater ecosystems have not been extensively explored (Griebler and Lueders, 2009). This environment is perceived to be relatively stable, characterized by long residence time (Brunke and Gonser, 1997; Griebler and Lueders, 2009) and subject to less frequent perturbations than surface systems (Odum et al., 1995). Factors controlling microbial spatial and temporal dynamics are poorly documented for most groundwater ecosystems (Storey et al., 1999; Simon et al., 2001; Griebler and Lueders, 2009; Lowell et al., 2009; Boulton et al., 2010).
At the Hanford Site 300 Area, the unconfined subsurface aquifer is subject to intrusion of organic and inorganic contaminants from a variety of sources associated with historical nuclear materials production and large-scale separations (Riley and Zachara, 1992). This region is environmentally sensitive due to its proximity to the Columbia River. The unconfined aquifer is an oligotrophic environment with low fluxes of external nutrient inputs; however, there are physical, chemical and hydrological complexities that could lead to increased spatial and temporal dynamics in the subsurface microbial community (Gilmore et al., 1993; Peterson and Connelly, 2004; Lin et al., 2012a). The sedimentary stratigraphy includes multiple transitions in sediment texture and redox conditions that result in a stratified distribution of microbial communities (Lin et al., 2012a). This aquifer is contained largely within Hanford formation sediments deposited under high-energy conditions during catastrophic Ice Age floods, resulting in lateral textural heterogeneity. The hydraulic gradient is affected by the stage height of the Columbia River, which can vary by as much as 3 m driven by the seasonal hydrological cycle and the operation of hydroelectric dams (Peterson and Connelly, 2004; Arntzen et al., 2006).
For marine and freshwater habitats, temporal variability has been used to provide insight into environmental factors that drive shifts in microbial community composition and select for functional traits (Ghiglione et al., 2005; Fuhrman et al., 2006; Kan et al., 2006; Sundberg et al., 2007; Anderson-Glenna et al., 2008; Lin et al., 2008; Nelson et al., 2008; Ayuso et al., 2009; Andersson et al., 2010; Gilbert et al., 2012). Comparable work in groundwater is rare (Simon et al., 2001; Anderson et al., 2003; Ayuso et al., 2009; Yagi et al., 2010). Therefore, we undertook this study of an oligotrophic subsurface habitat, where the effects of physical and hydrological heterogeneities across space and time could have a larger role in driving community composition dynamics.
The approach was to measure changes in groundwater microbial abundances and community composition over time and in distinct subsurface strata, to analyze system stability, where physical and hydrological heterogeneities could impact the oligotrophic system. Pyrosequencing analysis of both bacterial and archaeal 16S rRNA genes, as well as Sanger sequencing of 18S rRNA genes, was employed to give taxonomic information on the community components over a 10-month sampling period. Temporal shifts in microbial community composition were related to changes in groundwater geochemistry and water table elevation, revealing that subsurface microbial communities were considerably more dynamic over short intervals of time and space than anticipated.
Materials and methods
Sampling and chemical characterization
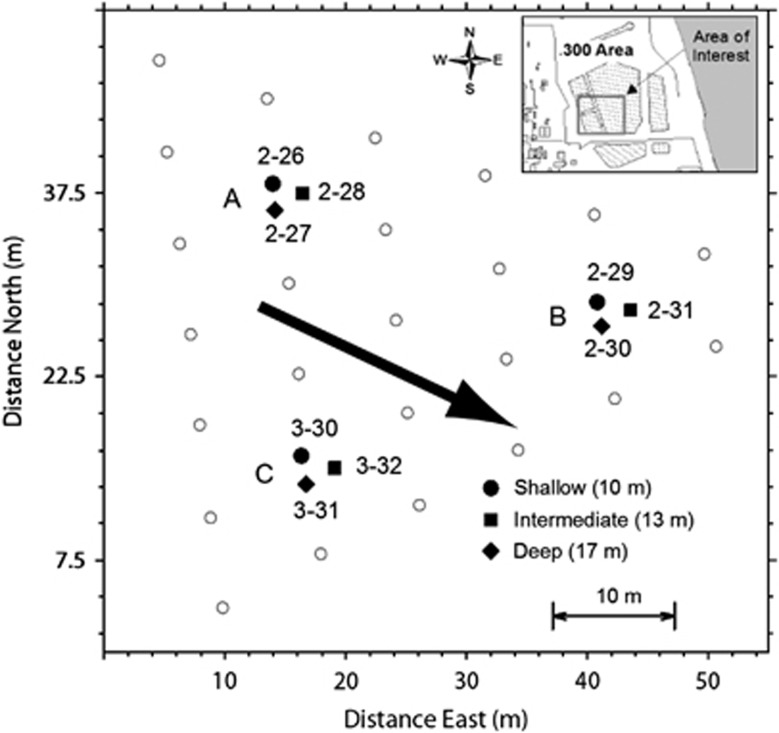
Samples were collected from three-well clusters (Cluster A: 2–26/2–27/2–28; B: 2–29/2–30/2–31; C: 3–30/3–31/3–32) in the Hanford Integrated Field Research Challenge (IFRC) site (46°22′15.80″N, 119°16′31.52″W), near the southern boundary of the Hanford Site in eastern Washington State. The wells were drilled 4″ in diameter. Each three-well cluster was completed at short (1 m) intervals within the Hanford formation: near the water table (10 m), at the middle of the Hanford formation (13 m) and near the contact with the underlying Ringold formation (17 m). The wells in each cluster were proximate (Figure 1), but the screened intervals did not overlap in depth, and each well annulus was filled with bentonite crumbles to prevent cross-stratum water sampling. A sampling pump was installed in the center of the screened interval. Sampling occurred from November 2009 until September 2010. This included periods in fall and winter when the river stage was relatively stable, late spring when the stage rose due to snowmelt from the river catchment and through the summer and into the early fall where river staged eclined. Approximately 380 samples were collected over 10 months. The sampling interval varied over the year: 10 samples within 15 days between 16 November to 1 December 2009; weekly sampling from 26 January to 10 March 2010; twice weekly sampling during the period of largest river stage changes (14 May to 20 July 2010); and biweekly sampling from 20 July to 24 September 2010. For samples collected from 1 May to 26 July, aliquots were analyzed for major anions and cations, inorganic carbon and uranium (U) concentrations. U concentrations were determined using a kinetics phosphorescence analyzer (KPA-11, Chemchek Instruments, Richland, WA, USA); anion concentrations were determined via anion chromatography (Dionex ICS2000, Thermo Scientific Co., Sunnyvale, CA, USA). Samples were filtered through a 0.2-μm pore Teflon filter and directly analyzed. During sampling, the water table elevation was recorded using an electrical depth tape. River water intrusion was estimated from nitrate concentrations according to a simple mixing calculation, based upon the relative nitrate concentrations of 0.4 and 25 mg nitrate l−1 in river and ground water. This calculation indicated that 16th June was the first day of detectable groundwater intrusion into the shallow wells.
Figure 1.
Three-well clusters for groundwater sampling. Each cluster (A, B or C) is comprised of three wells screened at shallow (∼10 m), intermediate (∼13 m) or deep (∼17 m) interval. The sites are approximately 250 m from the Columbia River shoreline. Although the local hydrological gradient varies due to river stage fluctuations, particularly during late spring, the regional gradient is from higher elevations to the west, and runs across the site at an approximate west to east bearing (black arrow).
Groundwater was pumped into a 4-l sterile polyethylene container from each well using the peristaltic pump and tubing installed for groundwater sampling after purging the tubing for 5 min at a pumped rate of 2 l min−1 (equivalent to five volumes of the PVC screening area). Cells were filtered onto a 0.2-μm pore-size 47 mm diameter polyethersulfone filter (Supor200, Pall Corporation, Cortland, NY, USA) that was stored at −80 °C. Genomic DNA from half of each membrane filter was extracted using a PowerSoil-htp 96-well DNA isolation kit according to the manufacturer's protocol (MoBio Laboratories, Carlsbad, CA, USA).
Quantitative real-time PCR
To estimate seasonal variations in abundance, quantitative real-time PCR assays were performed on a DNA extract from one sample per month in each Cluster A well. Bacterial 16S rRNA gene copy number was quantified according to Lin et al. (2012a). Eukaryotic 18S rRNA gene was measured by TaqMan Ribosomal RNA Control Reagents according to the manufacture's protocol (Applied Biosystems Inc., Carlsbad, CA, USA).
Bar-coded pyrosequencing
PCR amplification and amplicon preparation were similar to that described by Lauber et al. (2009) using primers targeting the V4-V5 regions of 16S small subunit ribosomal RNA gene of bacteria and archaea (Bates et al., 2011; Bergmann et al., 2011). Each sample was amplified in triplicate and included 1 μl genomic DNA, 30 μℳ forward and reverse primer in 1 × 5 PRIME Hot Master Mix using annealing temperature of 50 °C for 35 cycles (Fierer et al., 2008; Costello et al., 2009; Lauber et al., 2009) followed by PCR reaction clean up using the MoBio Ultra Clean PCR clean up kit (Carlsbad, CA, USA). Based on PicoGreen quantitation, equal amounts of PCR product for each sample were combined and sent to the Environmental Genomics Core Facility (Columbia, SC, USA) to be run on a Roche FLX 454 pyrosequencing machine (Branford, CT, USA).
Constructing 18S ribosomal RNA clone libraries
Six representative DNA clone libraries were constructed, including three winter samples (from shallow wells 2–26 and 2–29, and one deep well 2–27 on 24 February 2010) and three late spring samples (from the same wells on 21 June 2010). PCR amplification with the primer pair Euk360F/1492R (Medlin et al., 1988) was performed using Phusion High-Fidelity DNA polymerase (New England Biolabs Inc., Woburn, MA, USA) with an adenine (A) added using Taq polymerase (Qiagen, Santa Clarita, CA, USA) on a PTC-225 Peltier thermal cycler (MJ Research, Watertown, MA, USA). PCR products were purified using the Qiagen MinElute kit (Qiagen) and ligated into the pCRII-Topo Vector (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocols. After transformation, 48 randomly picked colonies were used for preparing glycerol stocks and sent to the Functional Biosciences Inc. (Madison, WI, USA) for plasmid isolation and sequencing. Sequencing was conducted with both M13f and M13r primers. Assembled forward and reverse reads were trimmed for vectors and checked for chimeras using Mallard (Ashelford et al., 2006). Eukaryotic operational taxonomic units (OTU) were defined at the level of 98% identity and picked in QIIME by the uclust clustering method (www.qiime.sourceforge.net; Caporaso et al., 2010). Representative 18S gene sequences from each OTU (GenBank JN705488-JN705542) were aligned to SILVA (Pruesse et al., 2007) databases (release 108) in the ARB package (Ludwig et al., 2004). Aligned fasta files were used for tree construction in MEGA4 using a neighbor-joining algorithm and a Kimura 2-parameter distance calculation (Tamura et al., 2007).
Processing of pyrosequencing data
Raw data was filtered using QIIME to exclude reads less than 200 nt; reads with low quality scores (<25); and those missing the 16S primer or with uncorrectable barcodes (Costello et al., 2009). The 12-bp barcode was used to assign reads to samples. The reads were then binned into OTUs at 97% sequence similarity using uclust followed by selection of a representative sequence. Chimeric sequences were identified using ChimeraSlayer (Haas et al., 2011) and removed. Taxonomy was assigned to each representative sequence using BLAST with a maximum e-value of 0.001 against the Greengenes core set and the Hugenholtz taxonomic nomenclature (DeSantis et al., 2006). For all OTU-based analyses, the original OTU table was rarefied to a depth of 500 sequences per sample, to minimize effects of sampling effort upon analyses.
Statistical analyses
Pairwise comparisons were computed as Bray–Curtis distances and were visualized using non-metric multidimensional scaling (NMS) in PRIMER v6 (www.primer-e.com) based on 500 iterations and maximally three dimensions. Temporal trajectories were connected based on sampling date to illustrate temporal shifts. Chao1 richness was calculated in QIIME. LOESS smoothing of Chao1 richness over time was performed in PAST (http://folk.uio.no/ohammer/past/) with default settings.
Community differences between groups of samples were examined using two-way nested ANOSIM in PRIMER v6 based on two levels of clustering: across wells or depths and time clusters nested within wells or depths. This permutation-based statistical analysis tested a null hypothesis that there are no differences among wells or among depths throughout the whole sampling duration. The analysis was based on the Bray–Curtis similarities between samples and produced a test statistic R, which could range from −1 to 1 and a significance level (P). A near-zero R implied no differences between samples.
Supervised classification using Random Forest was run in QIIME to identify OTUs important for classifying community composition based on defined time clusters (Knights et al., 2011). A list of discriminative OTUs was operationally defined as importance scores>1, where importance scores indicate the OTU's value in correct classification. This produced a list of 45 OTUs; their temporal shifts were hierarchically clustered based on Pearson correlation and visualized by a heatmap (http://www.tm4.org/mev/).
SIMPER analysis was used to identify OTUs contributing to differences in community composition before (14 May–15 June) and after (16 June–6 July) river water intrusion into the three shallowest wells. The average Bray–Curtis dissimilarity (AvDiss) between all pairs of samples and the contribution (Contrib%) of each OTU to total dissimilarity between communities before and after intrusion were calculated. OTUs with higher values of the AvDiss and Contrib% were useful for sample discrimination. For both Random Forest and SIMPER analyses, we used a genus-level OTU table (level six within Hugenholtz framework) and summed sequence counts from each genus-level assignment. In cases involving candidate divisions, sequences were summed to a defined clade of a higher taxonomic level. This data treatment was chosen based on the postulate that phylogenetically related bacteria had similar ecological roles (Andersson et al., 2010), although in practice there may be limits to this relationship.
An OTU-97 table with the 500 most abundant OTUs was used to analyze species-time relationships, and degree of temporal turnover (White et al., 2006b) using a completely nested method for the presence–absence (Sorensen) similarity matrix. Degree of temporal turnover was indicated by w in the species-time relationship power functions (S=/cTw). In this equation, c approximates the observed species richness (S) at specific time-scale (T) and w is the slope of the linearized species-time relationship on log-transformed axes (White et al., 2006b).
Linear correlation (R2) between bacterial and eukaryotic gene copy numbers was calculated by using SigmaPlot v11.0 (Systat Software Inc., Chicago, IL, USA).
Results
Seasonal variations in the water table and groundwater biogeochemistry
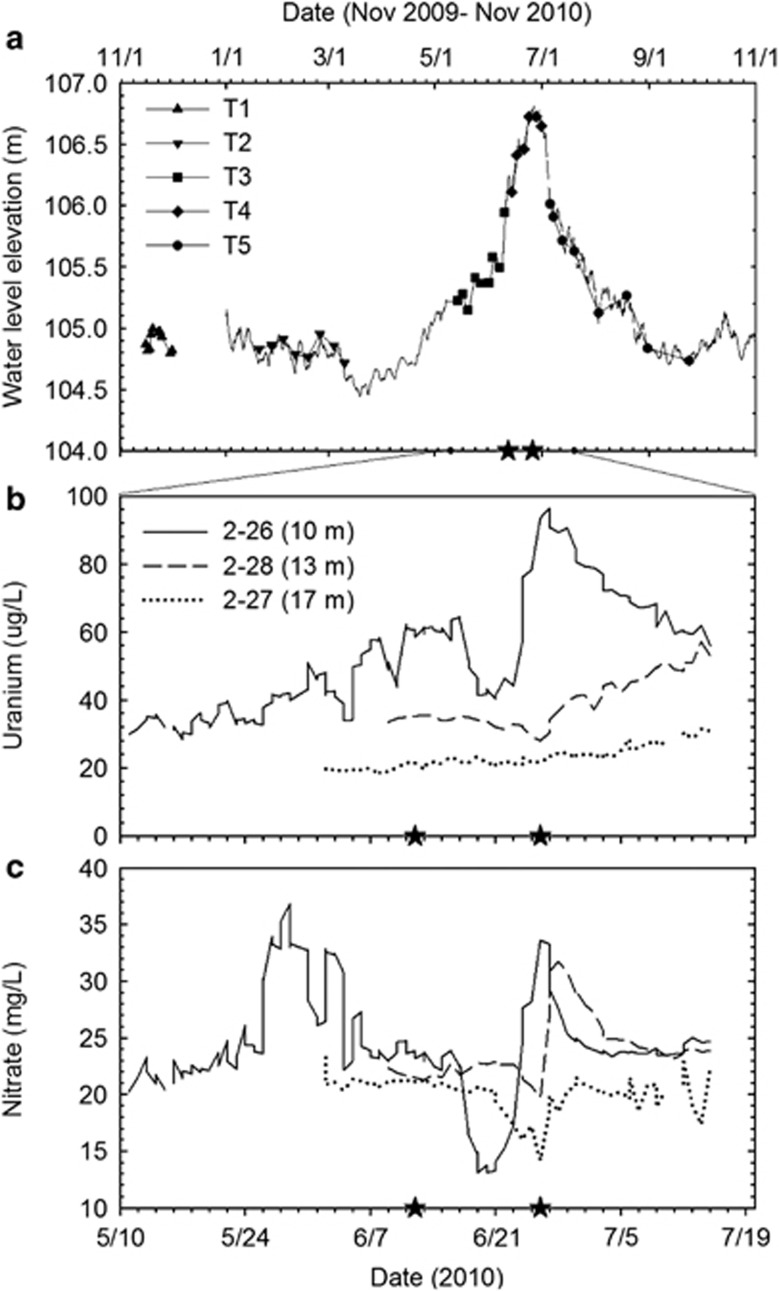
Water table elevation was relatively constant from November 2009 to April 2010 and from September to November 2010 (Figure 2a and Supplementary Figure S1). It rose from 105.5 m in May to a maximum of 106.8 m on 26 June, and then declined to105 m in September. Based on these changes, samples were grouped into five time clusters: T1 (16 November–1 December), T2 (26 January–10 March), T3 (14 May–14 June), T4 (15 June–8 July) and T5 (9 July–24 September). The breakpoint between T3 and T4 represents the detectable intrusion of Columbia River water into the IFRC well field (Figure 2a).
Figure 2.
Temporal variations in (a) water table elevation, estimated from river gauge data, (b) uranium or (c) nitrate concentrations in groundwater at 10, 13 or 17 m. Sampling time points were grouped into five time clusters: T1 (triangle), T2 (inverted triangle), T3 (square), T4 (diamond) and T5 (circle). T3 and T4 represent samples taken before and after the Columbia River water intrusion, respectively. Note that the time ranges presented in panels (b) and (c) are only a subset of that in (a). The stars on each x-axis indicates the date of detectable river water intrusion (15 June) and the peak of river water intrusion (26 June).
Analyses of groundwater uranium and nitrate were used to document how water table elevation affected the aquifer. The unsaturated sediments retain small amounts of desorbable U (J McKinley, unpublished observations); the water table rise produced a gradual increase in U concentrations from 30 μg l−1 in May to 64 μg l−1 on 17 June (Figure 2b). Groundwater nitrate concentrations were 24 mg l−1; decreases indicated the intrusion of low-nitrate river water. When the water table rose 0.4 m on 15 June, both nitrate and U concentrations first declined owing to dilution, and then increased abruptly owing to the desorption of nitrate and soluble U from the vadose zone over the following 10 days (Figures 2b and ). These hydrological effects were greatest in the shallow stratum (10 m), as wells screened at 13 and 17 m showed little variation in U concentration and a delayed change in nitrate concentration (Figures 2b and ).
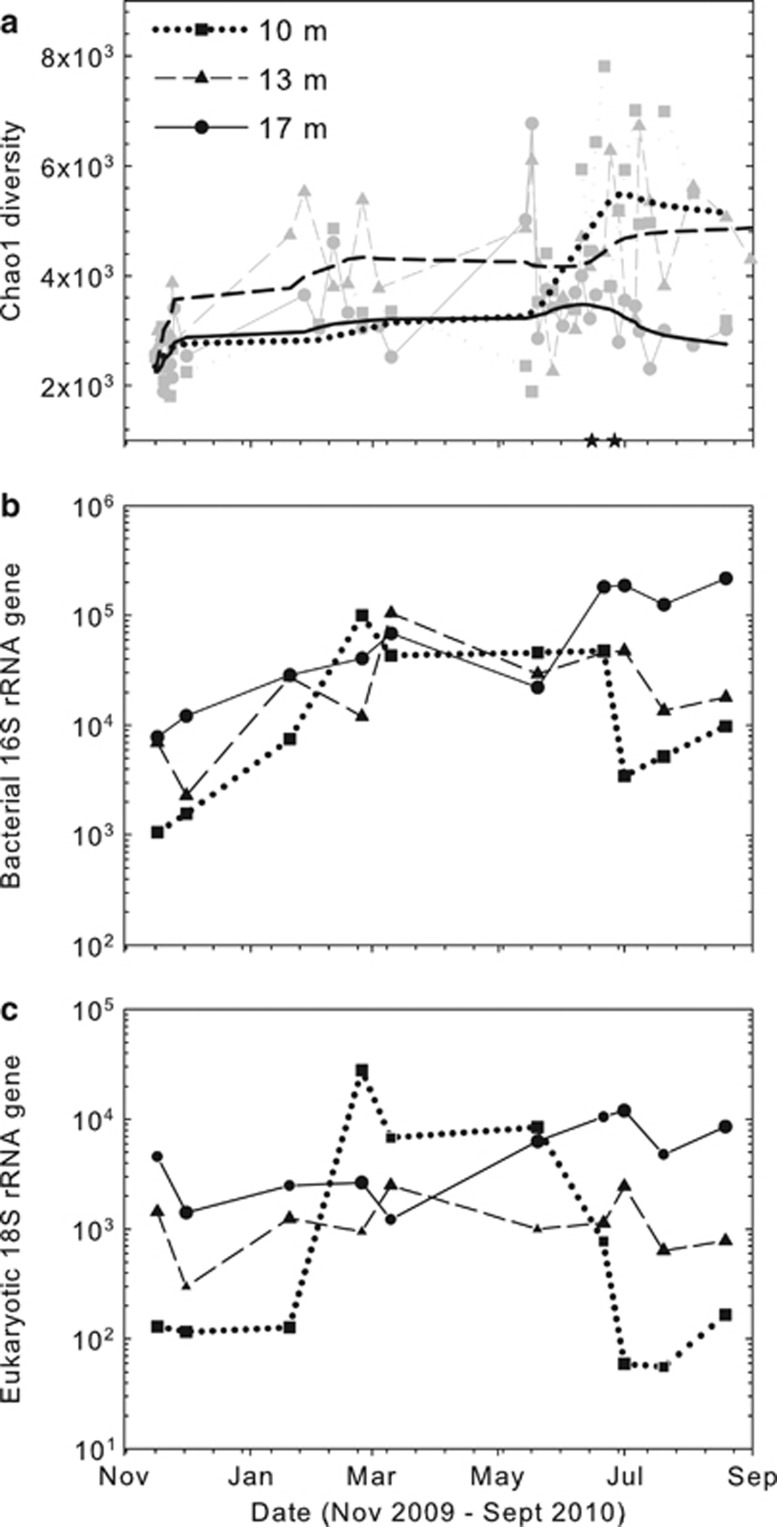
Bacteria plus Archaea OTU richness after river water intrusion (16 June) was twofold higher than during T1 and T2 at 10 m, but increases were much smaller at 13 and 17 m (Figure 3a). Bacterial 16S rRNA gene copies at all depths increased 10-fold from early T1 to late T2, and remained relatively constant during T3 and T4. After the water table subsided, there was a 10-fold decrease at 10 and 13 m by T5 (with a smaller decline in the deepest well) (Figure 3b). Similar temporal patterns were also observed for Eukarya, although the dynamic range in gene copy numbers was greater (100-fold) in the shallow well. Similar to bacteria at 17 m, the gene copy numbers for Eukarya were highest during T5. Positive correlations between bacterial and eukaryotic abundances were stronger at 10 m (R2= 0.83) than at 13 or 17 m (R2=0.64 and 0.63).
Figure 3.
Chao1 diversity (a), copy numbers of bacterial 16S rRNA gene (b) and eukaryotic 18S rRNA gene (c) at three depths of the well Cluster A. (a) Black lines were computed by LOWESS smoothing of the changes in Chao1 estimator (data points in grey).
Community temporal trajectories and degree of species turnover
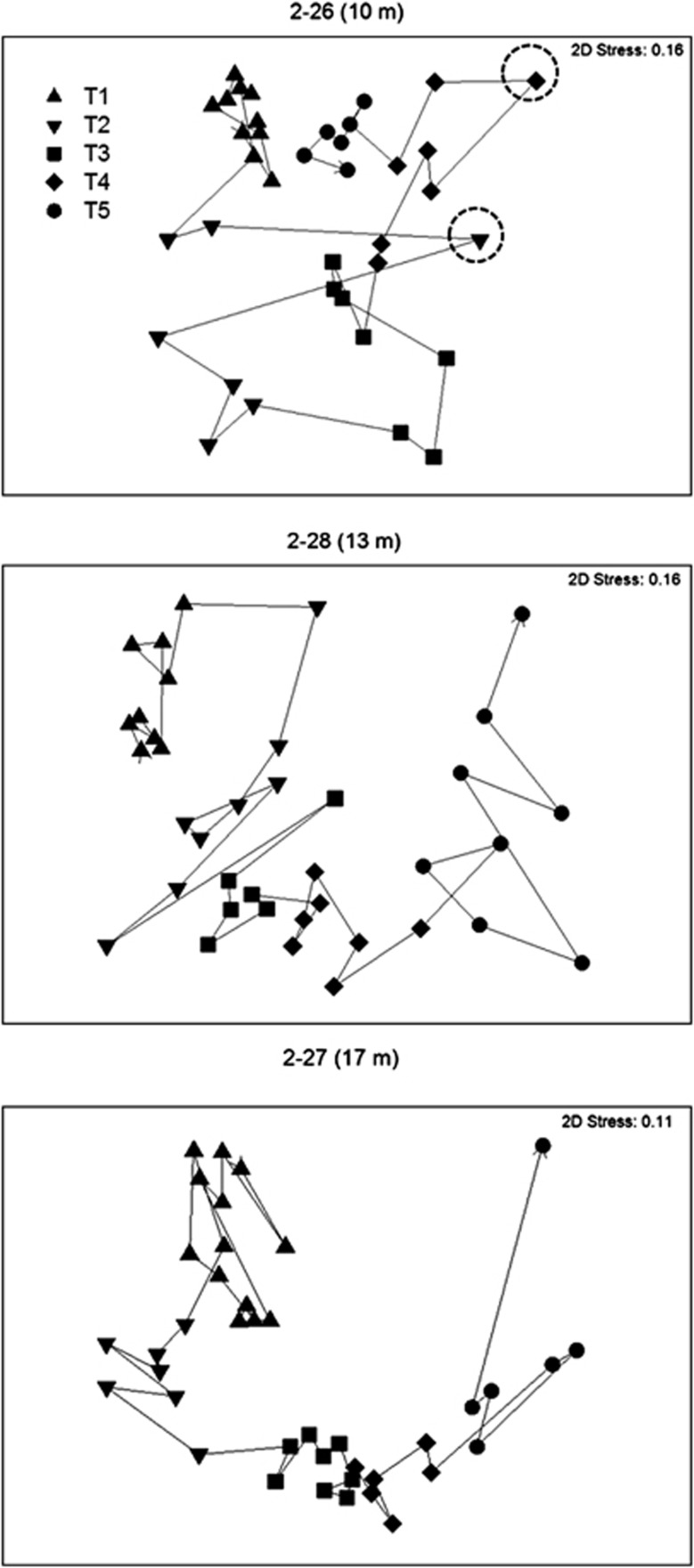
Pairwise comparisons of similarity through time indicated that communities exhibited similar trajectories at all three depths in Clusters A (Figures 4b and and Supplementary Figure S2a). A detrended correspondence analysis confirmed that the temporal trajectories observed in the NMS is not due to the artifact of horseshoe effect (Supplementary Figure S2b). Several abrupt changes in microbial community composition at particular times were observed (dashed circles, Figure 4 and Supplementary Figure S2a). These shifts were often related to changes in the relative abundance of specific OTUs (for example, Actinobacteria ACK-M1; see Supplementary Figure S5).
Figure 4.
Temporal trajectories in community composition, based on Bray–Curtis similarity and presented in the ordination space of non-metric multidimensional scaling analysis. Trajectories through trajectories were represented by arrows that sequentially connect sampling points. Circles highlighted two time points on 20 February and 24 June with abrupt change in microbial community composition.
Species-time relationships showed good fits with the power functions (Supplementary Figure S3 and Table 1). The w values for all samples ranged from 0.13 to 0.22, which was near the low end of the range observed for a broad variety of taxa and ecosystems, but close to the w values observed for marine bacterioplankton (White et al., 2006a; Sachdeva et al., 2010). The highest w values (0.21–0.39) were observed for samples taken from the shallowest wells in the late spring (Table 1), where and when the hydrological perturbations were greatest.
Table 1. Comparison of power function exponents (w) for the species-time relationship based on a complete-nested calculation method.
| Sampling time clusters |
10 m |
13 m |
17 m |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 2–26 | 2–29 | 3–30 | 2–28 | 2–31 | 3–32 | 2–27 | 2–30 | 3–31 | |
| All | 0.14 | 0.20 | 0.17 | 0.17 | 0.13 | 0.20 | 0.22 | 0.19 | 0.18 |
| T1 | 0.07 | 0.01 | 0.04 | 0.06 | 0.01 | 0.01 | 0.05 | 0.08 | 0.03 |
| T2 | 0.27 | 0.19 | 0.24 | 0.22 | 0.12 | 0.14 | 0.12 | 0.09 | 0.19 |
| T3+T4 | 0.39 | 0.29 | 0.21 | 0.15 | 0.16 | 0.09 | 0.14 | 0.09 | 0.11 |
Five time clusters were identified: T1 (16 November–1 December),T2 (26 January–10 March), T3 (14 May–14 June), T4 (15 June–8 July 8) and T5 (9 July–24 September). T3 and T4 were analyzed together to reflect species turnover across a period from the beginning of water level rise to the end of river water intrusion. T5 was omitted from discrete analysis because there were a small number of time points in this interval.
Temporal shift in microbial community composition at different strata
Proteobacteria comprised 50% of the total sequences that were retrieved. The next four most abundant phyla were Bacteroidetes (13%), Actinobacteria (6%), Acidobacteria (4%) and OP3 (3%) (Supplementary Figure S4). There were no clear differences in community composition between strata at this taxonomic level. Archaea averaged only 2.5% of the total microbial community, with a slightly higher contribution in shallow depths (3.9±1.7%) than other strata (1.9±0.8%) (Supplementary Figure S4).
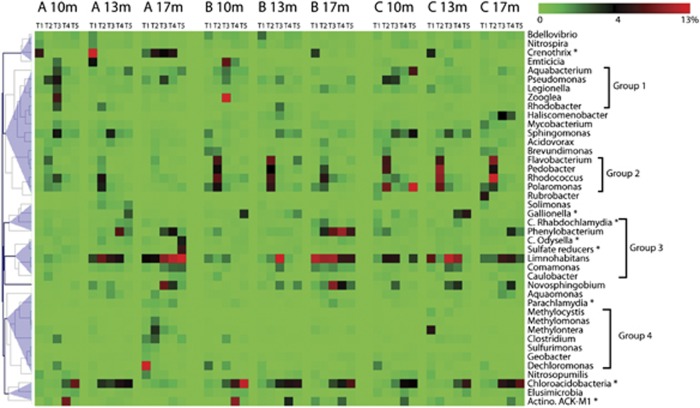
Temporal shifts in composition were analyzed at the genus level, employing Random Forest, a supervised machine learning tool in QIIME (Figure 5). In terms of sequence abundance, these discriminative genera averaged 35% of the total community. There were some clear vertical and temporal patterns, and hierarchical clustering identified in four major groups of organisms. Group-1 OTUs were prevalent at 10 m especially in T3; Group-2 OTUs were prevalent at all strata in T2; Group-3 OTUs had higher abundances at 13 and 17 m than at 10 m; this spatial pattern is most obvious for Limnohabitans; and Group-4 OTUs contained specific physiological groups—putative methane oxidizers (for example, Methylotenera and Methylomonas), Sulfurimonas (sulfur oxidizer), Geobacter and Nitrosopumilus (ammonia-oxidizing archaea). These Group-4 OTUs were most abundant at the base of the Hanford formation (17 m) at T1 and T2. In addition, methane-oxidizing Crenothrix were quite abundant in T1 at 10 and 13 m, and usually detected at 17 m in cluster A. Intracellular bacterial parasites of amoeba, such as Candidatus Rhabdochlamydia, Parachlamydia and Candidatus Odyssella, were found at several time points at all depths, consistent with detection of Amoebozoa in 18S clone libraries described below. In addition, the abundance of Chloracidobacteria, generally increased from winter to late summer.
Figure 5.
Relative abundance of genus-level OTUs (identified as feature OTUs by Random Forest analysis) at 10, 13 and 17 m in well clusters A, B or C. OTU abundances were averaged within each time cluster: T1 (16 November–1 December), T2 (26 January–10 March), T3 (14 May–14 June), T4 (15 June–8 July) and T5 (9 July–24 September). Stars highlight some taxa with distinctive distribution patterns.
Spatial variations in microbial community composition
Two-way nested ANOSIM was used to test whether significant differences in community composition occurred in space or time. There was no significant difference (P=0.2) among temporal clusters across shallow wells (Table 2). In contrast, the nested temporal analysis showed significant differences (P=0.002) across wells at 13 and 17 m (Table 2). Similar nested analysis illustrated that depth was an important factor discriminating community composition in each well cluster (P=0.001), except that the well Cluster C had relatively lower R values.
Table 2. Two-way nested ANOSIM analysis (R statistic and significance level (P)) to compare similarity between sample groups: A, time clusters nested in each well at the same depth; B, time clusters nested in each depth.
|
(A) Similarity across wells at the same depth | |||||
|---|---|---|---|---|---|
|
10 m |
13 m |
17 m |
|||
| R | P | R | P | R | P |
| 0.09 | 0.2 | 0.4 | 0.002 | 0.44 | 0.002 |
|
(B) Similarity across depths in each well cluster | |||||
|---|---|---|---|---|---|
| Cluster A | Cluster B | Cluster C | |||
| 0.58 | 0.001 | 0.59 | 0.001 | 0.21 | 0.049 |
Effects of the Columbia River water intrusion on community composition
SIMPER analysis was applied to identify genus-level OTUs that changed before and after Columbia River intrusion near the water table. Two groups of Actinobacteria (ACK-M1 and CL500-29) were not detected before intrusion, but constituted 12% of the organisms detected over the intrusion period, and comprised 38% at the highest water level (Table 3 and Supplementary Figure S5). In addition to this temporal effect, there was also a positional effect that suggested the appearance of these actinobacterial clades was related to river water intrusion. Well 3–30 is most distant from the river (and hence had the smallest intrusion of river water) and exhibited the smallest increase in Actinobacteria among the three shallow wells. Four other OTUs with increased abundances after river water intrusion included Chloracidobacteria, Sphingobacterium and those belonging to Candidate Divisions OP3 and ZB2. OTUs that decreased in abundance after river water intrusion were phylogenetically related to Zoogloea, Clostridium, Emticicia, Limnohabitans, Pseudomonas and Aquabacterium. These decreases also exhibited a positional effect, with smaller decreases in river-distal well 3–30 than river-proximal wells 2–26 and 2–29.
Table 3. SIMPER analysis identifying six most important OTUs contributing to the difference in community composition before (May-14 June) and after (15 June–6 July) the intrusion of Columbia River water into three shallow wells.
| Taxa |
2–26 (10 m) |
2–29 (10 m) |
3–30 (10 m) |
AvDiss | Contrib% | |||
| |
May–14 June |
15 June–6 July |
May–14 June |
15 June–6 July |
May–14 June |
15 June–6 July |
|
|
| OTUs with increased abundance after river water arrival | ||||||||
| ACK-M1 (Actinobacteria) | 0 | 8.6 | 0 | 9.6 | 0 | 2.4 | 3.4 | 4.8 |
| Chloracidobacteria | 0.6 | 2.6 | 2.6 | 6.6 | 2 | 4.2 | 1.5 | 2.2 |
| CL500-29 (Actinobacteria) | 0 | 2.8 | 0 | 2.4 | 0 | 1 | 1.0 | 1.4 |
| OP3 | 1 | 7.4 | 0.4 | 4.6 | 1.8 | 7.4 | 2.8 | 4.0 |
| Sphingobacterium | 1.8 | 3.6 | 4.2 | 4.8 | 1.8 | 2.8 | 1.2 | 1.8 |
| ZB2 | 0 | 0 | 3.6 | 6 | 1 | 2 | 2.0 | 2.7 |
| OTUs with decreased abundance after river water arrival | ||||||||
| Aquabacterium | 2.8 | 1.4 | 2.2 | 1.2 | 2 | 1.4 | 1.0 | 1.5 |
| Clostridium | 5.6 | 0.2 | 10.6 | 0.8 | 0 | 0 | 3.9 | 5.4 |
| Emticicia | 2.2 | 0 | 8 | 1.8 | 0 | 0 | 2.3 | 3.2 |
| Limnohabitans | 0 | 0 | 2.2 | 0.2 | 5.2 | 0.4 | 1.7 | 2.7 |
| Pseudomonas | 4.6 | 0.6 | 1.2 | 0.4 | 3 | 1 | 1.5 | 2.2 |
| Zoogloea | 6.8 | 0.4 | 13.6 | 0.6 | 0 | 0 | 4.9 | 6.7 |
Abbreviations: AvDiss, the average of the Bray–Curtis dissimilarity between all pairs of samples; Contrib%, the contribution of this OTU to the total dissimilarity between communities before and after the river water intrusion; OTU, operational taxonomic units. The gradual shift of these OTUs in time are graphically illustrated in Supplementary Figure S5.
Community composition of Eukarya
Clone libraries revealed that eukaryotic sequences primarily belong to microeukaryotes within kingdom/phyla Alveolates (Ciliophora and Dinophyceae), Amoebozoa, Euglenoza, Fungi, Katablepharidophyta and Stramenopiles (Supplementary Table S1). Mixotrophic and bacterivorous protists comprised 16–84% (44% on average) of total sequences in each clone library (Table 4). These protists were dominated by Amoebozoa (for example, Acanthamoeba) and heterotrophic flagellates such as Rhynchobodo, Bicosoeca, Dinobryon and Ochromonas.
Table 4. Percentages of heterotrophic and mixotrophic protists in 18S rRNA gene clone libraries from two shallow wells (2–26 and 2–29) and the deep well 2–27.
| Phyla | Closest relatives |
24 February |
21 June |
||||
|---|---|---|---|---|---|---|---|
| 2–26 | 2–29 | 2–27 | 2–26 | 2–29 | 2–27 | ||
| Ciliophora | Cyclidium glaucoma | 0.0 | 0.0 | 0.0 | 0.0 | 2.9 | 0.0 |
| Lembadion bullinum | 0.0 | 0.0 | 0.0 | 0.0 | 2.9 | 2.6 | |
| Tetrahymena tropicalis | 0.0 | 0.0 | 2.7 | 0.0 | 0.0 | 0.0 | |
| Vorticella convallaria | 0.0 | 5.0 | 0.0 | 0.0 | 5.7 | 0.0 | |
| Zosterodasys agamalievi | 0.0 | 5.0 | 0.0 | 2.9 | 2.9 | 2.6 | |
| Dinophyceae | Pfiesteria piscicida | 2.5 | 0.0 | 0.0 | 0.0 | 8.6 | 0.0 |
| Amoebozoa | Acanthamoeba sp. | 0.0 | 0.0 | 0.0 | 5.7 | 37.1 | 0.0 |
| Euglenoza | Rhynchobodo sp. | 2.5 | 5.0 | 0.0 | 0.0 | 0.0 | 2.6 |
| Stramenopiles | Bicosoeca petiolata | 0.0 | 0.0 | 29.7 | 17.1 | 0.0 | 0.0 |
| Dinobryon sp. | 15.0 | 5.0 | 5.4 | 5.7 | 17.1 | 2.6 | |
| Ochromonas sp. | 10.0 | 5.0 | 45.9 | 2.9 | 0.0 | 5.3 | |
| Total % in each clone library | 30.0 | 25.0 | 83.8 | 34.3 | 77.1 | 15.8 | |
Abbreviation: OTU, operational taxonomic units. These OTUs had >95% sequence similarity to their closest relatives
Discussion
The temporal dynamics of microbial community structure have important implications for understanding microbial interactions with the environment (Fuhrman et al., 2006; Fuhrman and Steele, 2008; Yagi et al., 2010). Differences between samples may merely be due to technical issues of sampling and analysis, or consequences of ecological or evolutionary factors (Preston, 1960; Torsvik et al., 2002; Storch et al., 2007). Over short timescales (from minutes to days), undersampling of community complexity would be the dominant factor for variability. Sorensen similarity values between samples taken 1 day apart were less than 70% (Supplementary Figure S3), this suggests that technical factors in sampling and pyrosequencing analysis could account for dissimilarities of up to 30%. To reduce the effects of low technical reproducibility (Zhou et al., 2011), we restricted analyses of beta-diversity to the 1000 most abundant OTUs that enabled us to resolve temporal gradients in community shifts (Supplementary Figure S3). The good fit of power functions to temporal changes suggested a relationship between community membership and time, and that community dynamics were being driven by environmental changes and/or ecologically neutral processes (Stegen et al., 2012).
Over a period of months, community dynamics can plausibly be ascribed to ecological processes that are driven by environmental variation and microbial dispersal. In this unconfined aquifer system, both groundwater temperature (16–18 °C) and major ion composition were relatively stable, except for ion composition in periods when the water table rose (J McKinley, unpublished observations). Hydrological perturbations were correlated with highest community turnover rates in the shallow wells in the late spring.
Spatial heterogeneities in community composition were found both laterally and vertically, especially at the intermediate and deep zones. The former could be a consequence of habitat heterogeneities imposed by local physicochemical regimes (Dechesne et al., 2007; Lehman, 2007; Vermeul et al., 2011), such as partially filled or unfilled voids between gravelly cobble within the Hanford formation (Lindberg and Bond, 1979), that would disperse nutrients and microbes. Groundwater samples were collected at depths where sediments were coarse-grained with silty sands filling voids between rounded river cobbles (Bjornstad et al., 2009). Within those sediments, differential measurements of hydraulic conductivity detected a zone of lower permeability within the central portion of the aquifer (Vermeul et al., 2011), where the intermediate-depth wells in each cluster were screened. Preferential groundwater flowpaths and regions of low sediment permeability could drive significant vertical variations in community composition. Relatively rapid flow near the water table would result in a rapid response to changes in river stage. Conversely, lower permeability deeper in the aquifer may have hindered community responses to changes in groundwater composition, owing to poor coupling of river-driven hydrological forces within deep sediments.
In the deeper groundwater depth (17 m), near the boundary between the Hanford and Ringold formations and about 1 m above a redox transition zone (Bjornstad et al., 2009), we observed a higher proportion of putative ammonia-oxidizing Archaea, methane oxidizers and metal reducers. Vertical geochemical profiles suggest that methane and other electron donors such as H2 and H2S could diffuse into the Hanford formation (Lin et al., 2012b). In addition, temporal differences in methane and sulfide concentrations have been measured near the Ringold oxic–anoxic interface (J McKinley, unpublished observations). The temporal dynamics in specific taxa near the base of the Hanford formation, coupled with seasonal changes in geochemistry provide motivation for more intensive analyses of temporally variable redox cycling in this portion of the aquifer.
Groundwater–river water interactions are of substantial ecological interest (Brunke and Gonser, 1997). However, most research has focused on effects of groundwater upon riverine ecology. Here, we demonstrate that Columbia River water intrusion affects microbial community composition in the aquifer, 250 m from the shoreline. The effects at this relatively large distance are consequences of the porous, hydrologically conductive sediments of the Hanford formation (Bjornstad et al., 2009), and the dramatic fluctuations in stage of the Columbia River that flows for 1900 km and drains more than 670 000 km2 in seven US states and Canada. At the highest water table elevation, the river water component accounted for 75% of groundwater–river water mixture. Well Cluster A contained a lesser component of river water than Cluster B, whereas Cluster C to the west site experienced fractional mixing of less than 15% river water (J McKinley, unpublished observations). The spatial heterogeneity of river water intrusion was consistent with spatial and temporal trajectories of several clades such as Actinobaceria ACK-M1, previously found in freshwater environments (Hahn, 2006) and which has been reported in the Columbia River (Crump et al., 1999). Actinobacteria ACK-M1 have neither been detected in the Hanford Site subsurface vadose sediments (Balkwill et al., 1998) nor in the saturated Hanford sediments near the water table (Lin et al., 2012a). However, we cannot rule out the possibility that the fluctuating water table may introduce vadose zone microbes into the saturated zone. The shift in community composition associated with the large dynamics in Actinobacteria ACK-M1 and other clades suggested that river water intrusion could have significant impacts on microbial community composition. Without analysis of riverine microbial populations, we cannot distinguish among immigration of riverine bacteria, elution from the lower vadose zone, or environmental selection of aquifer bacteria by the riverine nutrients. More work is needed to disentangle the specific effects of individual nutrient resources versus stochastic effects (Stegen et al., 2012).
Seasonal dynamics in many aquatic ecosystems are affected by top-down biological controls such as bacterivorous protozoa (Azam et al., 1983; Sherr and Sherr, 2002; Pernthaler, 2005). In pristine and contaminated subsurface habitats, high abundances (up to 104 individuals ml−1) of protozoan biomass have been reported (Sinclair et al., 1993; Ellis et al., 1998; Zarda et al., 1998; Kinner et al., 2002). This stimulated us to analyze the occurrence of microeukaryotes in the Hanford subsurface. The temporal dynamics in eukaryotic 18S rRNA gene copy numbers ranged over two orders of magnitude. However, these dynamics must be interpreted cautiously as they could also arise if there were large differences in rRNA gene copy number between the dominant taxa over time. The temporal pattern of microeukaryotes was positively correlated with Bacterial abundances, which would be consistent with the bacteria serving as a prey resource (Caron and Goldman, 1990; Pernthaler, 2005). Many of the detected taxa are known to be bacteriovores (Andersson et al., 1989; Bennett et al., 1990; Arndt et al., 2000; Boenigk et al., 2005). Thus, we hypothesize that protozoa exert grazing pressure on the aquifer microbial community, impacting temporal dynamics in community structure. Quantification of top-down controls on local microbial assemblages could reveal an important controlling element beyond the physical and chemical controls that affect contaminant fate and transportin the subsurface at Hanford and other locations.
In summary, spatial and temporal variation in microbial community structure in the Hanford unconfined aquifer is apparently governed not only by geophysical variables but also by hydrochemical and biotic ones. Fluctuations in ground water elevation impacted microbial community structure to the greatest extent near the water table, where river water mixing was greatest. A consequence of these hydrodynamic heterogeneities in space and time may be selection for different ecological strategies. Bacteria prevalent during periods of constant river stage (for example, Group-2 in Figure 5) or deeper in the aquifer (for example, Limnohabitans spp.) are targets for further analysis as autochthonous aquifer bacteria. The clades that became abundant after the intrusion of river water (such as Actinobacteria ACK-M1) could either represent allochthonous inputs into the aquifer system or aquifer opportunists that are stimulated by riverine nutrient additions. The ecological impacts of the river water intrusion can be positive (for example, influx of nutrient resources) or negative (for example, introduction of river-derived bacterial predators). The interaction between aquifer and fluvial systems has been recognized (Williams et al., 2010), but research interest has focused upon effects of groundwater upon stream ecology. In this work, we demonstrate that a large river system can impact the ecology (and presumably biogeochemistry) in the subsurface at distances more than 200 m from shore. A mechanistic understanding of these forces will require both additional field-based studies as well as ecophysiological studies in the laboratory of important clades that have been identified.
Acknowledgments
We thank Don Girvin, Micah Miller and David Kennedy for their assistance with the sampling, and Samantha B Reed for preparing transformation stocks of 18S rRNA gene clone libraries, and James Stegen for discussions. This research was supported by the US Department of Energy (DOE), Office of Biological and Environmental Research (BER), as part of Subsurface Biogeochemistry Research Program's Scientific Focus Area (SFA) and Integrated Field-Scale Research Challenge (IFRC) at the Pacific Northwest National Laboratory (PNNL). PNNL is operated for DOE by Battelle under contract DE-AC06-76RLO 1830.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anderson RT, Vrionis HA, Ortiz-Bernad I, Resch CT, Long PE, Dayvault R, et al. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl Environ Microbiol. 2003;69:5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson-Glenna MJ, Bakkestuen V, Clipson NJW. Spatial and temporal variability in epilithic biofilm bacterial communities along an upland river gradient. FEMS Microbiol Ecol. 2008;64:407–418. doi: 10.1111/j.1574-6941.2008.00480.x. [DOI] [PubMed] [Google Scholar]
- Andersson A, Falk S, Samuelsson G, Hagstrom A. Nutritional characteristics of a mixotrophic nanoflagellate, Ochromonas Sp. Microb Ecol. 1989;17:251–262. doi: 10.1007/BF02012838. [DOI] [PubMed] [Google Scholar]
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2010;4:171–181. doi: 10.1038/ismej.2009.108. [DOI] [PubMed] [Google Scholar]
- Arndt H, Dietrich D, Auer B, Cleven EJ, Gräfenhan T, Weitere M, et al. 2000Functional diversity of heterotrophic flagellates in aquatic ecosystemsIn BSC Leadbeater, JC Green (eds.)The Flagellates Taylor & Francis: London, UK; 240–268. [Google Scholar]
- Arntzen EV, Geist DR, Dresel PE. Effects of fluctuating river flow on groundwater/surface water mixing in the hyporheic zone of a regulated, large cobble bed river. River Res Appl. 2006;22:937–946. [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso SV, Guerrero MC, Montes C, Lopez-Archilla AI. Spatiotemporal distribution of microbial communities in a coastal, sandy aquifer system (Donana, SW Spain) Geobiology. 2009;7:66–81. doi: 10.1111/j.1472-4669.2008.00183.x. [DOI] [PubMed] [Google Scholar]
- Azam F, Fenchel T, Field JG, Gray JS, Meyerreil LA, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- Balkwill DL, Ghiorse WC. Characterization of subsurface bacteria associated with 2 shallow aquifers in oklahoma. Appl Environ Microbiol. 1985;50:580–588. doi: 10.1128/aem.50.3.580-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill DL, Murphy EM, Fair DM, Ringelberg DB, White DC. Microbial communities in high and low recharge environments: implications for microbial transport in the vadose zone. Microb Ecol. 1998;35:156–171. doi: 10.1007/s002489900070. [DOI] [PubMed] [Google Scholar]
- Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SJ, Sanders RW, Porter KG. Heterotrophic, autotrophic, and mixotrophic nanoflagellates—seasonal abundances and bacterivory in a eutrophic lake. Limnol Oceanogr. 1990;35:1821–1832. [Google Scholar]
- Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornstad BN, Horner JA, Vermeul VR, Lanigan DC, Thorne PD.2009Borehole Completion and Conceptual Hydrogeologic Model for the IFRC Well Field, 300 Area, Hanford Site http://ifchanford.pnl.gov/pdfs/18340.pdfPNNL-18340Pacific Northwest National Laboratory: Richland [Google Scholar]
- Boenigk J, Pfandl K, Stadler P, Chatzinotas A. High diversity of the 'Spumella-like' flagellates: an investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ Microbiol. 2005;7:685–697. doi: 10.1111/j.1462-2920.2005.00743.x. [DOI] [PubMed] [Google Scholar]
- Boulton AJ, Datry T, Kasahara T, Mutz M, Stanford JA. Ecology and management of the hyporheic zone: stream-groundwater interactions of running waters and their floodplains. JNABS. 2010;29:26–40. [Google Scholar]
- Brunke M, Gonser T. The ecological significance of exchange processes between rivers and groundwater. Freshwater Biol. 1997;37:1–33. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron DA, Goldman JC.1990Protozoan nutrient regenerationIn GM Capriulo (ed.)Ecology of Marine Protozoa Oxford University Press: New York; 283–306. [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Armbrust EV, Baross JA. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechesne A, Pallud C, Grundmann GL.2007Spatial distribution of bacteria at the microscale soilIn RB Franklin, AL Mills (eds.)The Spatial Distribution of Microbes in the Environment Springer: Dordrecht, The Netherlands; 87–107. [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elberling B, Askaer L, Jorgensen CJ, Joensen HP, Kuhl M, Glud RN, et al. Linking soil O(2), CO(2), and CH(4) concentrations in a wetland soil: implications for CO(2) and CH(4) fluxes. Environ Sci Technol. 2011;45:3393–3399. doi: 10.1021/es103540k. [DOI] [PubMed] [Google Scholar]
- Ellis BK, Stanford JA, Ward JV. Microbial assemblages and production in alluvial aquifers of the Flathead River, Montana, USA. JNABS. 1998;17:382–402. [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA. 2006;103:13104–13109. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA. Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat Microb Ecol. 2008;53:69–81. [Google Scholar]
- Ghiglione JF, Larcher M, Lebaron P. Spatial and temporal scales of variation in bacterioplankton community structure in the NW Mediterranean Sea. Aquat Microb Ecol. 2005;40:229–240. [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore TJ, Borghese JV, Newcomer DR. Effects of river stage and waste-water discharges on the unconfined Aquifer, Hanford, Washington. Ground Water Monit Remediat. 1993;13:130–138. [Google Scholar]
- Griebler C, Lueders T. Microbial biodiversity in groundwater ecosystems. Freshwater Biol. 2009;54:649–677. [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. The microbial diversity of inland waters. Curr Opin Biotechnol. 2006;17:256–261. doi: 10.1016/j.copbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Kan JJ, Wang K, Chen F. Temporal variation and detection limit of an estuarine bacterioplankton community analyzed by denaturing gradient gel electrophoresis (DGGE) Aquat Microb Ecol. 2006;42:7–18. [Google Scholar]
- Kinner NE, Harvey RW, Shay DM, Metge DW, Warren A. Field evidence for a protistan role in an organically-contaminated aquifer. Environ Sci Technol. 2002;36:4312–4318. doi: 10.1021/es020611m. [DOI] [PubMed] [Google Scholar]
- Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011;35:343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of Soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman RM.2007Microbial distributions and their potential controlling factors in terrestrial subsurface environmentsIn RB Franklin, AL Mills (eds.)The Spatial Distribution of Microbes in the Environment Springer: Dordrecht, The Netherlands; 135–178. [Google Scholar]
- Lin X, Kennedy D, Fredrickson J, Bjornstad B, Konopka A. Vertical stratification of subsurface microbial community composition across geological formations at the Hanford Site. Environ Microbiol. 2012a;14:414–425. doi: 10.1111/j.1462-2920.2011.02659.x. [DOI] [PubMed] [Google Scholar]
- Lin X, Kennedy D, Peacock A, Mckinley J, Resch CT, Fredrickson J, et al. Distribution of microbial biomass and the potential for anaerobic respiration in Hanford Site 300 area subsurface sediment. Appl Environ Microbiol. 2012b;78:759–767. doi: 10.1128/AEM.07404-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XJ, Scranton MI, Chistoserdov AY, Varela R, Taylor GT. Spatiotemporal dynamics of bacterial populations in the anoxic Cariaco Basin. Limnol Oceanogr. 2008;53:37–51. [Google Scholar]
- Lindberg JW, Bond FW.1979Geohydrology and Groundwater Quality Beneath the 300 Area, Hanford Site, WashingtonPNL-2949Pacific Northwest National Laboratory: Richland, WA [Google Scholar]
- Lowell JL, Gordon N, Engstrom D, Stanford JA, Holben WE, Gannon JE. Habitat heterogeneity and associated microbial community structure in a small-scale floodplain hyporheic flow path. Microb Ecol. 2009;58:611–620. doi: 10.1007/s00248-009-9525-9. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Kumar Y, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16s-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Minamikawa K, Nishimura S, Sawamoto T, Nakajima Y, Yagi K. Annual emissions of dissolved CO(2), CH(4), and N(2)O in the subsurface drainage from three cropping systems. Glob Change Biol. 2010;16:796–809. [Google Scholar]
- Nelson JD, Boehme SE, Reimers CE, Sherrell RM, Kerkhof LJ. Temporal patterns of microbial community structure in the Mid-Atlantic Bight. FEMS Microbiol Ecol. 2008;65:484–493. doi: 10.1111/j.1574-6941.2008.00553.x. [DOI] [PubMed] [Google Scholar]
- Odum WE, Odum EP, Odum HT. Nature's pulsing paradigm. Estuaries. 1995;18:547–555. [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol. 2005;3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- Peterson RE, Connelly MP. Water movement in the zone of interaction between groundwater and the Columbia River, Hanford Site, Washington. J Hydraul Res. 2004;42:53–58. [Google Scholar]
- Preston FW. Time and space and the variation of species. Ecology. 1960;41:611–627. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley RG, Zachara JM. Nature of Chemical Contamination on DOE Lands and Identification of Representative Contaminant Mixtures for Basic Subsurface Science Research. Subsurface Science Program OoER, US Department of Energy (ed.): Washington, DC; 1992. [Google Scholar]
- Sachdeva R, Shaw AK, Fuhrman J, Horner-Devine C. ISME Meeting. Seattle, WA; 2010. Temporal scaling of marine bacterioplankton assemblages. [Google Scholar]
- Sherr EB, Sherr BF. Significance of predation by protists in aquatic microbial food webs. Anton Leeuwenhoek Int J Gen M. 2002;81:293–308. doi: 10.1023/a:1020591307260. [DOI] [PubMed] [Google Scholar]
- Simon KS, Gibert J, Petitot P, Laurent R. Spatial and temporal patterns of bacterial density and metabolic activity in a karst aquifer. Arch Hydrobiol. 2001;151:67–82. [Google Scholar]
- Sinclair JL, Kampbell DH, Cook ML, Wilson JT. Protozoa in subsurface sediments from sites contaminated with aviation gasoline or Jet Fuel. Appl Environ Microbiol. 1993;59:467–472. doi: 10.1128/aem.59.2.467-472.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegen JC, Lin X, Konopka AE, Frederickson JK.2012Stochastic and deterministic assembly processes in subsurface microbial communities ISME Je-pub ahead of print 29 March 2012; doi: 10.1038/ismej.2012.22 [DOI] [PMC free article] [PubMed]
- Storch D, Marquet P, Brown J. Scaling Biodiversity. Cambridge University Press: Cambridge, UK; 2007. [Google Scholar]
- Storey RG, Fulthorpe RR, Williams DD. Perspectives and predictions on the microbial ecology of the hyporheic zone. Freshwater Biol. 1999;41:119–130. [Google Scholar]
- Sundberg C, Stendahl JSK, Tonderski K, Lindgren PE. Overland flow systems for treatment of landfill leachates—Potential nitrification and structure of the ammonia-oxidising bacterial community during a growing season. Soil Biol Biochem. 2007;39:127–138. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Ovreas L, Thingstad TF. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- Vermeul VR, McKinley JP, Newcomer DR, Mackley RD, Zachara JM. River-induced flow dynamics in long-screen wells and impact on aqueous samples. Ground Water. 2011;49:515–524. doi: 10.1111/j.1745-6584.2010.00769.x. [DOI] [PubMed] [Google Scholar]
- White EP, Adler PB, Lauenroth WK, Gill RA, Greenberg D, Kaufman DM, et al. A comparison of the species-time relationship across ecosystems and taxonomic groups. Oikos. 2006a;113:383–383. [Google Scholar]
- White EP, Adler PB, Lauenroth WK, Gill RA, Greenberg D, Kaufman DM, et al. A comparison of the species-time relationship across ecosystems and taxonomic groups. Oikos. 2006b;112:185–195. [Google Scholar]
- Williams DD, Febria CM, Wong JCY. Ecotonal and other properties of the hyporheic zone. Fundam Appl Limnol. 2010;176:349–364. [Google Scholar]
- Yagi JM, Neuhauser EF, Ripp JA, Mauro DM, Madsen EL. Subsurface ecosystem resilience: long-term attenuation of subsurface contaminants supports a dynamic microbial community. ISME J. 2010;4:131–143. doi: 10.1038/ismej.2009.101. [DOI] [PubMed] [Google Scholar]
- Zarda B, Mattison G, Hess A, Hahn D, Hohener P, Zeyer J. Analysis of bacterial and protozoan communities in an aquifer contaminated with monoaromatic hydrocarbons. FEMS Microbiol Ecol. 1998;27:141–152. [Google Scholar]
- Zhou J, Wu L, Deng Y, Zhi X, Jiang YH, Tu Q, et al. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J. 2011;5:1303–1313. doi: 10.1038/ismej.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.