Abstract
Leaf-litter decomposition is a central process in carbon cycling; however, our knowledge about the microbial regulation of this process is still scarce. Metaproteomics allows us to link the abundance and activity of enzymes during nutrient cycling to their phylogenetic origin based on proteins, the ‘active building blocks' in the system. Moreover, we employed metaproteomics to investigate the influence of environmental factors and nutrients on the decomposer structure and function during beech litter decomposition. Litter was collected at forest sites in Austria with different litter nutrient content. Proteins were analyzed by 1-D-SDS-PAGE followed by liquid-chromatography and tandem mass-spectrometry. Mass spectra were assigned to phylogenetic and functional groups by a newly developed bioinformatics workflow, assignments being validated by complementary approaches. We provide evidence that the litter nutrient content and the stoichiometry of C:N:P affect the decomposer community structure and activity. Fungi were found to be the main producers of extracellular hydrolytic enzymes, with no bacterial hydrolases being detected by our metaproteomics approach. Detailed investigation of microbial succession suggests that it is influenced by litter nutrient content. Microbial activity was stimulated at higher litter nutrient contents via a higher abundance and activity of extracellular enzymes.
Keywords: extracellular enzymes, leaf-litter decomposition, litter nutrient content, metaproteomics, microbial ecology, microbial succession
Introduction
Leaf-litter decomposition is the physical and chemical breakdown of dead plant material, a key ecosystem process that provides energy for microbial growth, releases nutrients for plant growth, influences ecosystem carbon (C) storage, and thus may have a long-term effect on the climate (Chapin et al., 2002). Investigations of litter decomposition at different geographic locations have shed light on the factors that influence decomposition rates, for example, that litter decomposition occurs above a certain threshold in mean annual temperature and depends on litter quality (Zhang et al., 2008; Prescott, 2010). In spite of the vast amount of litter decomposition studies, little is known about specific functions contributed by the different microbial groups involved in litter decomposition, mainly due to the lack of suitable methods available for in-depth studies of ecosystem functioning.
Leaf litter consists mainly of cellulose, but also lignin, hemicellulose, pectin and proteins (Yadav and Malanson, 2007), requiring numerous enzymes for degradation. The presence and activity of extracellular enzymes link environmental nutrient availability to microbial nutrient demand (Sinsabaugh et al., 2008), which in turn is determined by the elemental stoichiometry (that is, the C:N:P ratio) of the decomposer's biomass (Sterner and Elser, 2002). Within the microbial biomass, phylogenetic groups such as fungi and bacteria exhibit different nutrient demands and thereby influence decomposition and the carbon use efficiency within the ecosystem (Sinsabaugh et al., 2009; Keiblinger et al., 2010). Studies on extracellular enzyme activities may reflect the constraints on microbial biomass stoichiometry and enzyme relationships to litter decomposition, that is, for hydrolytic enzymes ratios of C, N and P acquisition activities have been assessed and they converged on 1:1:1 (Sinsabaugh et al., 2008). However, so far no attempt has been made to link enzyme activities to the respective protein abundances, their microbial origin, or the biogeochemistry of leaf litter.
In the last decade, metaproteomics has greatly advanced our understanding of microorganisms and their geochemical environment (reviewed in Newman and Banfield, 2002; Schneider and Riedel, 2010; Wilmes and Bond, 2009). In contrast to DNA and RNA, most proteins have an intrinsic metabolic function and can thus be used to relate specific microbial activities to defined organisms in multispecies communities. Therefore, the identification of the microbial proteins of a given habitat together with the analysis of their phylogenetic origin and their temporal distribution is expected to provide fundamentally new insights into the role of microbial diversity in biogeochemical processes. Recent large-scale characterisations of the entire protein complement of diverse microbial communities have proven useful to describe microbial functions in soil (Schulze et al., 2005; Wang et al., 2011), lake and ground water (Schulze et al., 2005; Benndorf et al., 2007, 2009), and leaf phyllosphere (Delmotte et al., 2009). We have recently employed a proteomics approach to investigate a model system, consisting of the litter degrading fungus Aspergillus nidulans and the bacterium Pectobacterium carotovorum grown on sterilized leaf litter (Schneider et al., 2010). In the current study, we expand our proteomics analyses to environmental litter samples.
The major goals of the present study were: (I) to link structure and function of microbial communities involved in leaf-litter decomposition and (II) to investigate the impact of leaf-litter nutrient content (stoichiometry) and season on the abundance and activity of the leaf-litter decomposers. We analyzed the metaproteome from beech (Fagus sylvatica L.) litter collected in February and May 2009 at four Austrian sampling-sites that differed in their nutrient content. The newly developed bioinformatics pipeline ‘PROteomics result Pruning & Homology group ANotation Engine' (PROPHANE, Schneider et al., 2011) was employed to assign proteins to their phylogenetic and functional origin. Phospholipid fatty acid (PLFA) analyses, enzymatic measurements and qualitative assessment of fungal communities were used for data validation.
Materials and methods
Litter sampling sites
Beech leaf litter was collected at four forest sites in Austria: (I) Achenkirch (AK), Tyrol; (II) Klausenleopoldsdorf (KL), Lower Austria; (III) Ort (OR), Gmunden, Upper Austria; and (IV) Schottenwald (SW), Vienna that differed in their nutrient content. AK is located in the Achental of the North Tyrolean limestone Alps. It is characterised by its location on calcareous bedrock that results in a high soil pH (Kitzler et al., 2006b). OR is situated at an elevation of about 700 m above sea level and northeast orientation. The forest consists mostly of beech, but also of various conifers, and has an average slope angle of 34%. The soil is a brown loam on carbonatic rock. SW is situated in direct vicinity of Vienna (Kitzler et al., 2006a) similar to KL that is located about 40 km south-west of Vienna (Kitzler et al., 2006a). The predominant vegetation, stand age, exposition and elevation, as well as soil type and texture of the different sampling sites are described in Table 1.
Table 1. Site and soil characteristics of the beech litter-sampling sites.
| Achenkirch (Wanek et al., 2010) | Klausenleopoldsdorf (Wanek et al., 2010) | Ort | Schottenwald (Wanek et al., 2010) | |
|---|---|---|---|---|
| Location | 47°35′N 11°39′E | 48°07′N 16°03′E | 47°51′N 13°42′E | 48°14′N 16°15′E |
| Vegetation | Spruce-fir, beech | Beech | Mainly beech, conifers | Beech |
| Stand age (years) | 127 | 62 | 90 | 142 |
| Exposition | N | NNE | NE | SE |
| Elevation (m a.s.l.) | 895 | 510 | 700 | 370 |
| Soil type | Rendzic leptosols/chromic cambisol | Dystric cambisol over sandstone | Cambisol | Dystric cambisol over sandstone |
| Soil texture | Loam | Loam-loamy clay | Loamy silt | Silty loam |
| Soil pH 0–7 cm | 6.4 | 4.6 | 3.7 | 4.4 |
Abbreviation: a.s.l., above sea level.
Litter sampling
At the respective sampling sites, 500 g of beech leaf litter was sampled in February and May 2009 (exact sampling dates are listed in Table 2). In February, all sites were covered by snow, whereas in May all sites were free of snow. Three biological replicates were collected from three plots of 10 × 10 m within an area of 50 × 50 m. Only the L-horizon was sampled carefully, but excluding mineral soil material and O-horizon. Litter samples consisted predominantly of last autumn's foliage, which means that leaves were almost intact as beech litter at the investigated sites is completely decomposed during the course of 1 year. The litter was transported to the laboratory at ambient temperature, sorted, that is, leaves other than beech leaves were removed, cut into <0.5 cm2 pieces using a kitchen knife and mixed thoroughly. Per sampling site, 10–50 g aliquots of cut leaf material from the three biological replicates per sampling site were stored at −20 °C until litter nutrient content had been analyzed, and at −80 °C until the metaproteome was analyzed.
Table 2. Different sampling sites and their mean air-temperature (T) and precipitation (Prec).
| Site | SamplingFeb | SamplingMay | TFeb (°C) | TMay (°C) | PrecFeb (mm) | PrecMay (mm) |
|---|---|---|---|---|---|---|
| Achenkirch | 4th | 18th | −1.66 | 5.88 | 55 | 65 |
| Klausenleopoldsdorf | 5th | 27th | −0.11 | 9.64 | 37 | 46 |
| Ort | 27th | 19th | 0.88 | 11.0 | 51 | 57 |
| Schottenwald | 5th | 27th | 1.32 | 11.39 | 44 | 55 |
Abbreviation: Feb, February.
Samples were collected at the given dates in 2009. Climatic data were collected over a 3-month time period before litter sampling. Mean air temperature and precipitation were calculated by averaging the climatic data of this period.
Determination of environmental parameters and litter nutrient quality
Air temperature and precipitation at the respective sampling sites were monitored for 3 months before litter collection; mean air-temperature (T) and mean precipitation (Prec) were calculated by averaging these values.
Water content (WC) was determined gravimetrically by drying the litter at 105°C for 24 h. Dry samples were ground with a mill (Retsch MM2000, Hanau, Germany) to a fine homogeneous powder. The total carbon (C) and nitrogen (N) contents of the litter were analyzed with an integrated oxidation and detection device (Leco CN2000, LECO corp. St Joseph, MI, USA). The ground samples were wet acidically oxidized (H2SO4+HNO3) in a microwave oven (CEM MARS Express), and elements (phosphorous (P), potassium (K), magnesium (Mg), manganese (Mn), calcium (Ca), iron (Fe)) were determined by inductively coupled plasma atomic emission spectrometry as described by Henschler (1988). Extractable nitrogen (N) in form of ammonium (NH4-N) and nitrate (NO3-N) and phosphorous (P) in form of phosphate (PO4-P) were determined from freshly shred litter samples, extracted with 1 M KCl at a ratio of 1:20, by shaking on a reciprocal shaker for 60 min at 200 oscillations. Extracts were stored at −20 °C until analysis. Ammonium was measured by the Berthelot reaction according to Schinner et al. (1996). Nitrate was determined with the vanadium chloride method as described by Hood-Nowotny et al. (2010). Phosphate was quantified photometrically based on the phosphomolybdate blue reaction (Schinner et al., 1996) in a microplate reader (μQuant mQx200, Bio-Tek Instruments, Winooski, VT, USA).
PLFA analysis
Lipids were extracted from 1 g sub-samples using a modified Bligh and Dyer technique (Hackl et al., 2005). Extracted PLFAs were analyzed with a HP 6890 Series GC-System and a 7683 series injector and auto sampler on a HP-5 capillary column and detected with a flame ionization detector. For identification of the fatty acid methyl esters, an external standard (bacterial acid methyl ester mix from SUPELCO) was used. For quantification of the peaks, methyl non-adecanoate fatty acid (19:0) was added. PLFA nomenclature is based upon Frostegard et al. (1993). The ratio fungal/bacterial PLFA was calculated by dividing the amount of 18:2ω6 through the amount of total bacterial PLFAs (Frostegard and Baath, 1996).
Cellulase and xylanase activity assay
Frozen cut leaf litter was ground in liquid nitrogen; 600 mg of the ground material were extracted with 4 ml extraction buffer (0.1 M sodium phosphate pH 6.0, 0.1% Triton X-100, 0.1% polyvinylpolyrrolidone). After shaking at 4 °C for 1 h, extracts were centrifuged for 5 min at 14 000 g. The resulting supernatants were filter-sterilized by cellulose acetate filters and directly subjected to enzyme activity assays.
Cellulase and xylanase activity were determined as described earlier (Riedel et al., 1998; König et al., 2002). 250 μl filter-sterilized supernatant were mixed with 250 μl 1% (w/v) carboxymethyl cellulose (Sigma, Buchs, Switzerland) or 1.5% (w/v) xylan (from oat spelts, Boehringer Ingelheim, Lot Nr. 06995) in 0.1 M sodium phosphate buffer, pH 6.0, and incubated for 3.5 h (cellulase) or 2 h (xylanase) at 40 °C. The enzymatic release of reducing groups was determined with the dinitrosalicylic acid reagent (Wood and Bhat, 1988). One unit of enzyme activity was defined as the amount of enzyme needed to release 1 μmol of glucose-equivalent reducing groups per minute.
Metaproteome analysis
From the cut and stored (−80 °C) litter material, 5 g were ground in liquid nitrogen and mixed with extraction buffer containing 1% SDS, 50 mM Tris/KOH, pH 7.0 in a 1:5 ratio (w/v). Samples were sonicated for 2 min, followed by boiling for 20 min and shaking at 4 °C for 1 h. To remove debris, extracts were centrifuged at 3000 g at 4 °C followed by 5 min centrifugation at 14 000 g and 4 °C. Supernatants were concentrated about 5-fold by vacuum-centrifugation (Eppendorf Vacuum Concentrator plus) at 30 °C. Concentrated supernatants of 25 μl were subjected to 1D SDS-PAGE (Laemmli, 1970) in a 12% polyacrylamide gel to clean samples from interfering substances (for example, humic acids) and to reduce sample complexity. Protein lanes were cut into seven slices, and gel slices were subjected to immediate in-gel tryptic digestion by employing sequencing grade-modified trypsin (Promega, Madison, WI, USA, reference V5111) (Shevchenko et al., 1996). The resulting peptide mixtures were analyzed on a hybrid LTQ-Orbitrap mass spectrometer (ThermoFischer Scientific, Waltham, MA, USA) as described earlier (Schneider et al., 2010).
Database searches
Using MASCOT (version no.2.2.04), MS and MS/MS data were searched against a database containing all proteins from UniRef100 (9808438 protein entries, downloaded from http://www.ebi.ac.uk/uniref/ at the 26 January 2010) and protein sequence information from a farm silage soil metagenome (Tringe et al., 2005) (184 374 entries, downloaded from http://img.jgi.doe.gov at 15 October 2009), as well as common contaminants such as keratin and trypsin (in total, 9 993 117 protein entries). The following search parameters were applied: (i) trypsin was chosen as protein-digesting enzyme and up to two missed cleavages were tolerated, (ii) carbamidomethylation of cysteine was chosen as fixed modification, and (iii) oxidation of methionine was chosen as variable modification. Searches were performed with a parent-ion mass tolerance of ±5 ppm and a fragment-ion mass tolerance of ±0.8 Da. A second database search was performed with the X!Tandem (version 2007.01.01.1, http://www.thegpm.org/TANDEM/) search engine (Craig and Beavis, 2004) with similar settings.
Data processing
Scaffold (version Scaffold 3.0, Proteome Software, Portland, OR, USA) was used to validate and quantify MS/MS-based peptide and protein identifications from both search engines. Protein identifications were accepted if they were established at >95% peptide probability and 90% protein probability with at least one peptide uniquely assigned to a respective protein in one of our samples. Proteins that were identified with the same set of peptides were grouped to protein clusters to satisfy the principles of parsimony. False-discovery rate was determined according to Elias and Gygi (2007) by searching against a composite version of our protein reference database, created by concatenating the target protein sequences with reversed sequences (19 986 234 entries). Starting from the Scaffold output files, all obtained protein hits were assigned to phylogenetic and functional groups and assignments were validated by the PROPHANE workflow (http://prophane.svn.sourceforge.net/viewvc/prophane/trunk/). Higher protein abundance is represented by a higher number of MS/MS spectra acquired from peptides of the respective protein. Thus, protein abundances were calculated based on the normalized spectral abundance factor (NSAF; Florens et al., 2006; Zybailov et al., 2006). This number allows relative comparison of protein abundances over different samples (Bantscheff et al., 2007).
Statistical analyses
The whole dataset was tested for normal distribution using the Shapiro–Wilk test and homogeneity of variances using Levene's test. When the assumption of normal distribution was violated, the data were transformed as indicated. Differences between sampling time and site were either checked by t-test or ANOVA, Tukey HSD. We performed single linear regression (SLR) analysis to evaluate which factors predict changes in microbial community structure and function. To identify key-parameters affecting structure and function of the decomposing community, we performed a PCA. All statistical analyses were conducted using PASW (version no. 18.0.0) statistical software packages.
Results
Litter from the sampling sites varied in nutrient content and environmental parameters
The four sampling sites AK, KL, OR and SW showed differences in their environmental conditions and leaf-litter quality. At all sampling sites, mean air temperature and precipitation increased from Feb to May. SW was the warmest of the four sites, whereas AK was the coldest (Table 2). Precipitation was highest in AK and lowest in KL at both sampling time points (Table 2).
As litter quality parameters, pH, WC and litter nutrient content were determined. WC was similar in litter of all sites in February (Table 3), which might be explained by the snow coverage of the litter. In May, WCs were generally lower, but showed site-specific differences. Litter pH was lowest in OR and highest in SW and KL. With respect to nutrient content, litter showed significant site-specific differences in C, N and P (Table 3). Carbon content was highest in AK and lowest in SW, N content was highest in SW and lowest in OR, and P content was highest in SW and lowest in AK. Huge differences were also found for the micronutrients Fe and Mn with highest values in SW and lowest in AK. Litter stoichiometry (C:nutrient and N:P ratios) differed strongly between sites. Generally, SW was the nutrient-richest litter (lowest C:N, C:P and N:P ratios), whereas AK was the nutrient-poorest litter (highest C:P, highest N:P).
Table 3. Beech leaf-litter water content, pH and nutrient content at the four sampling sites in February and May.
|
Achenkirch |
Klausenleopoldsdorf |
Ort |
Schottenwald |
|||||
|---|---|---|---|---|---|---|---|---|
| Feb | May | Feb | May | Feb | May | Feb | May | |
| Water content (%) | 73.07±0.6ab | 42.2±3.3c | 79.39±2.0a | 63.7±15.5ab | 57.0±4.1bc | 12.0±0.3d | 77.5±1.6a | 18.0±0.6d |
| pH | n.d. | 4.9±0.1cd | 4.7±0.1c | 5.3±0.1e | 4.5±0.1a | 4.6±0.1b | 5.2±0.1de | 5.2±0.1de |
| C (%) | 50.16±0.17d | 50.49±0.08d | 46.82±1.45bc | 46.12±0.68bc | 47.20±0.20bc | 47.70±0.26c | 45.47±0.39b | 43.44±1.13a |
| N (%) | 1.24±0.03d | 1.27±0.02d | 1.00±0.02b | 1.14±0.02c | 0.82±0.02a | 1.07±0.01c | 1.25±0.04d | 1.45±0.08e |
| C:N ratio | 40.31±0.76c | 39.87±0.63c | 46.76±2.32e | 40.64±0.97c | 57.75±1.25f | 44.42±0.49d | 36.43±1.19b | 29.96±1.30a |
| P (%) | 0.035±0.001a | 0.039±0.006a | 0.040±0.002a | 0.040±0.013a | 0.035±0.006a | 0.056±0.003b | 0.064±0.004b | 0.104±0.002c |
| C:P ratio | 1439.8±4.8d | 1318.6±197.9d | 1160.2±86.7cd | 1237.4±361.5d | 1361.2±200.7d | 853.24±42.42bc | 714.0±43.0ab | 418.6±17.3a |
| N:P ratio | 35.73±0.55e | 33.03±4.48de | 24.80±0.84bcd | 30.33±8.32cde | 23.62±3.86bc | 19.22±1.16ab | 19.59±0.55ab | 13.99±0.82a |
| PO4 (ng g–1 DW) | 11.51±6.15a | 14.09±1.55a | 14.87±4.97a | 9.87±4.14a | 22.73±5.39a | 90.30±6.93b | 9.38±2.48a | 113.66±6.29c |
| NH4 (μg g–1 DW) | 38.84±10.21b | 21.81±3.06a | 23.45±3.18a | 40.05±8.08b | 12.46±0.95a | 46.02±4.96b | 18.89±5.54a | 70.21±5.27c |
| NO3 (μg g–1 DW) | 4.39±1.84abc | 1.00±0.48ab | 7.31±1.87bc | 0.40±0.63a | 9.55±4.74c | 1.75±0.16ab | 5.09±2.14abc | 17.37±3.57d |
| K (%) | 0.102±0.005a | 0.087±0.008a | 0.185±0.018d | 0.151±0.007bc | 0.170±0.011cd | 0.140±0.003b | 0.163±0.006c | 0.260±0.009e |
| Ca (%) | 1.39±0.08ab | 1.32±0.001a | 1.42±0.03ab | 1.51±0.40ab | 1.35±0.08ab | 1.47±0.03ab | 1.76±0.06b | 1.37±0.01ab |
| Mg (%) | 0.209±0.013b | 0.173±0.002ab | 0.140±0.008ab | 0.160±0.069ab | 0.141±0.003ab | 0.116±0.002a | 0.182±0.012ab | 0.163±0.002ab |
| Fe (ppm) | 166.6±26.8a | 343.8±22.9a | 659.1±486.7a | 818.5±195.3a | 385.9±162.1a | 573.6±17.3a | 728.7±238.7a | 1703.9±391.9b |
| Mn (ppm) | 88.8±2.5a | 109.6±2.0a | 832.1±89.6b | 1025.2±417.2b | 1040.5±82.8b | 1172.3±16.5b | 1493.3±73.9c | 2140.7±63.7d |
| Zn (ppm) | 46.08±3.42b | 160.00±6.06d | 39.44±4.23b | 40.83±8.25b | 29.17±1.44a | 41.17±2.38b | 64.33±2.52c | 67.19±3.76c |
Abbreviations: Feb, February; n.d., not determined because of lack of material.
The different letters in a given line denote significant differences (P<0.05) between values as determined by ANOVA followed by Student–Newman–Keuls multiple range test.
Semi-quantitative metaproteome analysis
When litter samples prepared as described in the materials and methods section were analyzed by SDS-PAGE, no distinct bands were detected on the 1D-gels, but protein separation resulted in a smear covering the whole lane (Supplementary Figure 1). This might be a result of the high sample complexity and protein degradation in the litter material. Furthermore, humic substances, which interact with the Coomassie stain might be responsible for the intensive blue staining. Lanes were cut into seven equally sized gel slices and analyzed further by LC-MS/MS. Spectra were assigned to a total of 8895 proteins; protein hits that were identified based on the same set of peptides were subsequently merged to 1724 unique protein clusters. Decoy database searches resulted in a protein false-discovery rate below 1.5%. Protein assignments were validated using PROPHANE by testing cluster homology using multiple sequence alignment analyses. This resulted in 204 inconsistent clusters (amino-acid identity <50%), which were therefore excluded from further analyses.
Subsequently, PROPHANE assigned the remaining protein clusters to phylogenetic and functional groups and quantified cluster abundance by spectral counting based on the NSAF that is an indicator of relative differences in protein cluster abundances. All functional or phylogenetic group abundances presented in the figures are based on NSAFs. Proteins belonging to the respective clusters and protein identification parameters are listed in Supplementary Table 1. Supplementary Table 2 shows NSAF-based cluster abundance and a list of representative proteins assigned to the different phyla and functional categories. Supplementary Table 3 shows the sequences and charge states of all peptides that were assigned to proteins or protein clusters. The complete MASCOT results dataset including MS/MS spectra information is provided on the PRIDE database (Vizcaino et al., 2009) at http://www.ebi.ac.uk/pride/; accession number is 17171.
Litter microbial community differs between sampling sites and seasons
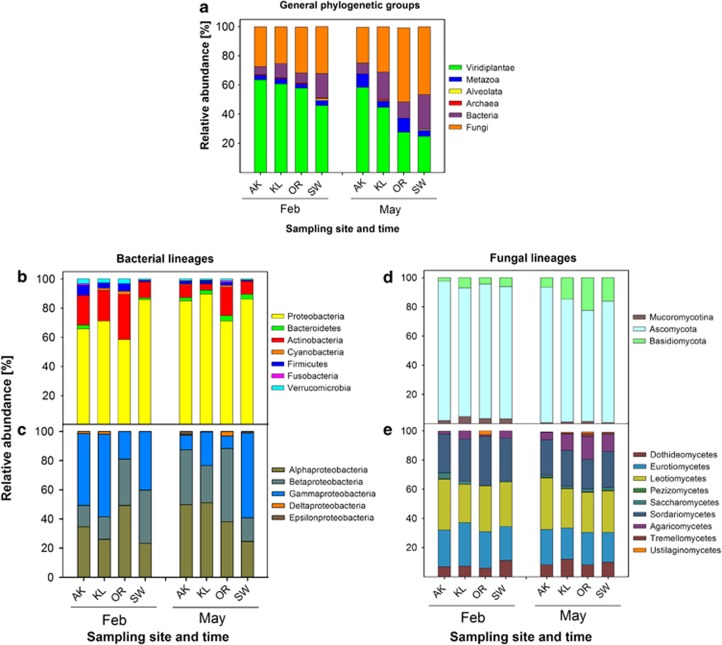
Independent of the sampling site, the majority of the spectra were assigned to three major phylogenetic groups: Viridiplantae (non-degraded leaf proteins, 45–64% of all assigned spectra), fungi (24–51%), and bacteria (5–24%). Moreover, spectra were assigned to Metazoa (3–9%), Alveolata (0–1%) and Archaea (0–0.8%) depending on sampling-site and -time (Figure 1a). We used the abundance of plant-derived spectra as a marker of the extend of litter decomposition. The obtained phylogenetic composition of the leaf-litter community is different from the proportions of database (DB) protein entries of the respective groups; in the DB, bacteria dominate the number of entries (62.2%) followed by Metazoa (16.7%), fungi (7.8%) and plants (6.3%).
Figure 1.
Assignment of spectra to taxonomic groups of organisms at different sampling times and sites. Relative abundances were calculated from the sum of NSAFs found for each group at the respective sampling sites and time. (a) General taxonomy, (b) Bacterial phyla, (c) Proteobacteria, (d) Fungal phyla, (e) Fungal classes. Groups are only presented if their relative abundance is >0.5% in the respective sample. Sampling sites are presented according to increasing mean air-temperature (from left to right).
When comparing samples collected in February and May, a significant decrease (t-test, t=2.64; P=0.019) of the spectra from plants was observed at all sampling sites. Vice versa, the number of fungal spectra increased significantly (t-test, t=−2.29; P=0.050). The number bacterial spectra also increased, but this was not significant.
A comparison of samples from the four sampling sites revealed site-specific differences of the community composition between February and May. The smallest decrease in the proportion of plant spectra was observed in AK (64 to 58%) followed by KL (60 to 45%), SW (45 to 25%) and OR (57 to 27% Figure 1a). Although the proportion of fungal spectra did not increase in AK, the proportion of fungi rose in KL from 25 to 31%, in SW from 32 to 46%, and in OR from 31 to 51%. A similar trend was observed for bacterial spectra (almost no changes in AK, 10 to 19% in KL, 7 to 11% in OR and 17 to 24% in SW). The ratio between the number of fungal and bacterial spectra (F/B-ratio) did not differ between February and May, but site-specific differences were observed. In February, the highest F/B-ratio (5.0) was found in AK and OR (4.5) followed by KL (2.6) and SW (1.9). In May the highest ratio was observed in OR (4.6) followed by AK (3.2), SW (2.4) and KL (1.6).
Fungi and bacteria are thought to be the main degraders of leaf litter and therefore their phylogeny was analyzed in more detail. Most of the bacterial spectra could be assigned to the Proteobacteria (between 59% and 90%) and the Actinobacteria (between 4% and 31% Figure 1b). In February, γ-Proteobacteria dominated in AK (50%), in KL (57%) and in SW (37%), whereas in OR, the majority of proteobacterial spectra assignments belonged to the α-Proteobacteria (49%, Figure 1c). A significant (t-test, t=−3.18; P=0.0066) absolute increase in α-proteobacterial spectra was observed from February to May in AK and KL, whereas in SW and OR, γ- and β-proteobacterial spectra dominated, respectively.
Detailed analysis of the fungal phylogeny revealed only minor differences in their community composition. A clear dominance (>80%) of ascomycotal spectra was observed at all sites and times. The proportion of basidiomycotal spectra increased slightly from February to May at all sites (Figure 1d) (t-test, t=−2.99; P<0.009). Moreover, a significant decrease in the number of Mucoromycotina-spectra was observed from February to May (t-test, t=3.37; P=0.007) at all sites. No significant changes were observed for fungal classes when different sites and times were compared (Figure 1e); most spectra fit data for the classes Leotiomycetes, Sordariomycetes and Eurotiomycetes.
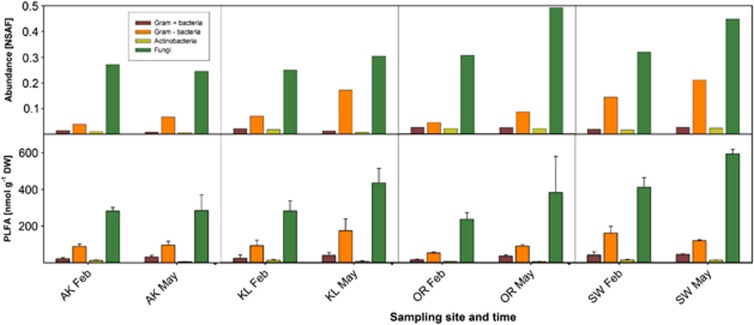
To validate data obtained from the metaproteomics analysis, PLFA analysis was performed and the phylogenetic origin of fungi present in the litter samples was investigated by cultivation and subsequent identification (Supplementary Information). To enable comparison of both datasets, spectra had to be assigned to the major taxonomic groups that can be distinguished by PLFA (Figure 2). A significant correlation of proteome and PLFA data for fungi (SLR, r2=0.380; P=0.014), total bacteria (SLR, r2=0.394; P=0.009) and Gram-negative bacteria (SLR, r2=0.352; P=0.015) was observed; Gram-positive bacteria and Actinobacteria showed similar but insignificant trends (Figure 2).
Figure 2.
Comparison of community structure based on NSAF and PLFA analyses. The upper part of the figure shows community structure based on NSAF. Data represent the mean of two biological replicates. The lower part of the figure depicts community structure based on PLFA analysis. Data represent the mean±SD of three biological replicates.
The assignment of spectra to different fungal groups was partly confirmed by cultivation experiments. The finding that the Ascomycota phylum dominates the cultivable fungi followed by Basidiomycota and Mucoromycotina is in good accordance with the metaproteome data (for details see Supplementary Information, Supplementary Table 4). Neither the metaproteome analysis nor the cultivation approach detected any members of the Glomeromycota. The absence of these arbuscular mycorrhizal fungi is in agreement with the assumption that they are only found in soil layers and in close association with plant roots (Domsch et al., 2007).
Fungi are the main producers of litter-degrading enzymes
Our metaproteomics analyses enabled us to link structure and function of the complex microbial community present in the leaf litter. The obtained spectra were classified into COG (prokaryotic proteins) and KOG (eukaryotic proteins) categories based on their respective protein assignments (Supplementary Figure 2). The majority of bacterial protein spectra were assigned to functional categories such as translation, cell wall/membrane/envelope biogenesis, post-translational modifications/protein turnover and conversion, as well as energy production and conversion. Dominant functional groups of fungal proteins were translation, chromatin structure and dynamics, post-translational modifications/protein turnover and conversion and energy production and conversion. The proportion of proteins related to transport functions was higher in fungi than in bacteria (Supplementary Figure 2). When comparing samples from February and May a significant increase of proteins assigned to post-translational modification and protein turnover was observed for both bacteria (t-test, t=−3.79; P=0.002) and fungi (t-test, t=−2.51; P=0.025).
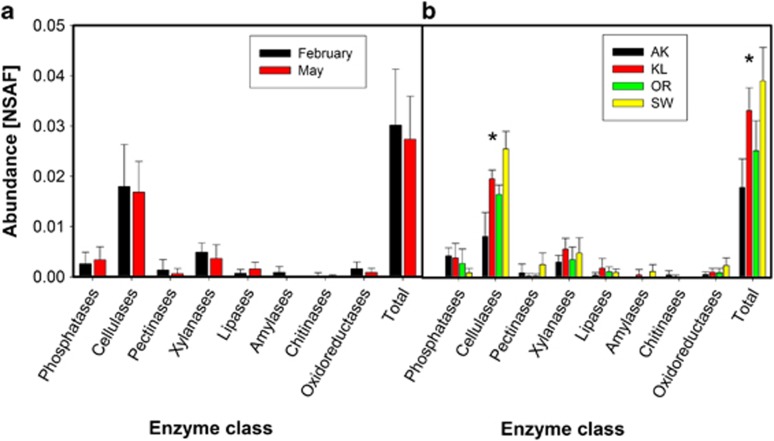
As we were mainly interested in litter-decomposing enzymes, the metaproteome was searched for proteins that were assigned as extracellular hydrolytic enzymes. Moreover, total cellulolytic and xylanolytic activities were measured by photometric assays. Strikingly, spectra could only be assigned to enzymes of fungal origin, including phosphatases, cellulases, pectinases, xylanases, lipases, amylases, chitinases and oxidoreductases (Figure 3). The most prominent hydrolases were cellulolytic enzymes, that is, exo- and endo-glucanases as well as β-glucosidases.
Figure 3.
Relative abundance of hydrolytic enzymes at different sampling times and sites. (a) Abundance of different enzymatic classes at different sampling times. Data represent mean±SD of eight samples collected at the respective sampling time. No significant differences among sites were detected by t-test analysis. (b) Abundance of different enzymatic classes at different sampling sites. Data represent mean±SD of four samples collected at the respective sampling site. *Represent significant differences, P<0.05, ANOVA followed by LSD-test.
The abundance of litter-degrading extracellular enzymes is affected by the sampling site but not by season
No significant changes were observed when the abundance of different fungal litter-degrading enzyme classes was compared between February and May (Figure 3a). However, significant differences in enzyme abundances based on the NSAF between the sampling sites were observed (ANOVA, F=10.32; P<0.01) (Figure 3b), which was particularly evident for cellulases (ANOVA, F=20.24; P<0.01). Cellulases were most abundant in SW followed by KL, OR and AK. Furthermore, obvious trends in the abundance of phosphatases were detected between the sampling sites; fungal phosphatases were most abundant in AK followed by KL, OR and SW.
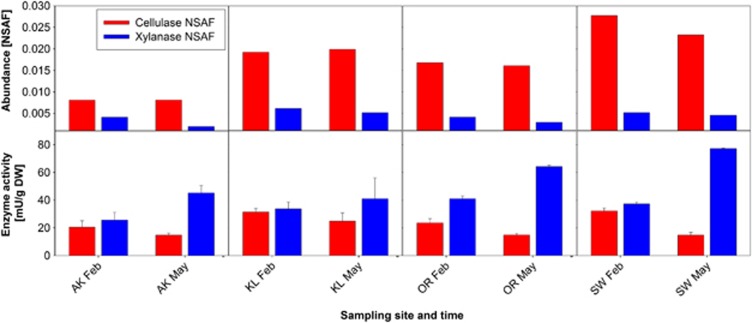
We did not observe any change in cellulolytic activity between February and May, which is in good agreement with cellulolytic enzyme abundance based on spectral counting (Figure 4). The comparison of cellulase abundance and total cellulolytic activity showed a significant but weak positive correlation (SLR, r2=0.285, P=0.033). In contrast, no correlation was observed when abundance and activity of xylanase(s) were compared (SLR, r2=0.009, P=0.279). Whereas xylanase abundance did not differ between seasons; xylanase activity was higher in May.
Figure 4.
Comparison of cellulase and xylanase abundance and total cellulolytic and xylanolytic activities. The upper part of the figure shows enzyme abundances based on NSAFs. Data represent the mean of two biological replicates. The lower part of the figure shows enzyme activities based on photometric assays. Data represent the mean±SD of three biological replicates.
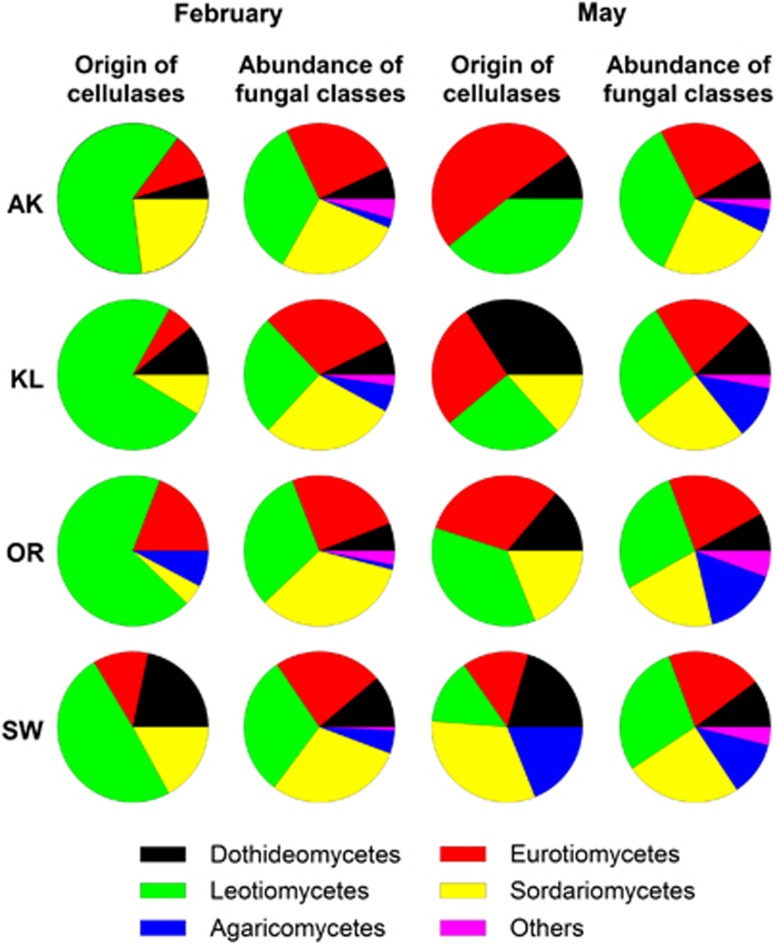
The phylogenetic origin of cellulase-producing fungi depends on the sampling time and site
Cellulases represented the most abundant enzyme class involved in litter degradation at the investigated sampling times; therefore, their phylogenetic origin was more precisely investigated (Figure 5). Although fungal community changed only slightly, dramatic changes in the cellulase-producing fungal taxa were observed when samples from February and May were compared. In February, Leotiomycetes were the main cellulase producers, whereas in May a general trend to a broader spectrum of cellulase producers was observed. Eurotiomycetes' cellulases were most abundant in AK, whereas Dothideomycetes' (KL), Leotiomycetes' (OR) and Sordariomycetes' (SW) cellulases dominated the other sampling sites.
Figure 5.
Phylogenetic origin of spectra assigned to fungal cellulases at different sampling times and sites compared with the overall distribution of fungal classes. Data represent the average of two biological replicates.
Community structure and function is influenced by season and leaf-litter nutrient content
SLR (Supplementary Figure 3) and PCA were performed to elucidate the influence of stoichiometry, season and leaf-litter nutrient content on community structure and functionality, as well as to identify the set of variables with the strongest relation to the decomposer community.
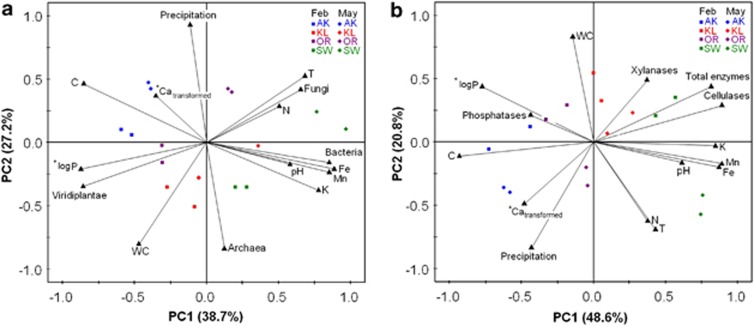
In our PCA biplots (Figures 6a, b and Supplementary Figures 4a–d), each axis (=principal component) is calculated from a set of variables, which together account for a high percentage of the variability of the data and can therefore be regarded as important drivers. The position of the sampling sites at the respective season within the biplot displays their relation to the measured variables. During litter decomposition, plant proteins decrease whereas fungal and bacterial proteins increase. On the x-axis (PC1) in Figure 6a, remaining plant proteins (‘Viridiplantae'), which can be considered as markers for the amount of remaining litter material, are allocated on the left/negative side together with the remaining C and Ca content. Bacterial proteins ordinate together with pH, K and the micronutrients Fe and Mn. Fungal proteins, temperature (T) and N content ordinate together. These variables are located opposite to the remaining plant proteins, indicating that they are related to a reduction in litter plant proteins. P content is also related to fungal growth as referred from the negative ordination of log P in the PCA (inversed direction results from log transformation). The y-axis (PC2) is dominated by the antagonistic variables precipitation and archaeal proteins. The positioning of sampling sites within the biplot and their shift from February to May towards the upper right reveals the effect of time/season factors on litter proteins: the reduction of plant proteins and the accumulation of (mainly) fungal proteins is positively correlated to warmer conditions. In addition, the individual sampling sites are separated according to their nutrient composition.
Figure 6.
PCA biplots of community structure and function obtained in the metaproteomics approach as well as enviromental data and leaf-litter nutrient content. (a) General taxonomy–community structure, (b) community function. Data points are from different sampling times (February—squares and May—diamonds) and four sampling sites. Stars indicate transformations of the variables to meet the assumption of normal distribution, P is log 10 transformed and Ca is (1/(1+log10(Ca)) transformed.; T, mean air-temperature; *indicates inversion of the vector orientation because of data transformation.
SLR (Supplementary Figure 3) and PCA were also performed to identify factors affecting enzyme abundance and activity at the different sampling sites. PCA of community function (represented by enzyme abundance) and time/season factors and nutrient content (Figure 6b) revealed relationships which support our findings on the community structure: the x-axis (PC1) can be regarded as ‘decomposition' axis with remaining, not yet respired, litter C content on the left/negative side and total enzymes, pH, K and micronutrients on the right side. Xylanases and cellulases—the main C-degrading enzymes—are ordinated opposite to litter C content. Also, Ca was inversely related to total enzyme abundance. Similarly, phosphatases were inversely related to P content. The y-axis (PC2) is dominated by season factors, such as WC and T, as well as by N content. Sampling sites are clearly separated along the x-axis due to their different nutrient and enzyme concentrations. The relation between time/season and nutrient release is indicated by an allocation of the May samples towards the warmer, drier conditions in the direction of the lower right in this plot. As enzyme abundances did not change with season, this movement is independent of enzyme ordination.
Interestingly, bacterial abundance correlated positively with the total abundance of extracellular hydrolytic enzymes (Supplementary Figure 3; r2=0.33, P=0.025) and in particular with cellulases (r2=0.44, P=0.007). However, no significant correlation between fungal abundance and extracellular hydrolytic enzymes was found (Supplementary Figure 3; r2=0.05, P=0.419 for cellulases; r2=0.02, P=0.619 for total enzymes). The bacterial relationship to cellulase abundance resulted in a negative correlation of the F/B ratio and cellulase abundance (Supplementary Figure 3; r2=0.42, P=0.002).
Discussion
Community structure of litter decomposers as revealed by metaproteomics
Metaproteomics can provide detailed information on the succession of the active part of the leaf litter inhabiting community (Nocker and Camper, 2009) and it thus enabled us to follow succession on a higher taxonomic resolution compared to e. g. PLFA (for a detailed review see Joergensen and Wichern, 2008). Ascomycota and Basidiomycota are commonly regarded as predominant fungal phyla in the soil-litter interface (Osono and Takeda, 2006). The metaproteomics data showed that the fungal community was dominated by Ascomycota and contained only a small proportion of Basidiomycota and Mucoromycotina at all sampling-sites and -times (Figure 1d). This in agreement with the results of our cultivation approach (Supplementary Information). For fungal succession it has been observed that Mucoromycotina belong to the first colonizers followed by Ascomycota (Torres et al., 2005). These fungi have limited ability to degrade lignin and are mainly regarded as cellulose decomposers or sugar fungi (Osono, 2007). Basidiomycota, with their ability to degrade the recalcitrant lignin-containing litter material, appear only later in the decomposition process (Osono, 2007; Lundell et al., 2010). Successional analysis of the fungal community showed a decrease of Mucoromycotina and Ascomycota and an increase of Basidiomycota from February to May (Figure 1d). This is in good agreement with the observed fungal succession described by Torres et al. (2005) and Osono (2007).
For bacterial succession, an increase in the proportion of Proteobacteria from winter to spring was observed, whereas that of Actinobacteria and Verrucumicrobia decreased (Figure 1b). Changes in the respective group abundances were validated by a PLFA analysis, which showed similar trends (Figure 2). A reduction of Actinobacteria was unexpected, because they are known to be involved in decomposition of organic materials, and thus are important for organic matter turnover and C cycle (Kirby, 2006). In other studies, an increase in the abundance of Actinobacteria has been shown during later stages of litter decomposition (Torres et al., 2005; Snajdr et al., 2011). The same accounts for the absence of Acidobacteria; members of this bacterial phylum can degrade various polysaccharides including cellulose and xylan (Ward et al., 2009). Based on RNA sequencing, Baldrian et al. (2012) found Acidobacteria to be the main bacterial group that was enriched in an active litter inhabiting community. The low number of acidobacterial proteins in our metaproteomics dataset might be explained by the limited number of Acidobacteria protein entries in the reference database (0.003% of database entries), which might have led to an underestimation of the contribution of Acidobacteria to the bacterial population.
Microbial succession is mainly influenced by leaf-litter quality
Previously, it has been shown that litter quality affects decomposing community and ecosystem processes (Prescott, 2010; Strickland and Rousk, 2010). A major factor in terrestrial ecosystems is pH (Sinsabaugh et al., 2008). Our results revealed that pH had an impact on the bacterial, but not on the fungal community, as seen from a strong negative correlation between pH and the F/B ratio. Bacterial abundance was higher at more basic pH values (Figure 6a, Supplementary Figures 3 and 4) as it has also been observed in other studies (Fierer and Jackson, 2006; Högberg et al., 2007). The lower impact of pH on fungal diversity might be explained by the fact that fungi can grow over a wide pH range (Penalva et al., 2008), whereas bacterial growth is restricted to a smaller pH range as has been shown in pure culture studies with isolates from natural soils (Baath, 1996).
Ecosystem processes are often controlled by the availability of N or P (Elser et al., 2007). Although litter N varied only slightly in our approach, clear differences between litter P values were present (Table 3). The SLR and PCA analysis revealed that P is a major factor influencing the abundances of all major fungal and bacterial groups with higher abundances at higher P (Figure 6a, Supplementary Figures 3 and 4). Phosphorous is needed for DNA replication and transcription and, therefore, fast growing bacteria are particularly affected under P-limiting conditions (Elser et al., 2000, 2003). Our data showing higher F/B ratios at nutrient and especially P poor sites (Supplementary Figure 3) are in good agreement with several earlier studies (Hieber and Gessner, 2002; van der Wal et al., 2006; Güsewell and Gessner, 2009).
The function of fungi and bacteria in the initial phase of litter decomposition
Fungi are thought to be key players during litter decomposition because of their ability to degrade recalcitrant compounds such as lignin and their dominance in the decomposition of cellulose and hemicellulose (de Boer et al., 2005; Meidute et al., 2008). However, our knowledge about the contribution of bacteria and fungi to this process is still scarce. Recently, the analysis of ligninolytic and chitinolytic enzymes by a targeted metatranscriptomics approach of a forest soil (Kellner et al., 2010) was performed with a focus on fungal enzymes, though neglecting the bacterial part of the community. Moreover, the analysis of a forest-soil metatranscriptome indicated that the active microbial community identified from RNA sequencing is dominated by Ascomycota and it is significantly different from the total community as indicated by DNA-based sequencing (Baldrian et al., 2012).
Our analysis of the entire community revealed that only fungi produced extracellular enzymes in the investigated environments, corroborating with earlier findings that dealt with a secretome study of two model organisms growing on beech litter as substrate (Schneider et al., 2010). Cellulases were the most abundant enzymatic class. Interestingly, the phylogenetic origin of cellulases changed over time (Figure 5) with fungal classes (Leotiomycetes, Dothideomycetes and Sordariomycetes of the Ascomycota) growing in association with leaves before litter fall as pathogens or endophytes dominating cellulase production in February. In May, the contribution of ascomycotal classes like the Eurotiomycetes and basidiomycotal classes derived from soil and known for their saprotrophic lifestyle (Domsch et al., 2007) increased. The generally low proportion of basidiomycotal cellulases might result from a low number of respective database entries. Only 21% of fungal cellulase entries are from Basidiomycota, whereas 73% belong to the Ascomycota and 6% to other fungal groups. Baldrian et al. (2012) were able to increase the number of Basidiomycota-derived transcripts of the exocellulase gene cbh1 by sequencing of Basidiomycota species with a previously unknown genome, supporting the hypothesis that basidiomycotal cellulase entries are underrepresented in public protein databases.
No enzymes of bacterial origin were detected; this is likely not a result of a low number of bacterial cellulases in the reference database (almost 60% of cellulase DB entries are of bacterial origin), but might rather be explained by a low abundance of bacterial enzymes indicating the minor importance of these enzymes in the overall decomposition process. During the investigated stages of litter decomposition, bacteria might proliferate on low molecular weight carbohydrates provided by fungal enzymes. This idea is supported by the observed strong positive correlation between bacterial abundance and the abundance of fungal enzymes (Supplementary Figure 3) and might be referred to as ‘cheating behaviour' (Velicer, 2003). Bacteria might contribute to the production of extracellular degrading enzymes later in the decomposition process as shown for Actinobacteria, a group known for its ability to utilize lignin-derived compounds (Kirby, 2006). Moreover, it was shown that bacteria dominate litter decomposition in particular micro niches where anaerobic or high temperature conditions prevail (Lynd et al., 2002).
Extracellular enzymes and their influence on decomposition
Our data indicated that the production of extracellular enzymes was influenced by litter stoichiometry. Cellulases and total enzyme abundances were highest at lowest C:P and C:N ratios (Supplementary Figure 3). C:P ratios correlated positively with the remaining plant proteins (Viridiplantae) and negatively with proteins assigned to many microbial groups (that is, Bacteroidetes, Ascomycota, Basidiomycota (Supplementary Figure 3), which shows that protein degradation and microbial community were strongly impacted by P. Sterner and Elser (2002) observed that the availability of P might be a rate-limiting factor in the synthesis of cellulolytic enzymes. The present study confirms this finding. Phosphatases ordinated opposite to P in our PCA (Figure 6b) supporting the idea that they are specifically produced when microbes have a certain demand for P (Sinsabaugh et al., 2008).
The same holds true for the release of C from litter material. Different models for C-acquisition propose a sequential decomposition of polysaccharides, starting with hemicellulose and cellulose degradation followed by the removal of lignin (Berg and Mcclaugherty, 2008; Snajdr et al., 2011). Recently, Klotzbücher et al. (2011) suggested that lignin is degraded preferentially in early phases of litter decomposition. In our approach, the dominance of xylanases and in particular cellulases indicates a decomposition phase in which hemicellulose and mainly cellulose can be considered as the major C-source. No insights into the degradation of highly recalcitrant phenolic lignin compounds were obtained because no lignolytic enzymes were detected. However, the increase of Basidiomycota, (known as main lignin decomposers) in May indicates that lignin degradation might start later in the decomposition process. Furthermore, our data imply that in litter with higher abundance of cellulose-degrading enzymes the amount of remaining degradable C is lower (Figure 6b) and the abundance of microorganisms is higher (Figure 6a, Supplementary Figure 3). This finding indicates that enzyme production might be regarded as the ‘bottleneck' of C-degradation/mineralization from litter. Similar hypotheses were postulated for soil organic-matter decomposition by Kuzyakov et al. (2009) and Schimel and Weintraub (2003).
Conclusion
After litter fall, microorganisms grow on the litter using easily accessible compounds. Microbes are affected by litter quality and environmental factors such as temperature and pH. The investigation of microbial succession revealed that Ascomycota dominated the fungal decomposer community with respect to abundance (total protein abundance) and activity (production of extracellular enzymes) especially in early stages of litter decomposition. In May, contribution of Basidiomycota to community functionality had increased in comparison with February. This strongly supports the assumption that the active part of microbial community changed over time. Only fungal, but no bacterial, polymer-degrading enzymes were detected at all sampling sites and -times, a finding that strongly supports our conclusion that fungi are the main players in litter decomposition at the investigated sites. Furthermore, a strong relationship between leaf-litter nutrient content and microbial abundance as well as cellulase production was found, indicating that high N and P content stimulate microbial decomposer activity. Accordingly, litter decomposition was fastest at sites with highest abundances of extracellular enzymes, suggesting that the production of extracellular enzymes is a rate-limiting step in the decomposition process.
We are fully aware that the metaproteomics-based approach suffers from certain limitations; the community structure and functionality assessed on the protein level might be influenced by the protein extraction efficiency, and hampered by false or missing assignments of MS and MS/MS data to protein entries in the reference database. Nevertheless, we believe that the metaproteome analysis, together with the validation by well-established complementary approaches, provides a deep insight into molecular details of the leaf-litter decomposition process. It opens up a new level of information in which both microbial succession and the activity of certain phylogenetic groups can be analyzed on the basis of their proteomes. The latter represent the active ‘building-blocks' in the ecosystem. Thus, further proteome analyses, for example, the investigation of later phases during litter decomposition or other litter types, will help to answer open questions in the field of litter decomposition: how the decomposer community further develops, who contributes to the actual degradation and which factors influence the process.
Acknowledgments
We thank Claus Beier and Ivan Ianssens for supporting the exchange grant for Katharina Keiblinger within the project ‘Climatic Change—Manipulation Experiments in Terrestrial Ecosystems' funded by the European Science Foundation (ESF). We thank Alfred Fürst from the Federal Research and Training Centre for Forests (BFW) for elemental analysis of beech litter samples and Simon Barkow–Österreicher for the help in designing the protein reference databases. Special thanks for litter sampling and discussions to Robert Jandl and Barbara Kitzler (BFW). We thank Barbara Schulz from the Technical University of Braunschweig for the critical reading of the manuscript. This research was performed within the National Research Network MICDIF (S100) of the Austrian Science Fund FWF (Project numbers S10001,2,3,4,6,7-B17). Katharina Keiblinger is a recipient of a DOC-fFORTE fellowship of the Austrian Academy of Sciences (ÖAW). The European Science Foundation (ESF, CLIMMANI Project) supported her scientific mission to Zurich.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Baath E. Adaptation of soil bacterial communities to prevailing pH in different soils. FEMS Microbiol Ecol. 1996;19:227–237. [Google Scholar]
- Baldrian P, Kolarik M, Stursova M, Kopecky J, Valaskova V, Vetrovsky T, et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012;6:248–258. doi: 10.1038/ismej.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Benndorf D, Balcke GU, Harms H, von Bergen M. Functional metaproteome analysis of protein extracts from contaminated soil and groundwater. ISME J. 2007;1:224–234. doi: 10.1038/ismej.2007.39. [DOI] [PubMed] [Google Scholar]
- Benndorf D, Vogt C, Jehmlich N, Schmidt Y, Thomas H, Woffendin G, et al. Improving protein extraction and separation methods for investigating the metaproteome of anaerobic benzene communities within sediments. Biodegradation. 2009;20:737–750. doi: 10.1007/s10532-009-9261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg B, Mcclaugherty CA.2008Plant litter: Decomposition, Humus Formation, Carbon Sequestration2nd edn.Springer: New York [Google Scholar]
- Chapin FS, Matson PA, Mooney HA. Principles of Terrestrial Ecosystem Ecology. Springer: New York; 2002. [Google Scholar]
- Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- de Boer W, Folman LB, Summerbell RC, Boddy L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, et al. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci USA. 2009;106:16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domsch KH, Gams W, Anderson TH.2007Compendium of Soil Fungi2nd edn.IHW-Verlag: Eching; p.672 [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Elser JJ, Acharya K, Kyle M, Cotner J, Makino W, Markow T, et al. Growth rate-stoichiometry couplings in diverse biota. Ecol Lett. 2003;6:936–943. [Google Scholar]
- Elser JJ, Bracken MES, Cleland EE, Gruner DS, Harpole WS, Hillebrand H, et al. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol Lett. 2007;10:1135–1142. doi: 10.1111/j.1461-0248.2007.01113.x. [DOI] [PubMed] [Google Scholar]
- Elser JJ, Sterner RW, Gorokhova E, Fagan WF, Markow TA, Cotner JB, et al. Biological stoichiometry from genes to ecosystems. Ecol Lett. 2000;3:540–550. [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frostegard A, Baath E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fertil Soils. 1996;22:59–65. [Google Scholar]
- Frostegard A, Tunlid A, Baath E. Phospholipid fatty-acid composition, biomass, and activity of microbial communities from 2 soil types experimentally exposed to different heavy-metals. Appl Environ Microbiol. 1993;59:3605–3617. doi: 10.1128/aem.59.11.3605-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güsewell S, Gessner MO. N:P ratios influence litter decomposition and colonization by fungi and bacteria in microcosms. Funct Ecol. 2009;23:211–219. [Google Scholar]
- Hackl E, Pfeffer M, Donat C, Bachmann G, Zechmeister-Boltenstern S. Composition of the microbial communities in the mineral soil under different types of natural forest. Soil Biol Biochem. 2005;37:661–671. [Google Scholar]
- Henschler G. Analysen im biologischen Material. Vol. 2. VCH Verlagsgesellschaft mbH: Weinheim; 1988. [Google Scholar]
- Hieber M, Gessner MO. Contribution of stream detrivores, fungi, and bacteria to leaf breakdown based on biomass estimates. Ecology. 2002;83:1026–1038. [Google Scholar]
- Högberg MN, Högberg P, Myrold DD. Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three. Oecologia. 2007;150:590–601. doi: 10.1007/s00442-006-0562-5. [DOI] [PubMed] [Google Scholar]
- Hood-Nowotny R, Umana NHN, Inselbacher E, Oswald-Lachouani P, Wanek W. Alternative Methods for Measuring Inorganic, Organic, and Total Dissolved Nitrogen in Soil. Soil Sci Soc Am J. 2010;74:1018–1027. [Google Scholar]
- Joergensen RG, Wichern F. Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol Biochem. 2008;40:2977–2991. [Google Scholar]
- Keiblinger KM, Hall EK, Wanek W, Szukics U, Hammerle I, Ellersdorfer G, et al. The effect of resource quantity and resource stoichiometry on microbial carbon-use-efficiency. FEMS Microbiol Ecol. 2010;73:430–440. doi: 10.1111/j.1574-6941.2010.00912.x. [DOI] [PubMed] [Google Scholar]
- Kellner H, Zak DR, Vandenbol M. Fungi unearthed: transcripts encoding lignocellulolytic and chitinolytic enzymes in forest soil. PLoS One. 2010;5:e10971. doi: 10.1371/journal.pone.0010971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby R. Actinomycetes and lignin degradation. Adv Appl Microbiol. 2006;58:125–168. [PubMed] [Google Scholar]
- Kitzler B, Zechmeister-Boltenstern S, Holtermann C, Skiba U, Butterbach-Bahl K. Nitrogen oxides emission from two beech forests subjected to different nitrogen loads. Biogeosciences. 2006a;3:293–310. [Google Scholar]
- Kitzler B, Zechmeister-Boltenstern S, Holtermann C, Skiba U, Butterbach-Bahl K. Controls over N2O, NOx and CO2 fluxes in a calcareous mountain forest soil. Biogeosciences. 2006b;3:383–395. [Google Scholar]
- Klotzbücher T, Kaiser K, Guggenberger G, Gatzek C, Kalbitz K. A new conceptual model for the fate of lignin in decomposing plant litter. Ecology. 2011;92:1052–1062. doi: 10.1890/10-1307.1. [DOI] [PubMed] [Google Scholar]
- König J, Grasser R, Pikor H, Vogel K. Determination of xylanase, beta-glucanase, and cellulase activity. Anal Bioanal Chem. 2002;374:80–87. doi: 10.1007/s00216-002-1379-7. [DOI] [PubMed] [Google Scholar]
- Kuzyakov Y, Blagodatskaya E, Blagodatsky S. Comments on the paper by Kemmitt et al. (2008) ‘Mineralization of native soil organic matter is not regulated by the size, activity or composition of the soil microbial biomass - A new perspective' [Soil Biology & Biochemistry 40, 61–73]: The biology of the Regulatory Gate. Soil Biol Biochem. 2009;41:435–439. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lundell TK, Makela MR, Hilden K. Lignin-modifying enzymes in filamentous basidiomycetes—ecological, functional and phylogenetic review. J Basic Microbiol. 2010;50:5–20. doi: 10.1002/jobm.200900338. [DOI] [PubMed] [Google Scholar]
- Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meidute S, Demoling F, Baath E. Antagonistic and synergistic effects of fungal and bacterial growth in soil after adding different carbon and nitrogen sources. Soil Biol Biochem. 2008;40:2334–2343. [Google Scholar]
- Newman DK, Banfield JF. Geomicrobiology: how molecular-scale interactions underpin biogeochemical systems. Science. 2002;296:1071–1077. doi: 10.1126/science.1010716. [DOI] [PubMed] [Google Scholar]
- Nocker A, Camper AK. Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol Lett. 2009;291:137–142. doi: 10.1111/j.1574-6968.2008.01429.x. [DOI] [PubMed] [Google Scholar]
- Osono T. Ecology of ligninolytic fungi associated with leaf litter decomposition. Ecol Res. 2007;22:955–974. [Google Scholar]
- Osono T, Takeda H. Fungal decomposition of Abies needle and Betula leaf litter. Mycologia. 2006;98:172–179. doi: 10.3852/mycologia.98.2.172. [DOI] [PubMed] [Google Scholar]
- Penalva MA, Tilburn J, Bignell E, Arst HN., Jr Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 2008;16:291–300. doi: 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Prescott CE. Litter decomposition: what controls it and how can we alter it to sequester more carbon in forest soils. Biogeochemistry. 2010;101:133–149. [Google Scholar]
- Riedel K, Ritter J, Bauer S, Bronnenmeier K. The modular cellulase CelZ of the thermophilic bacterium Clostridium stercorarium contains a thermostabilizing domain. FEMS Microbiol Lett. 1998;164:261–267. doi: 10.1111/j.1574-6968.1998.tb13096.x. [DOI] [PubMed] [Google Scholar]
- Schimel JP, Weintraub MN. The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: a theoretical model. Soil Biol Biochem. 2003;35:549–563. [Google Scholar]
- Schinner F, Öhlinger R, Kandeler E, Margesin R. Methods in Soil Biology. Springer Verlag: Berlin; 1996. [Google Scholar]
- Schneider T, Gerrits B, Gassmann R, Schmid E, Gessner MO, Richter A, et al. Proteome analysis of fungal and bacterial involvement in leaf litter decomposition. Proteomics. 2010;10:1819–1830. doi: 10.1002/pmic.200900691. [DOI] [PubMed] [Google Scholar]
- Schneider T, Riedel K. Environmental proteomics: analysis of structure and function of microbial communities. Proteomics. 2010;10:785–798. doi: 10.1002/pmic.200900450. [DOI] [PubMed] [Google Scholar]
- Schneider T, Schmid E, de Castro Junior JV, Cardinale M, Eberl L, Grube M, et al. Structure and function of the symbiosis partners of the lung lichen (Lobaria pulmonaria L. Hoffm.) analyzed by metaproteomics. Proteomics. 2011;11:2752–2756. doi: 10.1002/pmic.201000679. [DOI] [PubMed] [Google Scholar]
- Schulze WX, Gleixner G, Kaiser K, Guggenberger G, Mann M, Schulze ED. A proteomic fingerprint of dissolved organic carbon and of soil particles. Oecologia. 2005;142:335–343. doi: 10.1007/s00442-004-1698-9. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Sinsabaugh RL, Hill BH, Follstad Shah JJ. Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature. 2009;462:795–798. doi: 10.1038/nature08632. [DOI] [PubMed] [Google Scholar]
- Sinsabaugh RL, Lauber CL, Weintraub MN, Ahmed B, Allison SD, Crenshaw C, et al. Stoichiometry of soil enzyme activity at global scale. Ecol Lett. 2008;11:1252–1264. doi: 10.1111/j.1461-0248.2008.01245.x. [DOI] [PubMed] [Google Scholar]
- Snajdr J, Cajthaml T, Valaskova V, Merhautova V, Petrankova M, Spetz P, et al. Transformation of Quercus petraea litter: successive changes in litter chemistry are reflected in differential enzyme activity and changes in the microbial community composition. FEMS Microbiol Ecol. 2011;75:291–303. doi: 10.1111/j.1574-6941.2010.00999.x. [DOI] [PubMed] [Google Scholar]
- Sterner RW, Elser JJ. Ecological Stoichiometry: The Biology of Elements from Molecules to the Biosphere. Princeton University Press: Princeton; 2002. [Google Scholar]
- Strickland MS, Rousk J. Considering fungal:bacterial dominance in soils - methods, controls, and ecosystem implications. Soil Biol Biochem. 2010;42:1385–1395. [Google Scholar]
- Torres PA, Abril AB, Bucher EH. Microbial succession in litter decomposition in the semi-arid Chaco woodland. Soil Biol Biochem. 2005;37:49–54. [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- van der Wal A, van Veen JA, Smant W, Boschker HTS, Bloem J, Kardol P, et al. Fungal biomass development in a chronosequence of land abandonment. Soil Biol Biochem. 2006;38:51–60. [Google Scholar]
- Velicer GJ. Social strife in the microbial world. Trends Microbiol. 2003;11:330–337. doi: 10.1016/s0966-842x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Cote R, Reisinger F, Foster JM, Mueller M, Rameseder J, et al. A guide to the proteomics identifications database proteomics data repository. Proteomics. 2009;9:4276–4283. doi: 10.1002/pmic.200900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanek W, Mooshammer M, Blochl A, Hanreich A, Richter A. Determination of gross rates of amino acid production and immobilization in decomposing leaf litter by a novel N-15 isotope pool dilution technique. Soil Biol Biochem. 2010;42:1293–1302. [Google Scholar]
- Wang HB, Zhang ZX, Li H, He HB, Fang CX, Zhang AJ, et al. Characterization of metaproteomics in crop rhizospheric soil. J Proteome Res. 2011;10:932–940. doi: 10.1021/pr100981r. [DOI] [PubMed] [Google Scholar]
- Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol. 2009;75:2046–2056. doi: 10.1128/AEM.02294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes P, Bond PL. Microbial community proteomics: elucidating the catalysts and metabolic mechanisms that drive the Earth's biogeochemical cycles. Curr Opin Microbiol. 2009;12:310–317. doi: 10.1016/j.mib.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Wood TM, Bhat KM.1988Methods for measuring cellulase activitiesIn: Wood WA, Kellog ST (eds). Methods in Enymology Academic Press: San Diego; 87–112. [Google Scholar]
- Yadav V, Malanson G. Progress in soil organic matter research: litter decomposition, modelling, monitoring and sequestration. Prog Phys Geog. 2007;31:131–154. [Google Scholar]
- Zhang D, Hui D, Luo Y, Zhou G. Rates of litter decomposition in terrestrial ecosystems: global patterns and controlling factors. J Plant Ecol. 2008;1:85–93. [Google Scholar]
- Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.