Abstract
The quest to ‘forward-engineer’ and fabricate biological machines remains a grand challenge. Towards this end, we have fabricated locomotive “bio-bots” from hydrogels and cardiomyocytes using a 3D printer. The multi-material bio-bot consisted of a ‘biological bimorph’ cantilever structure as the actuator to power the bio-bot, and a base structure to define the asymmetric shape for locomotion. The cantilever structure was seeded with a sheet of contractile cardiomyocytes. We evaluated the locomotive mechanisms of several designs of bio-bots by changing the cantilever thickness. The bio-bot that demonstrated the most efficient mechanism of locomotion maximized the use of contractile forces for overcoming friction of the supporting leg, while preventing backward movement of the actuating leg upon relaxation. The maximum recorded velocity of the bio-bot was ~236 µm s−1, with an average displacement per power stroke of ~354 µm and average beating frequency of ~1.5 Hz.
A ‘biological machine’ or cellular system can be defined as a set of sub-components consisting of living cells and cell-instructive micro-environments that interact to perform a range of prescribed tasks1. Examples of prescribed tasks include sensing, information processing, transport, protein expression, and actuation. By combining clusters of different cell types, complex biological machines can possibly be created for specific applications in health, security, and the environment. Exemplary biological machines include organ mimics for drug testing, biological robots for replication and repair, and implantable systems for drug sensing, synthesis, and release2. An intelligent and instructive micro-environment is critical in these efforts to understand and design biological machines. The cells have to thrive, communicate, and proliferate in such a micro-environment while performing their designated functions. These engineered 2D and 3D micro-environments should form the scaffolding of biological machines, and should have spatially controlled mechanical and chemical properties to control their functionalities3. The development of enabling technologies that can fabricate the desired intelligent scaffold will greatly expedite the development of biological machines4,5,6,7,8.
Progress in developing biological actuators powered by mammalian cells has been limited to just a few reports. Montemagno and colleagues9 cultured cardiomyocytes on a patterned film of chromium and gold attached to a thin silicon beam. After releasing the microdevice, muscle contractions caused the beam to break from the rest of the structure and bend/stretch in a walking motion, traveling at a maximum speed of 38 µm s−1. Park and colleagues10 created a microrobot with grooved cantilever beams by micromolding PDMS and aligning primary cardiac cells on the grooves to enhance their contractility. Relative to flat beams, an increase in force (88%) and bending (40%) was recorded, with an average walking speed of 140 µm s−1. Finally, Parker and colleagues11 seeded cardiomyocytes on 2D thin films of PDMS, which were cut into various shapes. When released, the thin films curled or twisted into 3D conformations that performed customized functionalities of gripping, pumping, walking (133 µm s−1), and swimming functions (400 µm s−1). Recently, these thin film devices were used to reverse engineer jellyfish-like constructs with comparable functional performance12.
In this study, we built a biological machine with an actuation module for locomotion, which we refer to as an autonomous “bio-bot”. Actuation produced by a cluster of muscle cells when appropriately designed can be used to power the bio-bot. Our central hypothesis is that by integrating 3D lithographic technology with appropriate biomaterials, we can ‘forward-engineer’ spatially organize contractile cardiac cells on a bio-bot with desired geometry, mechanics, and cell adhesion molecules for optimal and robust locomotion. A stereo-lithographic apparatus (SLA) is a rapid prototyping tool13,14 used to produce 3D models, prototypes, and patterns by repetitive deposition and processing of individual layers15,16. It uses an ultraviolet laser (325 nm) to directly write on and polymerize photosensitive liquid materials based on a computer-aided design (CAD)-based digital blueprint, sliced into a collection of 2D cross-sectional layers, and processed into a real 3D part using layer-by-layer polymerization. The automated, high-throughput process can be particularly useful for the development of cellular systems due to its multi-material capability17,18,19, which has been used with photopolymerizable hydrogels20,21 to pattern cells or proteins at precise locations on the structure22,23. Cell adhesion domains, growth factors, and hydrolytic and proteolytic sequences can be incorporated into the backbone. With this technology, we expect to build biological machines with a number of prescribed tasks.
Here, we build on our earlier work of developing cell-based biohybrid actuators19 by incorporating it into the design of our autonomous bio-bots for locomotion. We show that the initial shape and degree of curvature of the bio-bot cantilever can be precisely defined by adjusting its thickness during fabrication. The tension generated by self-organizing cardiomyocytes into cell sheets on the cantilever results in the final curvature. Based on its radii of curvature, the residual stress and cell-induced surface stresses on the cantilevers can be simulated and used in future designs. By harnessing an asymmetric design and the synchronous contraction of the cardiac cell sheet, we demonstrated a walking motion of the bio-bot. Locomotion was predicated on varying friction between the bio-bot legs and the substrate during the course of contraction. We examined how altering the cantilever curvatures can change the surface area of contact, and hence, the friction, and its effect on net forward movement of the bio-bot. Together, these studies demonstrate a unique approach to the design and fabrication of a walking, autonomous bio-bot with hydrogels and cardiomyocytes, and explain the mechanisms underlying the directional actuation.
Results
Bio-Bot fabrication
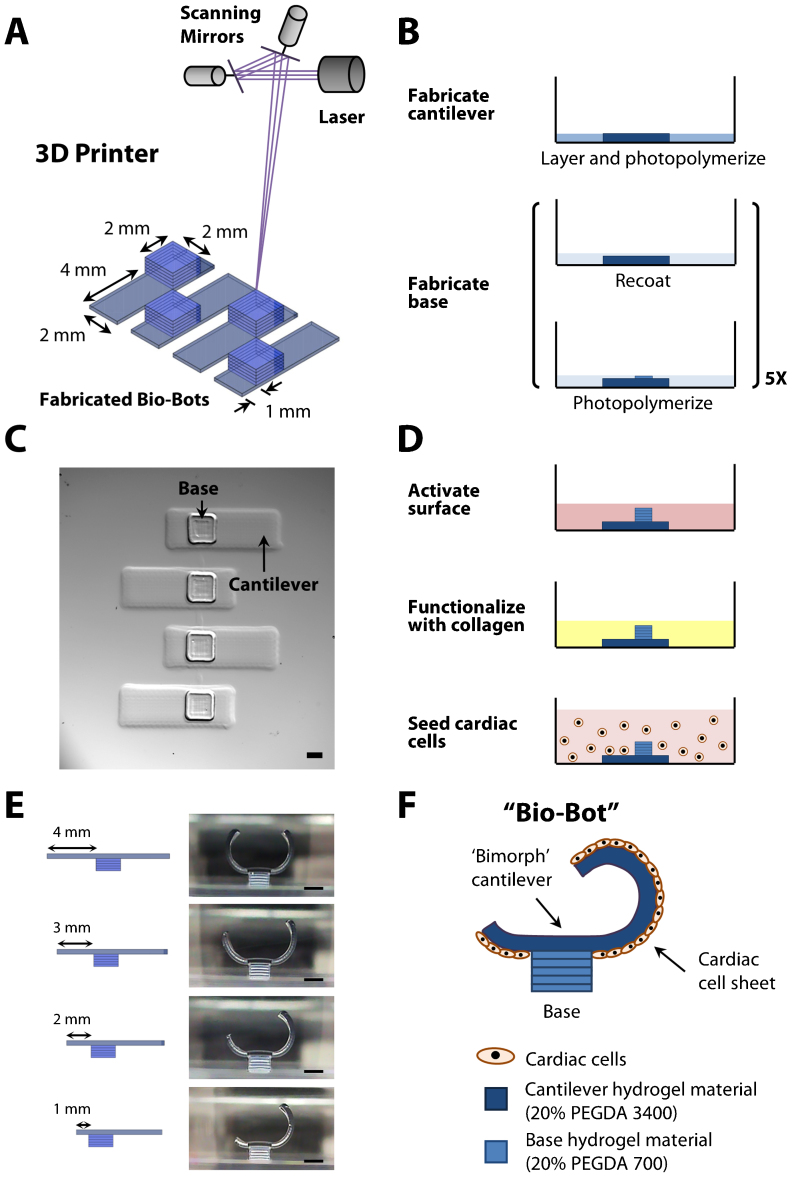
A 3D stereo-lithographic printer was used to build an array of multi-material bio-bots (Fig. 1a). Each bio-bot consisted of ‘biological bimorph’ cantilever structures and a base structure as its main components. The cantilevers were fabricated first; a pattern of the cantilevers was traced onto the surface of a thin layer of PEGDA (Mw 3400) hydrogel precursor solution with an ultraviolet (UV, 325 nm) laser beam. The part was then recoated with a thin layer of PEGDA (Mw 700) hydrogel precursor solution, and the laser traced a pattern of the bases. This layer-by-layer process was repeated until the base structures were complete (Fig. 1b). The cantilever material was chosen to mimic the elasticity of the rat heart muscle (22–50 kPa)24, while the base material was chosen to be rigid (~500 kPa) and to retain its shape19. A top down image of an array of bio-bots with cantilevers (7×2 mm) attached to their corresponding bases (2×2 mm) is shown (Fig. 1c). Since PEG is relatively inert, it requires surface modification with extracellular matrix proteins to enable subsequent cell attachment. We coated type I collagen onto the surface of the cantilever through addition reactions of light-activated Sulfo-SANPAH. Cardiomyocytes derived from neonatal rat hearts were extracted and seeded onto the cantilevers (Fig. 1d). The cardiomyocytes attached to the cantilevers, increased in size, and exhibited spontaneous contractile activity. Over several days, they produced a confluent culture and formed a cardiac cell sheet that contracted in synchrony.
Figure 1. Fabrication and cell seeding methods.
(a) Diagram of 3D stereo-lithographic printer consisting of a HeCd laser at 325 nm and galvanometer scanning mirrors. The mirrors are computer-controlled, and a stage lowers the part a specified distance after each layer. (b) Process flow diagram for high-throughput array of bio-bots. (c) Representative top-down images depicting an array of fabricated bio-bots with cantilever and base structures. (d) Process flow diagram for functionalization of the cantilevers with collagen and seeding of cardiac cells. (e) Representative cross-sectional images depicting iterative bio-bot designs from symmetry-to-asymmetry. (f) Diagram of final bio-bot design consisting of biological bimorph cantilever with seeded cardiac cell sheet. All scale bars are 1 mm.
Design considerations for locomotion
Our primary experimental approach was to use the 3D printer to generate precise and iterative designs of a bio-bot that would demonstrate and help understand the basic mechanical principles for simple locomotion. The first condition was to design a bio-bot that could produce asymmetric actuation. Actuation was a result of the contractile forces of a cardiac cell sheet to retract a cantilever structure (a power stroke). A bio-bot with appropriate asymmetry would maximize the use of a power stroke for net forward movement. To support this argument, we created a symmetric bio-bot in which the base was positioned at the center of the cantilever. While the bio-bot actuated with each power stroke, it did not generate any net forward movement (Supplementary Movies 1). Consequently, we re-positioned the base away from the center of the cantilever to establish an asymmetric structure. To accomplish this, the length of the cantilever was shortened on one side of the base. Cross-sectional images showing several iterations of the bio-bot were taken after rotating it onto its base (Fig. 1e). The last iteration was used as the final bio-bot design, as illustrated by a schematic image with attached cardiac cell sheet (Fig. 1f).
The second condition was to optimize the shape of the bio-bot, which was determined by the curvature of the cantilever, in order to regulate the friction between bio-bot “legs” and substrate. Friction is necessary to generate propulsive forces for locomotion. As it relates to the surface area of contact, friction can be optimized by using a cantilever with proper curvature. The residual surface stress generated during photopolymerization was employed to introduce a well-controlled curvature on the hydrogel cantilever following hydrogel swelling. The final curvature of the cantilever was a combined effect of the residual surface stress in the hydrogel fabrication process and a cell-induced surface stress originated from the cardiac cell sheet. By varying the thickness of the cantilever, we were able to control the initial and final curvature of the cantilever as well as the locomotive mechanism of the bio-bots. The thickness of the cantilever can be easily controlled by adjusting the volume of PEGDA (MW 3400) hydrogel precursor solution that was added during fabrication (Supplementary Fig. S1, Supplementary Text).
Extraction of residual and cell-induced surface stresses
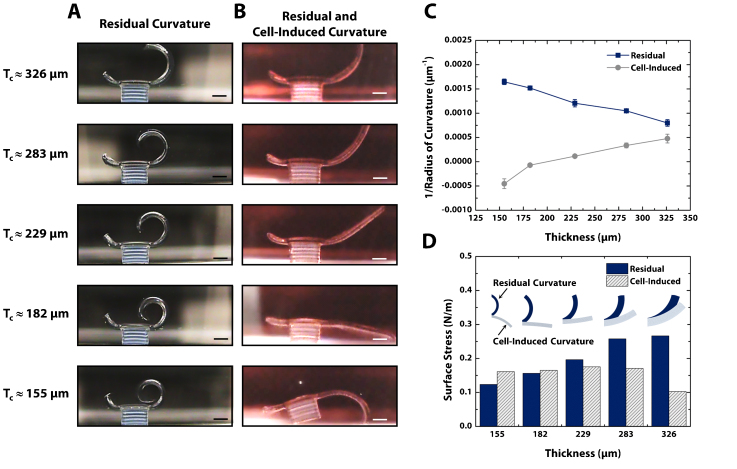
For quantitative analysis of the bending mechanism of the cantilever, bio-bots with five different cantilever thicknesses were fabricated and imaged before seeding the cardiomyocytes (Fig. 2a) and after formation of the cardiac cell sheet (Fig. 2b). The cantilevers before cell seeding were bent upward by the residual surface stress on the top side of the cantilever and the curvature of the cantilever increased with decreasing cantilever thickness. After the formation of the cardiac cell sheet, it was clearly shown that the cell-induced surface stress on the bottom side of the cantilever decreased the curvature of the cantilever or even bent the cantilever downward.
Figure 2. Design of bio-bots through residual and cell-induced surface stresses.
(a) Representative cross-sectional images of bio-bots with varying cantilever thicknesses. After overnight swelling, the residual stresses cause the cantilevers to curl upward, depending on the thickness of the cantilever. (b) Representative cross-sectional images of bio-bots three days after cardiac cell seeding on the cantilever side facing the base. Cytoskeletal tension from the cells causes the cantilever to curl downward to a final bio-bot shape. (c) Plot of inverse radius of curvature vs. cantilever thickness for residual (pre-seeded) and residual + cell-induced curvature (post-seeded). (d) Plot of surface stress vs. cantilever thickness for residual and residual + cell-induced curvature. All scale bars are 1 mm.
The radii of curvature (ROC) of the cantilevers were measured from the cross-sectional images (Supplementary Text) and presented as the inverse of the measured ROC (Fig. 2c). From the measured ROC of the cantilevers, the intrinsic residual surface stress and the cell-induced surface stress were extracted using finite element analysis (Supplementary Fig. S2 and Fig. 2d). The intrinsic residual surface stress was 200 ± 62 mN·m−1, and decreased with decreasing cantilever thickness. The origin of the residual surface stress can be attributed to the changes of laser intensity through the thickness of the cantilever during photopolymerization. As the laser penetrates the hydrogel precursor solution, its intensity decreases exponentially as described by the Beer-Lambert law. It is speculated that varying the intensity of the laser affected the mechanical properties of the hydrogel during photopolymerization, and that the variation in the swelling properties would lead to the generation of the surface stress19. The dependence of the residual surface stress on the cantilever thickness can be explained in a similar manner. As the cantilever becomes thicker, the difference in laser intensity increases and the cantilever experiences a larger variation in hydrogel swelling, resulting in higher residual surface stress. The final curvature of the cantilever was also affected by the surface stress induced by cytoskeletal tension of the cardiac cell sheet on the bottom side of the cantilever. As the cardiac cells adhered to the cantilever surface and formed a sheet, cell traction forces25 from that sheet generated a compressive surface stress on the bottom side of cantilever surface and forced the cantilever to bend downward. This cell-induced surface stress was found to be 155 ± 30 mN·m−1.
Demonstration of locomotion
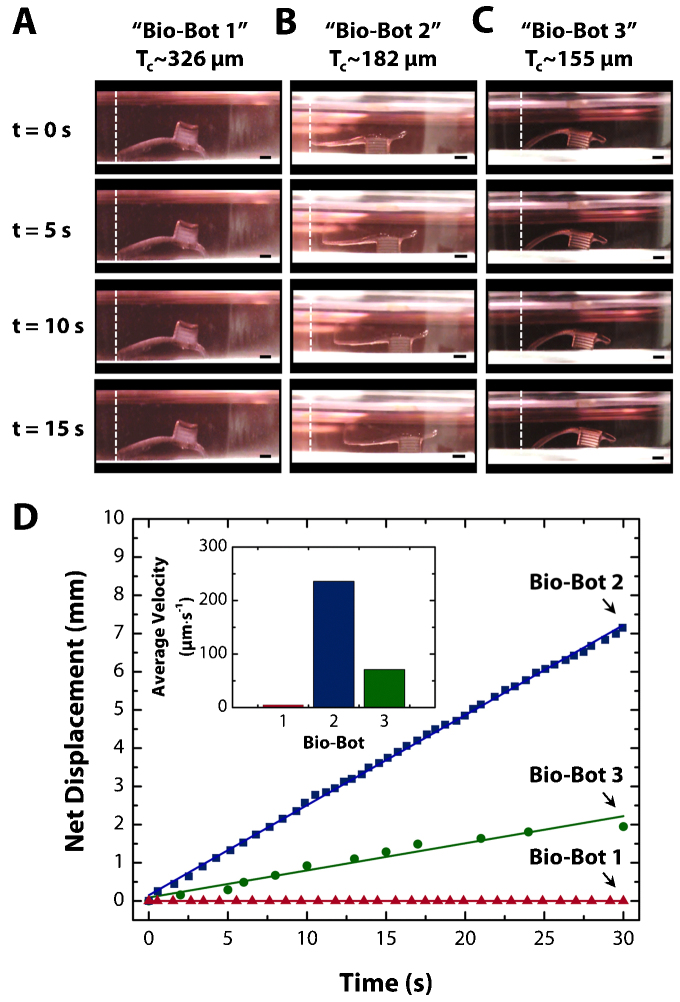
By varying the thickness and the curvature of the cantilever, three distinctive locomotive mechanisms were demonstrated (Fig. 3). Bio-bot 1 with 229~326 µm thick cantilever exhibited the first locomotive mechanism, in which the bio-bot was turned upside-down with an up-facing base so that the actuating leg would be in contact with the substrate. The 182 µm thick cantilever would result in bio-bot 2. When relaxed, the actuating leg of bio-bot 2 was not in contact with the substrate, but the leg retracted against the substrate during its power stroke. Bio-bot 3 was implemented with the 155 µm thick cantilever. The locomotive mechanism of bio-bot 3 was similar to that of bio-bot 2, except that the actuating leg was always in contact with the substrate. The movements of the bio-bots with the following cantilever thicknesses, 326 ± 17 (bio-bot 1, Supplementary Movie 2), 182 ± 31 (bio-bot 2, Supplementary Movie 3), and 155 ± 9 (bio-bot 3, Supplementary Movie 4) µm were further evaluated by quantitative characterization.
Figure 3. Demonstrations of bio-bot locomotion.
(a–c) Time course of net forward motion for “bio-bot 1” (326 μm thick), “bio-bot 2” (182 μm thick), and “bio-bot 3” (155 μm thick) over 5 second intervals for a period of 15 seconds. (d) Plot of net displacement vs. time for all three bio-bot designs. The inset is a plot of average velocity vs. bio-bot design, which is extracted from the plot of net displacement vs. time. All scale bars are 1 mm.
Time course images show net displacement of the bio-bots (Figs. 3a–c). We measured the net displacement of the bio-bots over 10 power strokes (Supplementary Fig. S3). The cumulative net displacement over 10 power strokes was 0, 3.37, and 1.82 mm, with an average displacement per power stroke of 0, 337, and 182 µm for bio-bot 1, 2, and 3, respectively. High R-squared values (above 0.999) shows that the locomotive mechanism produces a stable and identical movement of the bio-bots for each power stroke. The average beating frequencies of the cardiac cell sheets were found to be 1.14, 1.50, and 0.39 Hz for bio-bots 1, 2, and 3, respectively, at the time of recording (Supplementary Fig. S4a). The beating frequencies showed fluctuations during different measurement periods (Supplementary Fig. S4b). The cumulative net displacement over 30 seconds was 0, 7.15, and 1.95 mm, with an average velocity of 0, 236, and 71 µm·s−1 for bio-bots 1, 2, and 3, respectively (Fig. 3d).
Evaluation of the locomotive mechanisms
The driving force for locomotion of our bio-bot is the ‘biological bimorph’ cantilever consisting of a cardiac cell sheet and a variably-thick hydrogel layer. Spontaneous contraction of the cell sheet causes the cantilever to actuate and produce a power stroke. We will refer to the cantilever as the actuating leg. The power stroke of the actuating leg must exceed all opposing forces which are dominant at these length scales, including drag force and friction in an aqueous medium. Friction between the actuating leg and substrate must be present. If it is too low, the actuating leg will slide; if it is too high, it will prevent the bio-bot from moving forward. Friction between the supporting leg and substrate is also needed to balance the relaxation force of the actuating leg; otherwise, the bio-bot will slide back to its original position. Therefore, friction between the legs and substrate need to be adjusted to maximize the power stroke for net forward movement.
Friction (F) is proportional to the actual area of contact (A): 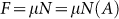 , where µ is the coefficient of friction and N is the normal force26. There are two forms of friction to consider here: kinetic and static. The applied force needed to overcome static friction of a stationary bio-bot is greater than that to keep it sliding, or kinetic friction. The angle of repose method was used to determine the coefficients of static (µs) and kinetic (µk) friction by raising and lowering the incline plane until the bio-bot started and stopped sliding, respectively. The measured angles, θs = 39.7° for static friction and θk = 18.9° for kinetic friction, were used to calculate their coefficients of friction:
, where µ is the coefficient of friction and N is the normal force26. There are two forms of friction to consider here: kinetic and static. The applied force needed to overcome static friction of a stationary bio-bot is greater than that to keep it sliding, or kinetic friction. The angle of repose method was used to determine the coefficients of static (µs) and kinetic (µk) friction by raising and lowering the incline plane until the bio-bot started and stopped sliding, respectively. The measured angles, θs = 39.7° for static friction and θk = 18.9° for kinetic friction, were used to calculate their coefficients of friction: 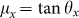 , which were µs = 0.83 and µk = 0.34. Relative friction forces were generated for each of the legs against the substrate by multiplying the area of contact by the normalized ratio of µx/µk depending on the motion state, as assessed through video capture.
, which were µs = 0.83 and µk = 0.34. Relative friction forces were generated for each of the legs against the substrate by multiplying the area of contact by the normalized ratio of µx/µk depending on the motion state, as assessed through video capture.
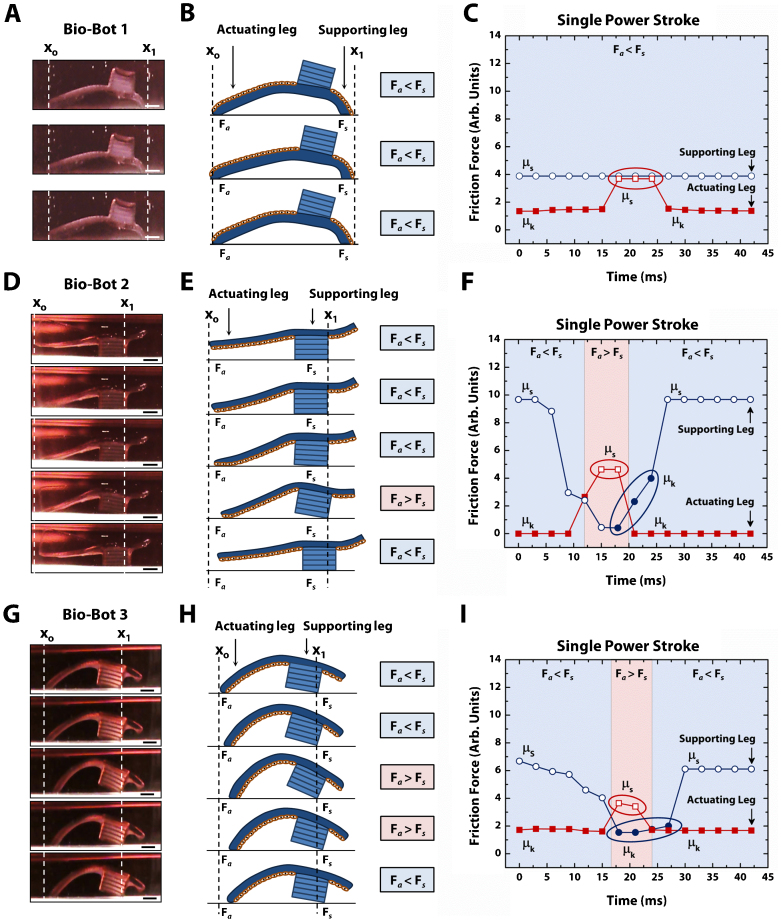
By changing the ROC of the cantilevers, we were able to generate three different iterations of bio-bots with distinct locomotive mechanisms. To evaluate how these bio-bots moved, we analyzed their representative power strokes using a digital camcorder at 30 frames per second (fps). A single power stroke spanned a total of 39, 27, and 30 ms each for bio-bots 1, 2, and 3, respectively. Bio-bot 2 produced power strokes with the shortest time and farthest distance of all three bio-bot designs. The sequence of a single power stroke for each bio-bot was revealed in several characteristic steps (Fig. 4 and Supplementary Fig. S5). We examined each of these power strokes as it relates to the aforementioned principles to evaluate the efficiency of their locomotive mechanisms:
Figure 4. Mechanisms of bio-bot locomotion.
(a) Representative cross-sectional images of a bio-bot 1 power stroke. (b) Step-by-step diagram of bio-bot 1 power stroke depicting no change between the friction forces of the actuating (Fa) and supporting legs (Fs), which results in no net forward motion. (c) Plot of friction force vs. time for a single power stroke of bio-bot 1 showing the change between the coefficients of static friction (µs, open fill) and kinetic friction (µk, solid fill). (d) Representative cross-sectional images of a bio-bot 2 power stroke. (e) Step-by-step diagram of bio-bot 2 power stroke depicting changes between Fa and Fs, which results in net forward motion. (f) Plot of friction force vs. time for a single power stroke of bio-bot 2 showing changes in µs (open fill) and µk (solid fill). (g) Representative cross-sectional images of a bio-bot 3 power stroke. (h) Step-by-step diagram of bio-bot 3 power stroke depicting changes between Fa and Fs, which results in net forward motion. (i) Plot of friction force vs. time for a single power stroke of bio-bot 3 showing changes in µs (open fill) and µk (solid fill). All scale bars are 1 mm.
Bio-bot 1 – Generally, there was no net forward movement from bio-bot 1. The actuating leg slid back and forth between power strokes and subsequent relaxation periods (Figs. 4a and b). This would indicate that there was not enough friction between the actuating leg and substrate to generate enough propulsion force to overcome the friction between the supporting legs and substrate (Fig. 4c). Without displacement of the supporting leg, the actuating leg returned to its original position. As it relates to friction, similar surface areas of contact between both legs and the substrate seem to support this argument. Without a dominant power stroke, bio-bot 1 would not able to overcome the opposing friction of the supporting leg to produce net forward movement. However, there was the rare case that bio-bot 1 moved very slowly (27 µm·s−1) (Supplementary Movies 5 and 6).
Bio-bot 2 – The mechanism of locomotion for bio-bot 2 was the most efficient and produced the largest net displacement. During the power stroke, the actuating leg bent against the substrate, which caused a conformational change in its shape (Fig. 4d and e). As a result, there was an increase in contact area of the actuating leg. The supporting leg tilted upward, causing a decrease in its area of contact. During relaxation, the bio-bot reverted back to its original conformation. Because the friction of the actuating leg was greater than the friction of the supporting leg, it resulted in net forward motion (Fig. 4f). After returning to its original conformation, the contact area (and thus, the friction) of the supporting leg increased, which prevented the bio-bot from moving backwards.
Bio-bot 3 – The mechanism of movement for bio-bot 3 worked similarly, but less efficiently. The actuating leg was already in contact with the substrate. During the power stroke, it retracted, causing a similar forward tilt of the supporting leg (Fig. 4g and h). However, the friction of the actuating leg was smaller and did not change much. The supporting leg friction decreased as a result of the tilt, causing net forward movement during relaxation. However, during relaxation, the friction of the supporting leg was not high enough to withstand the momentum from relaxation of the actuating leg, which caused the bio-bot to slide back slightly. Therefore, the net displacement of bio-bot 3 was smaller than that of bio-bot 2 (Fig. 4i).
Discussion
While various non-biological mechanical devices have been reported for locomotion on surfaces27,28 and in aquatic environments29, locomotion using biological elements has remained a major challenge. Inspired from biological micromachines, we have developed precise and repeatable methodologies for realizing autonomous, walking biological machines using a 3D stereo-lithographic printer. This technology enables convenient design and fabrication of bio-bots for actuation and locomotion. The 3D printer is particularly useful for testing numerous iterations of designs because of the reduction in cycle time for fabricating new constructs and the ease with which to do so by simply editing a CAD model. In contrast, micro-fabrication techniques require master molds patterned by photolithography. This conventional process requires clean room facilities and equipment, which limits the complexity of multi-layer constructs and the ability to make changes rapidly. Furthermore, a multi-material component can be added to the 3D printer for changing the hydrogel composition or inserting cells or proteins at precise locations in the structure. Thus, mechanical (substrate elasticity) or material (cell adhesiveness) properties of the hydrogels can be specified depending on the desired structure and function of the bio-bot to maximize force output. Hydrogels also have the unique capability of being used simultaneously as sensors, actuators, and delivery systems for therapeutic agents. Kong and colleagues demonstrated that reservoirs of living ‘cell factories’ can be embedded in a numerically-designed, 3D-printed hydrogel to produce and deliver growth factors in a sustained and targeted manner. These cell factories were used to actively stimulate and pattern new blood vessels30. Shinohara and colleagues demonstrated that glucose oxidase can be combined with pH-sensitive hydrogels to sense glucose and regulate insulin release. The presence of glucose and its subsequent conversion to gluconic acid by glucose oxidase enhanced insulin permeability through the hydrogel because of the lower pH31. These and other intelligent hydrogels, which respond to stimuli such as pH, temperature, enzyme, antigen, glucose, and electrical stimulation, can eventually be integrated into the bio-bots to add new tasks and functionalities32.
It will be necessary to further improve net displacement and longevity of the bio-bots, as well as to implement control mechanisms for developing more complex biological machines. Improved net displacement will be dependent on an optimized bio-bot design and proper formation of the cardiac cell sheet to include alignment33 and appropriate ratios of cardiomyocytes-to-fibroblasts. Increased longevity is important for long-term function of complex biological machines. The ‘biological bimorph’ cantilevers of our bio-bots perform optimally for 3–5 days before the contraction force of the power stroke decreases significantly. This is also seen in other cell-based biohybrid actuators9,10. A decrease in performance can be attributed to enzymatically-isolated cardiac myocytes reverting to a less differentiated phenotype by losing many of their myofibrils and other ultrastructural properties when cultured in monolayers. Furthermore, while a heterogeneous cell composition is necessary for the alignment, elongation, and network formation of cardiac myocytes and the secretion and degradation of ECM components34, high fractions of proliferative noncardiac cell populations can be detrimental to the performance by forming high-resistance junctions with cardiac myocytes that act as passive current sinks35. It has also been found that the ultrastructure closely resembles the adult ultrastructure if the culture cells are placed into media containing low serum concentrations which inhibits cell division and favors differentiation. By optimizing factors such as plating density, embryonic/neonatal age of the hearts, composition of the culture medium, and period in culture while minimizing damage during cell isolation, we can improve its long-term performance. Cultivation of cardiac myocytes in 3D biomaterial scaffolds along with electrically-induced stimulation rather than relying on spontaneous contraction may also promote more differentiated cellular phenotype and function36.
Future work to control when the bio-bot starts and stops or changes speed or direction can be implemented for more complex locomotion. For example, influence of various drug treatments and other control mechanisms are available37. The addition of isoproterenol (β-agonist) or carbamylcholine chloride (cholinergic agonist) will increase or decrease contraction frequency of the cardiac cell sheet, respectively. Heptanol is a gap junction blocker that will reversibly stop synchronous contraction. Pulsatile electrical stimulation can also be used to induce and pace the muscle cells38. Recently, light-induced stimulation of genetically-engineered muscle cells that express the light-activated cation channels, halorhodopsin or channelrhodopsin-2 was demonstrated39,40,41,42. Integration of these engineered muscle cells to optically drive the bio-bots offers another exciting future possibility. Furthermore, we envision that neural networks can be integrated into our bio-bots to regulate muscle contraction through neuromuscular junctions43,44. Clusters of sensory neurons can be engineered to sense and monitor the concentration of a particular species, drive the synthesis of a neurotransmitter, and communicate it to the processing neurons. The processing neurons can then initiate locomotion by translating these sensory events into action potentials that activate muscle contraction. One particular task that the bio-bots could be instructed to perform is to sense and move toward a chemical toxin and subsequently release chemicals to neutralize the toxin. Many challenges would still need to be addressed, including the development of in vitro neuromuscular junctions, neuronal circuitry to control the actuation of the muscle strips, coordination of the biological actuators for net movement, sensing of the target neurotoxins, and release of the neutralization agents by cell factories embedded in the hydrogels. Overall, the potential applications for these bio-bots can be truly transformative in the areas of biosensing, drug delivery, energy production, environmental remediation, and the development of artificial immune systems. In summary, we believe that our work provides a path to design and produce intelligent multi-cellular bio-bots for applications in health, security, and the environment.
Methods
Preparation of 3D printer and pre-polymer solution
The 3D printer, SLA 250/50 (3D Systems), was modified as described previously23. Briefly, an 18×18 mm square No. 1 cover glass (0.13–0.17 mm thick, Corning) was attached to the center of a 35 mm culture dish (Corning) using two 1×1 mm squares of double-sided tape. The dish was positioned at the center of the printing platform. Poly(ethylene glycol) diacrylate (PEGDA) of Mw 700 (Sigma-Aldrich) or 3400 (Laysan Bio) was dissolved in sterilized distilled water (dH2O) to form a 20% (w/v) pre-polymer solution. The photoinitiator, 1-[4-(2-hydroxyethoxy)-phenyl]-2-hydroxy-2-methyl-1-propane-1-one (Irgacure 2959, Ciba), was dissolved in dimethyl sulfoxide (DMSO, Fisher Scientific) at 50% (w/v) stock solution and added to the pre-polymer solution to form 0.5% (w/v) of photoinitiator.
Bio-bot fabrication
CAD models of bio-bot arrays were generated using AutoCAD 2012 (Autodesk) and exported to 3D Lightyear v1.4 (3D Systems) in .stl format. The models were sliced into thin 2D cross-sectional layers. PEGDA MW 3400 pre-polymer solution was pipetted into the culture dish at carefully characterized volumes (Supplementary Fig. S1). An ultraviolet laser beam was used to selectively crosslink the bio-bot cantilever structures at 150 mJ/cm2. The elevator controlled by the SLA 250/50 was lowered by a distance equal to the thickness of the layer. Uncrosslinked PEGDA MW 3400 was removed, and replaced with PEGDA MW 700 to begin building the bio-bot bases. PEGDA MW 700 was pipetted into the culture dish at the same volume that PEGDA MW 3400 was removed. The laser was used to selectively crosslink the bio-bot base. The part was recoated with 250 µL of PEGDA MW 700, and the process was repeated until completion of the bio-bot base structures. The bio-bot arrays were rinsed with dH2O in a shaker to remove uncrosslinked pre-polymer solution.
Surface functionalization with collagen
The bio-bot constructs invariably swelled and lifted-off the glass coverslips because of their hydrogel material. Each construct was transferred using a small spatula onto individual 12 mm circular glass coverslips (Fisher Scientific) with cantilevers down (as fabricated). Sufosuccinimidyl-6-(4′-azido-2′-nitrophenylamino)-hexanoate (Sulfo-SANPAH; Pierce Biotechnology) was prepared at 0.5 mg/mL in sterilized 50 mM HEPES, pH 8.5 immediately before use. Approximately 100 µL volume of Sulfo-SANPAH solution was added to cover the bio-bot cantilever structures. The solution was exposed to a 302 nm ultraviolet lamp (UVP Blak-Ray) at a distance of 3 inches for 5 min. The solution was aspirated out and the bio-bots were rinsed with HEPES solution to eliminate excess solution. Collagen was prepared at a concentration of 0.10 mg/mL in 0.1 N acetic acid. It was added to cover the bio-bot cantilever structures and incubated overnight at 37°C. The solution was then aspirated and rinsed with sterilized water and PBS, pH 7.4.
Rat heart dissection and cell sheet formation
Complete growth medium consisted of Dulbecco's modified Eagle's medium (DMEM, Corning cellgro), 10% horse serum (HS, Lonza), 2.5% fetal bovine serum (FBS, Atlanta Biologicals), 100 U mL−1 penicillin, and 100 µg mL−1 streptomycin (Gibco). Neonatal hearts obtained from 0–5 day old Sprague-Dawley rats were dissected according to the Institutional Animal Care and Use Committee (IACUC) at the University of Illinois, Urbana-Champaign, under an approved protocol #11160. Briefly, whole hearts were excised from the rats and immediately placed in ice-cold HBSS buffer. Ventricles were separated from the hearts and minced into 1 mm3 pieces with small scissors in ice-cold HBSS. The minced ventricles were gently rocked in sterilized 0.1% (w/v) purified trypsin (Worthington Biochemical) in ice-cold HBSS for 18 hours at 4°C. Complete growth medium was added to inhibit trypsin digestion and warmed in the water bath at 37°C for 5 min. The tissue was then digested in warm 0.1% (w/v) purified type II collagenase (Worthington Biochemical) in HBSS on a rocker for 45 min at 37°C. The tissue was gently triturated to mechanical loosen cells and passed through a 75 µm cell strainer to filter undigested connective tissue. Cells were pelleted by centrifugation at 150 x g for 5 minutes to remove the supernatant with trypsin and type II collagenase. After re-suspension in warm complete growth medium, the cells were pre-plated twice for 30 minutes each to enrich for cardiac myocytes. The cells were counted and seeded on the bio-bot cantilever structures at 1,800 cells mm−2. The cell-laden bio-bots were incubated overnight at 37°C and 5% CO2 before changing the medium.
Experimental setup and analysis
Side-view images and movies of the bio-bots were taken with a Sony Handycam DCR-SR68 camcorder at 30 frames per second (fps). The camcorder was fixed on a custom-made multi-axis stage (Thorlabs). Top-view images and movies of the bio-bots were taken with digital microscope cameras (Flex and RT3, SPOT) and software on either an inverted microscope (IX81, Olympus) or stereomicroscope (MZ FL III, Leica). Movies were recorded at approximately 11 fps. Net displacement of bio-bot forward motion was measured using the Measure Tool in Photoshop software (CS5.1, Adobe) by superimposing consecutive frames. Frames (3 ms) were extracted in Movie Maker (Windows) software. Contact area of the bio-bot legs to the substrate was also measured with side-view movies in Movie Maker and Photoshop, with the assumption that the contact was uniform into the frame.
Finite element simulation of cantilever surface stresses
The residual and cell-induced surface stresses were extracted with finite element analysis software (ANSYS). The radius of curvature (ROC) of the cantilever with varying surface stress (0~0.5 N/m) on its bottom side was simulated and the residual surface stress was interpolated from the simulation results with the measured ROC from the experimental images (Fig. 2a, Residual Curvature). The cell-induced surface stress was obtained in similar manner. The ROC of the cantilever with the previously obtained residual surface stress on the bottom side and varying surface stress (0~0.5 N/m) on the top side, as simulated and the cell-induced stress was interpolated with the experimentally measured ROC (Fig. 2b, Residual and Cell-Induced Curvature).
Author Contributions
V.C. and R.B. designed the experiments. V.C. and M.B.C. performed the experiments. V.C., K.P., T.A.S. and R.B. analyzed the data and contributed to discussion. V.C., H.K., T.A.S. and R.B. wrote and edited the manuscript.
Supplementary Material
Supplementary Information
Bio-bot 1 (326 microns)
Symmetric Bio-bot
Bio-bot 2 (182 microns)
Bio-bot 3 (155 microns)
Bio-bot 3 (155 microns) - slow back movement
Bio-bot 3 (155 microns) - slow front movement
Acknowledgments
We thank Brian Williams and Wylie Ahmed at the University of Illinois at Urbana-Champaign (UIUC) for their help with the angle of repose measurements. We thank Elise Corbin, Larry Millet, and Professor K. J. Hsia at UIUC, and Professors H. Harry Asada, Ron Weiss, and Roger D. Kamm at the Massachusetts Institute of Technology (MIT) for their helpful discussions. This project was funded by the National Science Foundation (NSF), Science and Technology Center (STC), and Emergent Behaviors in Integrated Cellular Systems (EBICS) Grant CBET-0939511 and by a cooperative agreement that was awarded to UIUC and administered by the U.S. Army Medical Research & Materiel Command (USAMRMC) and the Telemedicine & Advanced Technology Research Center (TATRC), under Contract #: W81XWH0810701.
References
- Basu S., Gerchman Y., Collins C. H., Arnold F. H. & Weiss R. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130–1134 (2005). [DOI] [PubMed] [Google Scholar]
- Kamm R. D., Nerem R. M. & Hsia K. J. Cells into systems. Mech. Eng. 30–34 (2010). [Google Scholar]
- Lutolf M. P. & Hubbell J. A. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 23, 47–55 (2005). [DOI] [PubMed] [Google Scholar]
- Chung S., Lee J. H., Moon M. W., Han J. & Kamm R. D. Non-lithographic wrinkle nanochannels for protein preconcentration. Adv. Mater. 20, 3011–3016 (2008). [Google Scholar]
- Grzybowski B. A. & Bishop K. J. Micro- and nanoprinting into solids using reaction diffusion etching and hydrogel stamps. Small 5, 22–27 (2009). [DOI] [PubMed] [Google Scholar]
- Lee S. J., Kang T., Rhie J. W. & Cho D. W. Development of three-dimensional hybrid scaffold using chondrocyte-encapsulated alginate hydrogel. Sens. Mater. 19, 445–451 (2007). [Google Scholar]
- Ling Y. et al. A cell-laden microfluidic hydrogel. Lab Chip 7, 756–762 (2007). [DOI] [PubMed] [Google Scholar]
- Park K. et al. ‘Living cantilever arrays’ for characterization of mass of single live cells in fluids. Lab Chip 8, 1034–1041 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J., Schmidt J. J. & Montemagno C. D. Self-assembled microdevices driven by muscle. Nat. Mater. 4, 180–184 (2005). [DOI] [PubMed] [Google Scholar]
- Kim J. et al. Establishment of a fabrication method for a long-term actuated hybrid cell robot. Lab Chip 7, 1504–1508 (2007). [DOI] [PubMed] [Google Scholar]
- Feinberg A. W. et al. Muscular thin films for building actuators and powering devices. Science 317, 1366–1370 (2007). [DOI] [PubMed] [Google Scholar]
- Nawroth J. C. et al. A tissue-engineered jellyfish with biomimetic propulsion. Nat. Biotech. 30, 792–797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola S. M., Melchels F. P. W., Grijpma D. W. & Kellomaki M. A review of rapid prototyping techniques for tissue engineering purposes. Ann. Med. 40, 268–280 (2008). [DOI] [PubMed] [Google Scholar]
- Burg T., Cass C. A. P., Groff R., Pepper M. & Burg K. J. L. Building off-the-shelf tissue-engineered composites. Philos. Transact. A Math Phys. Eng. Sci. 368, 1839–1862 (2010). [DOI] [PubMed] [Google Scholar]
- Mechels F. P. W., Feijen J. & Grijpma D. W. A review on stereolithography and its applications in biomedical engineering. Biomaterials 31, 6121–6130 (2010). [DOI] [PubMed] [Google Scholar]
- Jacobs P. F. Rapid prototyping and manufacturing: Fundamentals of StereoLithography (Society of Manufacturing Engineers, Dearborn, 1992). [Google Scholar]
- Arcaute K., Mann B. K. & Wicker R. B. Stereolithography of spatially controlled multi-material bioactive poly(ethylene glycol) scaffolds. Acta Biomater. 6, 1047–1054 (2010). [DOI] [PubMed] [Google Scholar]
- Zorlutuna P., Jeong J. H., Kong H. & Bashir R. Stereolithography-based hydrogel microenvironments to examine cellular interactions. Adv. Func. Mater. 21, 3642–3651 (2011). [Google Scholar]
- Chan V. et al. Multi-material bio-fabrication of hydrogel cantilevers and actuators with stereolithography. Lab Chip 10, 2062–2070 (2012). [DOI] [PubMed] [Google Scholar]
- Drury J. L. & Mooney D. J. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials 24, 4337–4351 (2003). [DOI] [PubMed] [Google Scholar]
- Slaughter B. V., Khurshid S. S., Fisher O. Z., Khademhosseini A. & Peppas N. A. Hydrogels in regenerative medicine. Adv. Mater. 21, 3307–3329 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcaute K., Mann B. K. & Wicker R. B. Stereolithography of three-dimensional bioactive poly(ethylene glycol) constructs with encapsulated cells. Ann. Biomed. Eng. 34, 1429–1441 (2006). [DOI] [PubMed] [Google Scholar]
- Chan V., Zorlutuna P., Jeong J. H., Kong H. & Bashir R. Three-dimensional photopatterning of hydrogels using stereolithography for long-term cell encapsulation. Lab Chip 10, 2062–2070 (2010). [DOI] [PubMed] [Google Scholar]
- Wang J. H. C. & Lin J. S. Cell traction force and measurement methods. Biomechan. Model Mechanobiol. 6, 361–371 (2007). [DOI] [PubMed] [Google Scholar]
- Bhana B. et al. Influence of substrate stiffness on the phenotype of heart cells. Biotechnol. Bioeng. 105, 1148–1160 (2010). [DOI] [PubMed] [Google Scholar]
- Krim J. Friction at the atomic scale. Sci. Am. 275, 74–80 (1996). [Google Scholar]
- Shepherd R. F. et al. Multigait soft robot. Proc. Natl. Acad. Sci. USA 108, 20400–20403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan B., Ji S., Kovea C. & Hosoi A. E. Mechanical devices for snail-like locomotion. J. Int. Mater. Sys. Struct. 18, 111–116 (2007). [Google Scholar]
- Sfakiotakis M., Lane D. M. & Davies J. B. C. Review of fish swimming modes for aquatic locomotion. IEEE J. Oceanic Eng. 24, 237–252 (1999). [Google Scholar]
- Jeong J. H. et al. ‘Living’ microvascular stamp for patterning of functional neovessels; orchestrated control of matrix property and geometry. Adv. Mater. 24, 58–63 (2012). [DOI] [PubMed] [Google Scholar]
- Ishihara K., Kobayashi M., Ishimaru N. & Shinohara I. Glucose induced permeation control of insulin through a complex membrane consisting of immobilized glucose oxidase and a poly(amine). Polymer J. 16, 625–631 (1984). [Google Scholar]
- Ravichandran R., Subramanian S., Jayarama R. V., Shayanti M. & Seeram R. Advances in polymeric systems for tissue engineering and biomedical applications. Macromol. Biosci. 12, 286–311 (2012). [DOI] [PubMed] [Google Scholar]
- Chan V. et al. Directed cell growth and alignment on protein-patterned 3D hydrogels with stereolithography. Virtual Phys. Prototyp. 7, 219–228 (2012). [Google Scholar]
- Luna J. I. et al. Multiscale biomimetic topography for the alignment of neonatal and embryonic stem cell-derived heart cells. Tissue Eng. Part C Methods 17, 579–588 (2011). [DOI] [PubMed] [Google Scholar]
- Bursac N. et al. Cardiac muscle tissue engineering: toward an in vitro model for electrophysiological studies. Am. J. Physiol. Heart Circ. Physiol. 277, H433–H444 (1999). [DOI] [PubMed] [Google Scholar]
- Sperelakis N. Cultured heart cell reaggregate model for studying cardiac toxicology. Environ. Health Perspect. 26 243–267 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira-Schweitzer K., Habib M., Gepstein L. & Seliktar D. A photopolymerizable hydrogel for 3-D culture of human embryonic stem cell-derived cardiomyocytes and rat neonatal cardiac cells. J. Mol. Cell Cardiol. 46, 213–224 (2009). [DOI] [PubMed] [Google Scholar]
- Berger H. J. et al. Continual electric field stimulation preserves contractile function of adult ventricular myocytes in primary culture. Am. J. Physiol. 266, H341–H349 (1994). [DOI] [PubMed] [Google Scholar]
- Bruegmann T. et al. Optogenetic control of heart muscle in vitro and in vivo. Nat. Methods 7, 897–900 (2010). [DOI] [PubMed] [Google Scholar]
- Arrenberg B., Stainier D. Y., Baier H. & Huisken J. Optogenetic control of cardiac function. Science 330, 971–974 (2010). [DOI] [PubMed] [Google Scholar]
- Asano T., Ishizua T. & Yawo H. Optically controlled contraction of photosensitive skeletal muscle cells. Biotechnol. Bioeng. 109, 199–204 (2012). [DOI] [PubMed] [Google Scholar]
- Sakar M. S. et al. Formation and optogenetic control of engineered 3D skeletal muscle bioactuators. Lab Chip 12, 4976–4985 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach J. A., Adams K. L., Gundersen C. B. & Novitch B. G. Functional neuromuscular junctions formed by embryonic stem cell-derived motor neurons. PLoS One 7, e36049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Randolph M. A., Gröger A. & Winograd J. M. Embryonic stem cell-derived motor neurons form neuromuscular junctions in vitro and enhance motor functional recovery in vivo. Plast. Reconstr. Surg. 123, 139S–148S (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Bio-bot 1 (326 microns)
Symmetric Bio-bot
Bio-bot 2 (182 microns)
Bio-bot 3 (155 microns)
Bio-bot 3 (155 microns) - slow back movement
Bio-bot 3 (155 microns) - slow front movement