Abstract
Tubby and Tubby-like proteins (TLPs) were first discovered in mammals, where they are involved in the development and function of neuronal cells. Due to their importance as plasma membrane (PM)-tethered transcription factors or mediators of vesicle trafficking, their lack causes obesity and other disease syndromes. Phosphatidylinositol 4,5-bisphosphate binding of the carboxyl-terminal Tubby domain attaches these proteins to the PM and vesicles and is essential for function. TLPs are conserved across eukaryotic kingdoms including plants, suggesting fundamental biological functions of TLPs. Plant TLPs possess an amino-terminal F-box domain that distinguishes them from other eukaryotic TLPs. Arabidopsis (Arabidopsis thaliana) encodes 11 AtTLPs that fall into six phylogenetic clades. We identified the significance of AtTLPs for root colonization of Arabidopsis by the mutualistic fungus Piriformospora indica. Our results further indicate conserved phosphatidylinositol 4,5-bisphosphate-binding sites in the Tubby domains that are required for PM anchoring of AtTLPs. More detailed studies revealed phospholipase C-triggered release of AtTLP3 from the PM, indicating a conserved mechanism as reported for mammalian Tubby and TLP3. We further show that hydrogen peroxide stimulates the release of AtTLP3 from the PM, presumably for activating downstream events. Different from mammalian homologs, the amino-terminal part of almost all AtTLPs has nucleocytosolic and plastidial localization patterns. Thus, it is tempting to assume that TLPs translate reactive oxygen species currents into signaling not only for transcriptional regulation in the nucleus but also affect plastid-associated functions after release from the PM.
Plants have to cope with a variety of abiotic and biotic stresses that can impair their development or even result in plant lethality. They have thus developed diverse strategies to counteract stress situations and evolved stress perception systems that activate adaptive stress responses. The plant immune system is highly effective in the recognition of nonself molecules, termed microbe-associated molecular patterns (MAMPs), such as chitin of fungal cell walls or flagellin of bacterial flagella. So-called pattern recognition receptors specifically recognize these MAMPs. The resulting pattern recognition receptor-mediated immune response is known as MAMP-triggered immunity and is highly effective in stopping invasions by plant pathogens (Boller and Felix, 2009). Abiotic stress sensing obviously does not rely on specific receptors. However, alterations in phospholipid properties of the plasma membrane (PM) initiate stress signaling, which is associated with Ca2+ influx and the production of reactive oxygen species (ROS; Huang et al., 2012). The PM of Arabidopsis (Arabidopsis thaliana) constitutes about 50% of phospholipids, with phosphatidylcholine and phosphatidylethanolamine as principal constituents (Uemura et al., 1995). Less than 10% of the PM consists of phosphatidylinositol (PI). A diverse set of PI kinases is able to phosphorylate the inositol group of PIs at different positions to synthesize a set of PI monophosphates and diphosphates (Munnik and Nielsen, 2011). In plants, phosphorylated PIs make less than 1% of all membrane phospholipids, among which PI diphosphate levels are particularly low (30- to 100-fold lower than PI monophosphates; Munnik and Testerink, 2009). Phosphorylated PIs such as phosphatidylinositol 4,5-bisphosphate (PIP2) are discussed to affect cell signaling by binding target proteins or enzymes and to serve as substrates for phospholipases to generate bioactive inositolphosphates (Munnik and Nielsen, 2011).
Plant hormones such as salicylic acid, jasmonate, ethylene, abscisic acid (ABA), auxin/indole-3-acetic acid, brassinosteroids, and GA substantiate and modify immune and stress signaling (Robert-Seilaniantz et al., 2011). Among these hormones, ABA takes a central role in abiotic stress signaling and in mediating stress tolerance in plants (Huang et al., 2012). ROS (e.g. hydrogen peroxide [H2O2]) are versatile messenger molecules that are produced by metabolic processes or external cues and activate stress response pathways (Mittler et al., 2011). In addition, hormone and ROS metabolism are intertwined. Hormone signaling regulates H2O2 synthesis, while, in turn, H2O2 affects hormonal cross talk (Mittler et al., 2011). However, it is almost unknown by which means cells sense ROS and how ROS currents are translated into specific signaling outputs.
Recent studies revealed the participation of Tubby-like proteins (TLPs) in plant stress signaling (Cai et al., 2008). The Tubby protein as the founding member of TLPs was first identified in mice, where its mutation causes late-onset obesity and neurosensory deficits. Tubby and other vertebrate TLPs are essential for neuronal cell development (Tubby, TLP1), spinal cord development during embryogenesis (TLP3), and skeletal development (Boggon et al., 1999; Gilissen et al., 2010; Mukhopadhyay and Jackson, 2011). TLPs are conserved among eukaryotes and carry a conserved C-terminal Tubby domain, while the N terminus is highly variable and probably specifies downstream function. In mouse (Mus musculus) cells, Tubby and TLP3 are suggested to act as stress-responsive PM-tethered transcription factors (Boggon et al., 1999; Santagata et al., 2001). PM attachment is achieved by PIP2 binding. After ligand binding, stimulation of G-protein-coupled receptors results in the activation of phospholipase C-β, which cleaves off Tubby and TLP3 from the PM, allowing their translocation into the nucleus. Since Tubby has DNA-binding activity, it is postulated to regulate stress-responsive gene expression. TLP1 might conduct similar functions, as it has DNA-binding activity (Boggon et al., 1999). In contrast, the function of TLP2 is currently unknown. In addition to TLP4, Tubby and TLP3 also participate in vesicle trafficking in the cytoplasm in cilia of mouse and human (Homo sapiens) cells (Gilissen et al., 2010; Mukhopadhyay et al., 2010). Cilia function as microtubule-based antennae. Tubby and TLPs act as bridging molecules between the intraflagellar transport-A complex and vesicles, thereby influencing the receptor composition at the PM of cilia (Mukhopadhyay et al., 2010). It is currently unknown how the function of Tubby and TLP3 in the regulation of gene transcription and vesicle trafficking is spatiotemporally organized. However, both functions crucially depend on the PIP2-binding properties of the Tubby domain (Mukhopadhyay and Jackson, 2011).
In comparison with four TLPs and one Tubby identified in vertebrates (Lai et al., 2012), TLP families are expanded in plants. While 11 TLPs have been identified in the Arabidopsis and poplar (Populus spp.) genomes, 14 rice (Oryza sativa) genes encode TLPs (Yang et al., 2008). This expansion is the result of segmental duplication (rice, poplar) and random translocation and insertion (Arabidopsis; Yang et al., 2008) and indicates a significant role of TLPs in plants. Studies from the last decade have indicated a role of OsTLP2 in plant immunity and of AtTLP9 in drought resistance and ABA signaling (Lai et al., 2004; Ko et al., 2006; Cai et al., 2008). However, the functions of other TLPs as well as the signaling mechanisms behind the characterized TLPs are less understood. The amino acid sequence of plant Tubby domains is quite divergent but contains four conserved blocks (Yang et al., 2008). Despite this partial conservation, it is currently unknown whether plant TLPs bear functional sites and activities, as reported for their mammalian relatives. The structure of the N terminus of plant TLPs is highly conserved. Similar to other plants, Arabidopsis TLPs, except AtTLP8, contain an F-box (FB) domain, which is not found in mammalian TLPs. A highly conserved linker sequence connects the FB to the Tubby domain (Lai et al., 2004; Yang et al., 2008). With about 700 members, FB proteins constitute the largest family of ubiquitin E3 ligases in Arabidopsis and are part of the SCF complex, which functions in the degradation of proteins, thereby regulating various biological processes. This complex consists of Cullin1, Arabidopsis Suppressor of Kinetochore Proteins (ASKs), Ring-Box1/Regulator of Cullin1, and FB proteins. The latter determine substrate and thus protein degradation specificity (Lechner et al., 2006). SCF-mediated degradation of negative regulators or repressor proteins is essentially required for plant development (e.g. lateral root formation) and physiological responses to hormone signaling (Lechner et al., 2006; Robert-Seilaniantz et al., 2011).
In addition to the evolution of effective stress perception and adaptation systems, plants gain protection by establishing symbioses with mutualistic microbes. Such beneficial interactions enhance biotic and abiotic stress resistance of plants. Mutualistic microbes commonly possess a restricted range of potential host plants. By contrast, an apparently unlimited number of plants is able to establish mutualistic symbioses with the fungus Piriformospora indica (Qiang et al., 2012) that initially establishes root symbioses with living cells. This colonization phase lasts until 3 d after inoculation (dai) and is followed by a cell death-dependent colonization phase (more than 3 dai; Jacobs et al., 2011). While analyzing plant factors determining root colonization by P. indica, we observed the significance of AtTLPs for P. indica colonization. Therefore, we examined the subcellular localization of AtTLPs and searched for stimuli that affect their localization in order to uncover signaling processes regulated by AtTLPs. We identified localization patterns and translocation mechanisms that show the conservation of those reported for mammalian Tubby and TLP3. By analyzing AtTLP3 in more detail, we detected a putative role of this protein in abiotic stress and ROS signaling.
RESULTS
TLPs Support Colonization by the Mutualistic Fungus P. indica But Do Not Affect Pathogenic Interactions
Microarray analysis revealed the induction of a TLP gene during the colonization of barley (Hordeum vulgare) roots by P. indica (Schäfer et al., 2009). Recent studies demonstrated that P. indica colonizes Arabidopsis roots in a similar manner to that observed before for barley (Deshmukh et al., 2006; Schäfer et al., 2009; Jacobs et al., 2011). In order to examine systematically the effect of TLPs on P. indica colonization, we used Arabidopsis attlp transferred DNA (T-DNA) insertion mutants for our studies. TLPs of Arabidopsis fall into six clades: AtTLP1, -5, and -10 (clade A1), AtTLP2 and -6 (clade A2), AtTLP3, -9, and -11 (clade A3), AtTLP7 (clade A4), AtTLP4 (clade B), and AtTLP8 (clade C; Yang et al., 2008). We selected two representative members of each clade A1 (AtTLP5 and -10) and A3 (AtTLP3 and -9), while mutant lines attlp7 and attlp8 were chosen as sole members of clades A4 and C, respectively. In the case of clade A2, we were unable to obtain knockout mutants of AtTLP6; therefore, we only analyzed the colonization of attlp2. Except for AtTLP7, for which no second independent insertion line was available, we analyzed two independent mutant lines per gene. AtTLP4 was considered a pseudogene (Lai et al., 2004). Consistent with this, we could not amplify any transcript of AtTLP4 by PCR and did not include attlp4 mutants in our studies. All T-DNA insertion lines used showed a reduction or a lack of target TLP transcripts (Supplemental Fig. S1).
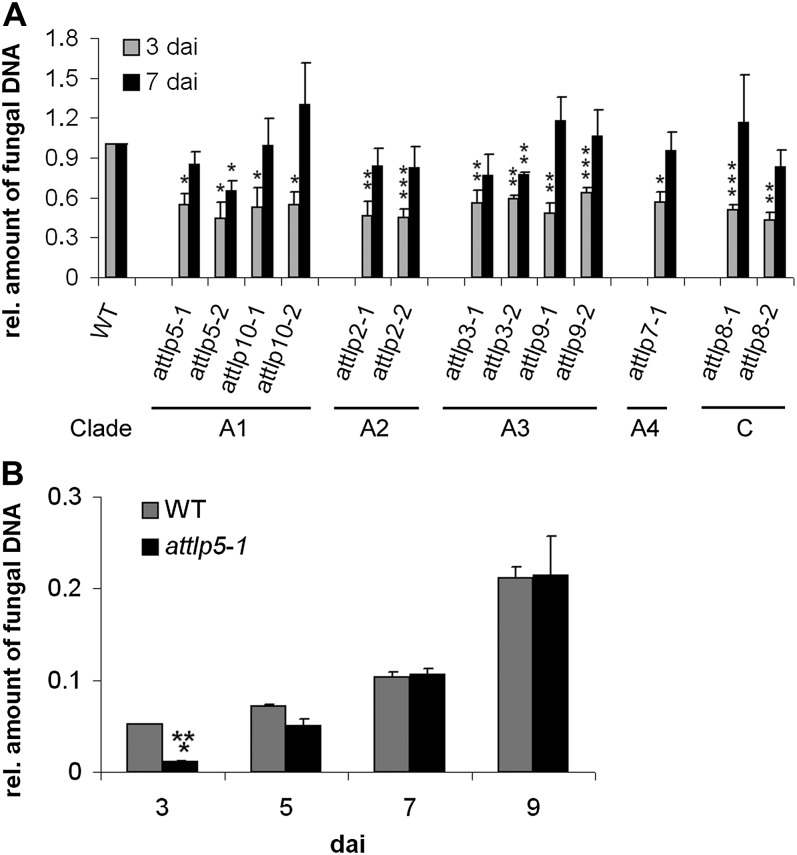
The amount of fungal DNA, as an indicator for root colonization, was quantified in mutant and wild-type roots by quantitative real-time (qRT)-PCR. All lines showed a significantly reduced colonization at 3 dai (biotrophic phase) with P. indica, although the colonization of mutant roots reached wild-type levels at 7 dai (cell death-dependent phase). Only attlp3-2 and attlp5-2 showed significantly reduced colonization at the later time point (Fig. 1A). As the delayed colonization patterns observed in most attlp mutants might indicate an improved colonization at later stages (more than 7 dai), we monitored root colonization of the representative mutant line attlp5-1 at 3, 5, 7, and 9 dai. This analysis confirmed the delayed colonization phenotype in attlp5-1, which was indicated by a reduced colonization at 3 dai, while no differences were detected at 7 and 9 dai (Fig. 1B). We further compared the expression of AtTLP2, -3, -5, -7, -8, -9, and -10 in P. indica-colonized and noncolonized wild-type roots at 1, 3, and 7 dai, but none of these genes showed a strong differential regulation during fungal colonization (Supplemental Fig. S2).
Figure 1.
Delayed colonization of attlp mutants by P. indica. Three-week-old plants were inoculated with P. indica, and fungal biomass was determined during biotrophic (3 dai) and cell death-dependent (7 dai) colonization (A) or at 3, 5, 7, and 9 dai (B) by qRT-PCR. A, Clades indicate the membership of tested mutants. The relative amount of fungal biomass in mutant roots was related to the wild type (WT; set to 1). Data are averages from at least three independent experiments, and error bars represent se. B, This experiment was performed twice with similar results. se values are from three technical replicates of one biological experiment. For both experiments, 200 plants were analyzed per mutant or wild type and per time point. Asterisks indicate significance at P < 0.05 (*), P < 0.01 (**), or P < 0.001 (***) as analyzed by Student’s t test.
P. indica suppresses root immunity in order to colonize roots (Jacobs et al., 2011). Delayed colonization might thus be due to enhanced activation of immune signaling. Therefore, we examined flg22-triggered seedling growth inhibition and P. indica’s ability to suppress it in attlp mutants. MAMP (flg22)-triggered seedling growth inhibition was virtually identical in attlp mutants and the wild type, while P. indica successfully suppressed flg22-induced seedling growth inhibition in all the mutants (Supplemental Fig. S3). To see whether the lack of AtTLPs generally affects host immunity, we tested attlp mutants for pathogenesis of the leaf pathogens Erysiphe cruciferarum and Botrytis cinerea as well as the root pathogen Phytophthora parasitica. The susceptibility of attlp2-1, -3-1, -5-1, or -8-1 to B. cinerea (Supplemental Fig. S4A), of attlp2-1, -3-1, -5-1, or 9-1 to P. parasitica (Supplemental Fig. S4B), and of attlp3-1, -3-2, or -5-2 to E. cruciferarum (Supplemental Fig. S4C) was almost indistinguishable from the respective wild-type plants. In addition, attlp3 roots normally expressed immune marker and stress-related genes when compared with the wild type with and without P. indica colonization (Supplemental Fig. S5). These results suggest that the delayed colonization of attlp mutants was not caused by a general activation of plant immunity but might indicate a lack of compatibility.
Conserved Three-Dimensional Protein Structure of TLPs
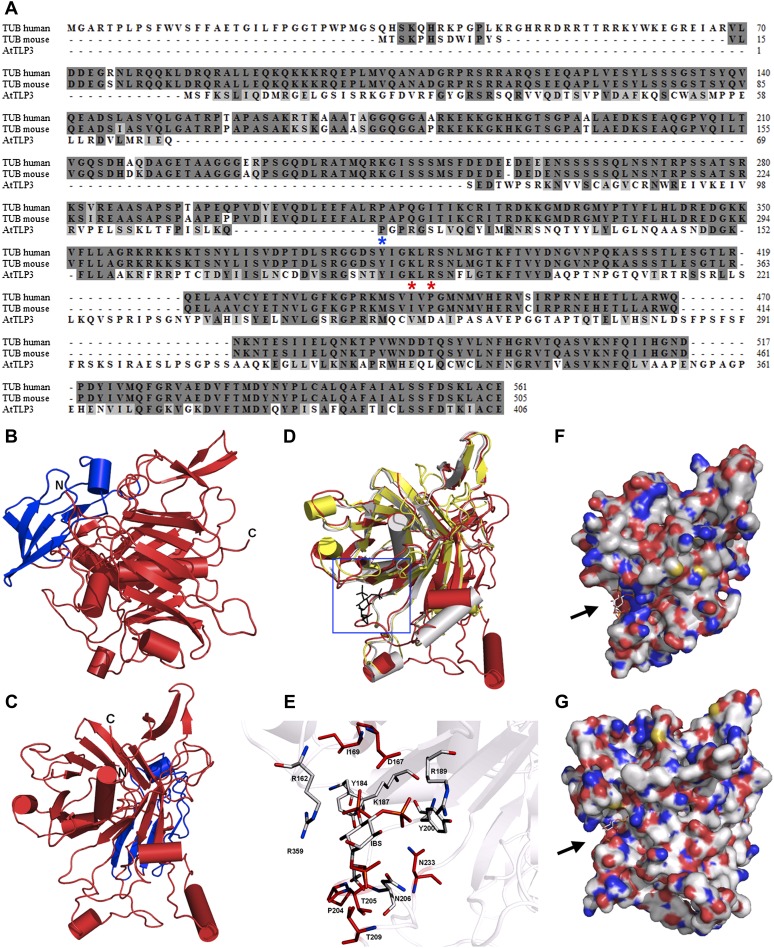
We performed three-dimensional (3D) homology modeling to determine the protein structure of AtTLP3, as a representative member of AtTLPs. AtTLP3 showed 35% to 38% amino acid conservation in comparison with mammalian TLPs (Fig. 2A). The mouse Tubby protein has been crystallized and structurally resolved by Boggon et al. (1999; Protein Data Bank [PDB] identifier 1C8Z) and served as a template for 3D modeling, as did the crystal structure of the human TLP1 isoform a (PDB identifier 1S31). The 3D alignment with both structures revealed that AtTLP3 may possess a highly conserved 3D orientation of α-helices and β-sheets (Fig. 2, B–D) of the Tubby domain, while some other secondary structure elements and the FB domain are unique features for a plant TLP such as AtTLP3 (Fig. 2, B–D). Moreover, the studies confirmed conservation of the PIP2-binding site (Fig. 2D, blue box). The superposition with the crystal structure of the mouse Tubby protein bound to the PIP2 analog l-α-glycerophospho-d-myoinositol 4,5-bisphosphate (PDB identifier 1I7E; Santagata et al., 2001) showed high positional conformity of the amino acid residues (Fig. 2E) that are essential for inositol lipid binding. The charged amino acid residues AtTLP3 Lys-187/Arg-189 show high positional overlaps with the mouse Tubby protein residues Lys-330/Arg-332 or human TLP1 Lys-386/Arg-388, which represent the determinants of PIP2 binding of mammalian TLPs (Fig. 2E). We observed the most significant difference in the PIP2-binding cavity at the amino acid residues Pro-204/Thr-205 in AtTLP3 and the corresponding residues in the mouse protein, Gly-346/Val-347, which results in a change of the run of the backbone (Fig. 2, E–G). Importantly, the 3D modeling did not show conservation of the DNA-binding cavity (arrows in Fig. 2, F and G) within the Tubby domain of the mouse Tubby protein and AtTLP3, which became apparent by the comparison of the amino acid charges on their surfaces (Fig. 2, F and G). However, the overall structures of the Tubby domains of human and mouse TLPs and AtTLP3 are highly conserved.
Figure 2.
Sequence and 3D structure of AtTLP3 and its Tubby domain. A, Sequence alignment for human (accession no. 19923167) and mouse (accession no. 11230782) Tubby (TUB) protein with AtTLP3 (accession no. 30690823). Identical and similar amino acids are shaded in dark gray and light gray, respectively. The two amino acids that are necessary for PIP2 binding (Santagata et al., 2001) are indicated (red asterisks) underneath those amino acids. The blue asterisk underneath one amino acid indicates the start of the Tubby domain. B and C, 3D homology model of AtTLP3 showing the FB domain (blue) and the Tubby domain (red). The N and C termini are highlighted. D, Superposition of the structural model of AtTLP3 (red) with the crystal structure of the human Tubby protein (PDB identifier 1S31; yellow) and the crystal structure of mouse brain Tubby protein (gray) bound to PIP2 (black, blue frame). E, Magnification of the frame in D shows the highly conserved inositol lipid-binding domain of AtTLP3. A comparison of AtTLP3, mouse, and human Tubby proteins was done. The PIP2 analog l-α-glycerophospho-d-myoinositol 4,5-bisphosphate (IBS) was used to show the conservation of binding sites. Amino acid residues that are similar in type and position (conformation) in all three proteins are colored in gray, and amino acids that are specific for the AtTLP3 protein are marked in red. F and G, Surface representation of the mouse Tubby protein with bound IBS (F) and structural model of AtTLP3 (G) with modeled IBS. The groove of highly positive charge (blue regions) is more significant in the mouse protein, and the dimension of the groove differs in the AtTLP3 structural model.
Subcellular Localization of TLPs
We adopted a biolistic transformation assay to examine the subcellular localization of AtTLPs fused to GFP. Full-length versions of AtTLP2, -3, and -7 under the control of the cauliflower mosaic virus 35S promoter fused either at the N or C terminus to GFP were not detectable by confocal laser-scanning microscopy in Arabidopsis leaf epidermal cells. As failed detection might reflect cell death occurrence, the absence of cell autofluorescence and the clear detection of cobombarded mCherry rather suggests the instability of the AtTLP-GFP fusion proteins.
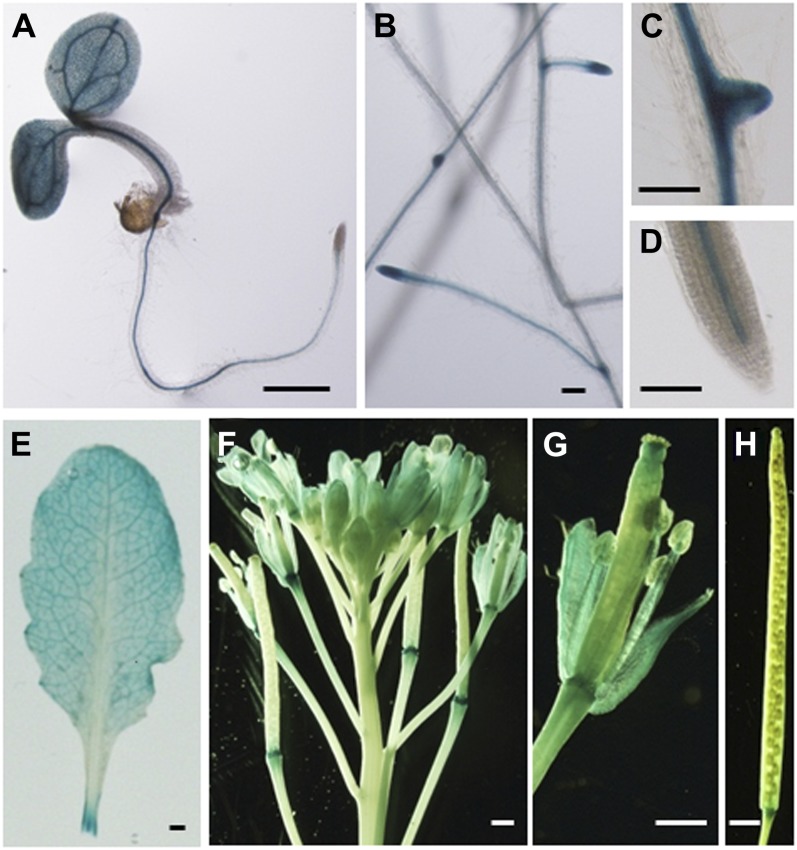
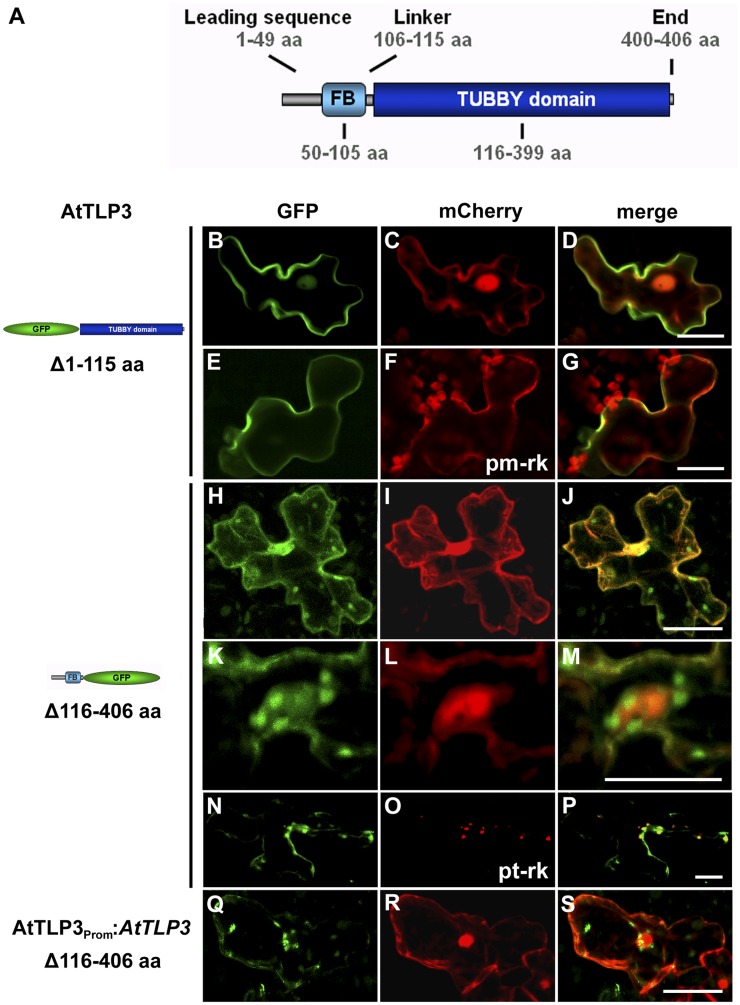
For further localization studies, we decided to detect the C terminus (CT) and N terminus (NT) of AtTLP3 separately. In Arabidopsis lines expressing AtTLP3PROM:GUS, GUS was detected in most organs (except mature siliques), including roots (especially in the stele, root primordia, root tips) and leaves of seedlings as well as adult plants and anthotaxies (e.g. petals, stamens; Fig. 3). These results are in agreement with observations made by Lai et al. (2004), who studied the expression of AtTLP3 using PCR. AtTLP3 consists of a leading sequence (1–49 amino acids), the FB domain (50–105 amino acids), a linker sequence (106–115 amino acids), the Tubby domain (116–399 amino acids), and an end sequence (400–406 amino acids; Fig. 4A). All AtTLPs except AtTLP8, which lacks a FB, have a highly similar protein structure (Yang et al., 2008). NT-AtTLP3 (Δ116–406) included the FB domain, while CT-AtTLP3 (Δ1–115) contained the Tubby domain. We fused N- and C-terminal variants to GFP and shot them into Arabidopsis leaves. Soluble mCherry was cobombarded and served as cytosolic and nucleoplasmic markers. GFP-CT-AtTLP3 accumulated at the PM of transformed cells, which was distinct from the cytosolic and nucleoplasmic localization of coexpressed mCherry (Fig. 4, B–D). We confirmed the PM localization of GFP-CT-AtTLP3 by cotransformation of cells with the PM marker pm-rk (Nelson et al., 2007; Fig. 4, E–G). For the N-terminal parts of AtTLPs, in silico protein localization analyses using WoLF PSORT (Horton et al., 2007) predicted plastidial localization for all of them except AtTLP2 (cytosol), AtTLP7 (mitochondria), and AtTLP8 (nucleus; Supplemental Table S1). Although full-length AtTLP3-GFP was not detectable, NT-AtTLP3-GFP was detected in plastids, nuclei, and cytosol of transformed cells (Fig. 4, H–P). Interestingly, plastids often accumulated around the nucleus (Fig. 4, K–M). We confirmed the plastidial localization of NT-AtTLP3-GFP by cobombardment with the plastidial marker pt-rk (Nelson et al., 2007; Fig. 4, N–P). In order to exclude localization artifacts due to cauliflower mosaic virus 35S-driven overexpression, we transformed cells with NT-AtTLP3-GFP under the control of its own promoter (AtTLP3PROM:AtTLP3 Δ116–406). This fusion protein also displayed plastidial localization (Fig. 4, Q–S). To further substantiate these results, the subcellular localization of the C terminus and N terminus of AtTLP2, -7, -8, and -10 as representative members of each clade was determined. Similar to AtTLP3, the Tubby domains of AtTLP2, -7, and -10 showed PM localization, while the FB domains showed plastidial localization (Supplemental Fig. S6). By contrast, NT-AtTLP8-GFP and GFP-CT-AtTLP8 showed cytoplasmic and nuclear localization (Supplemental Fig. S6).
Figure 3.
Organ- and tissue-specific expression of AtTLP3 in AtTLP3Prom:GUS plants. Four-day-old seedling (A), roots of 18-d-old plants (B–D), rosette leaf (E), anthotaxy (F), single flower with some sepals and petals removed (G), and mature silique (H) are shown. Bars = 1 mm in A and E to H and 0.1 mm in B to D.
Figure 4.
Subcellular localization of the FB and Tubby domains of AtTLP3. A, Model of AtTLP3 with the N-terminal FB domain and the C-terminal Tubby domain. Numbers indicate the lengths and positions of domains within AtTLP3 in amino acids (aa). For all experiments below, leaf cells were cotransformed with the GFP-CT-AtTLP3 (Δ1–115) (B and E) or NT-AtTLP3-GFP (Δ116–406) (H, K, and N) fusions under the control of the 35S promoter (B–P) or the native promoter (Q) and cytosolic and nucleoplasmic marker mCherry (C, I, L, and R), PM marker pm-rk (F), or plastidial marker pt-rk (O) by biolistic transformation. D, G, J, M, P, and S are merged images indicating the subcellular localization of domains. Yellow color indicates the colocalization of green- and red-fluorescing proteins. Constructs used for transformation are indicated on the left. B to D, PM localization of GFP-CT-AtTLP3. E to G, GFP-CT-AtTLP3 colocalizes with the PM marker pm-rk at the PM. H to S, Plastidial and nucleocytosolic localization of NT-AtTLP3-GFP. K to M, Accumulation of plastids around the nucleus in a cell expressing NT-AtTLP3-GFP. N to P, Plastidial localization was confirmed by cotransformation of the NT-AtTLP3-GFP fusion with the plastidial marker pt-rk. Q to S, Cells transformed with NT-AtTLP3-GFP under the control of its endogenous promoter (AtTLP3PROM) also showed plastidial, cytosolic, and nucleoplasmic localization. Bars = 20 µm.
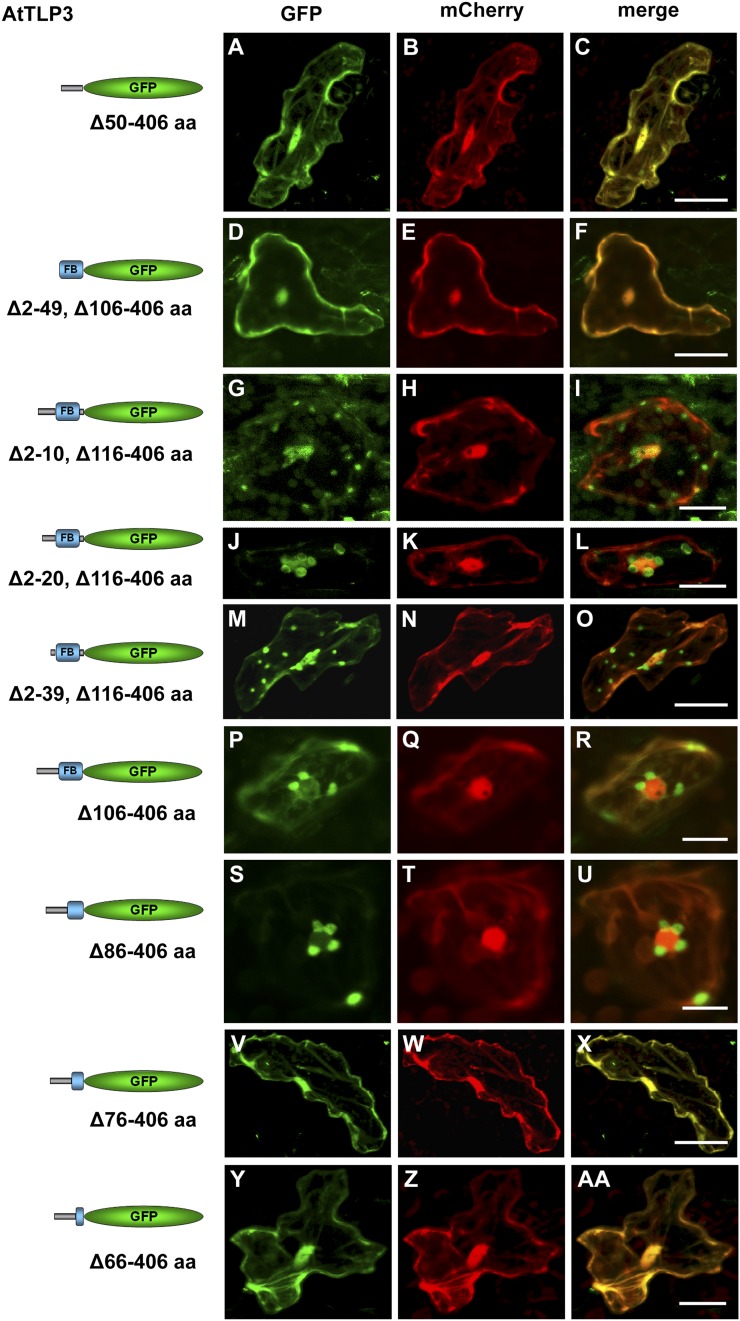
A Stretch of the Leading Sequence and FB Domain Are Required for Plastidial Localization of the N Terminus of AtTLP3
In silico analyses did not reveal a plastid-targeting peptide in AtTLP3. Therefore, we examined which part of the N-terminal sequence is critical for the plastidial localization of NT-AtTLP3 and generated a series of truncated NT-AtTLP3 versions used for biolistic transformation. Plastidial localization was neither exclusively determined by the leading sequence (NT-AtTLP3 Δ50–406-GFP; Fig. 5, A–C) nor the FB domain (NT-AtTLP3 Δ2–49, Δ106–406-GFP; Fig. 5, D–F), as indicated by the nucleocytosolic localization of the truncated GFP fusions. Next, we generated a series of NT-AtTLP3 constructs with successive truncations within the leading sequence (Fig. 5, G–O). Even the elimination of the first 39 amino acids (Δ2–39, Δ106–406) resulted in plastidial localization of NT-AtTLP3. Elimination of the linker sequence (Δ106–406) also did not hinder plastidial localization (Fig. 5, P–R). Further successive truncations at the C-terminal part of the FB domain (without changing the leading sequence) resulted in a loss of plastidial localization (Fig. 5, S–AA). While a truncated protein version lacking amino acids 86 to 406 (Δ86–406) showed plastidial localization (Fig. 5, S–U), a loss of amino acids 76 to 406 (Δ76–406) or 66 to 406 (Δ66–406) resulted in nucleocytosolic localization (Fig. 5, V–AA). These results suggest the presence of a target sequence for plastidial localization, which lies mainly within the FB domain.
Figure 5.
Parts of the leading sequence and FB domain determine plastidial localization of the N terminus of AtTLP3. A, D, G, J, M, P, S, V, and Y, Arabidopsis leaf cells were transiently transformed with truncated versions of the N terminus of AtTLP3 fused to GFP. Constructs used for transformation are indicated on the left. B, E, H, K, N, Q, T, W, and Z, Leaf cells were cotransformed with the cytosolic and nucleoplasmic marker mCherry. C, F, I, L, O, R, U, X, and AA, Merged images indicate the subcellular localization of truncated versions. Yellow color indicates the colocalization of green- and red-fluorescing proteins. All truncated versions lack amino acids (aa) of the C terminus (Tubby domain; amino acids 116–406) of AtTLP3, while the linker sequence (amino acids 106–115) is either missing (Δ116–406) or included (Δ106–406). In addition, various amino acids of the N terminus are missing as indicated. A to F, Cytosolic and nucleoplasmic localization of GFP fused to the leading sequence (Δ50–406; A–C) or the FB domain (Δ2–49, Δ106–406; D–F). G to O, Plastidial and nucleocytosolic localization of GFP fused to truncated N-terminal versions lacking amino acids 2 to 10 (Δ2–10, Δ106–406; G–I), 2 to 20 (Δ2–20, Δ106–406; J–l), or 2 to 39 (Δ2–39, Δ106–406; M–O). P to R, Plastidial localization of GFP fused to an N-terminal version lacking the linker sequence and Tubby domain (Δ106–406). S to U, Plastidial localization of GFP fused to an N-terminal version lacking the linker sequence, Tubby domain, and the last 20 amino acids of the FB (Δ86–406). V to AA, Cytosolic and nucleoplasmic localization of GFP fused to an N-terminal version lacking the linker sequence, Tubby domain, and the last 30 (Δ76–406; V–X) or 40 (Δ66–406; Y–AA) amino acids of the FB. Experiments were repeated three times with similar results. Bars = 20 µm.
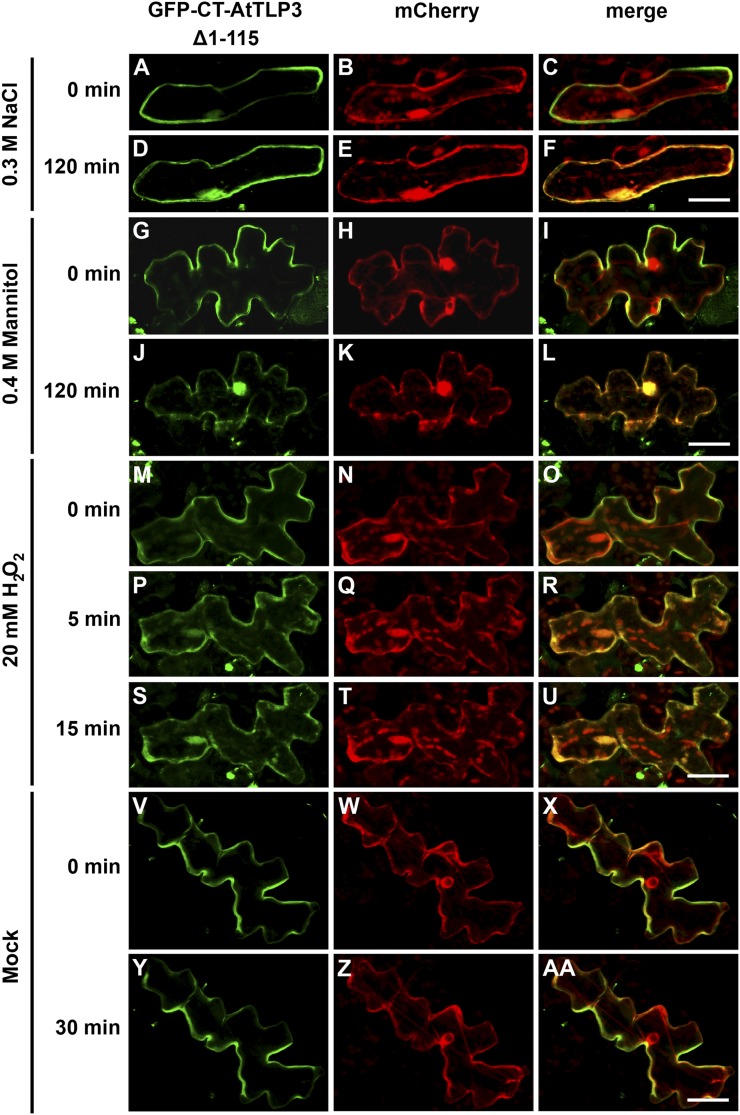
Abiotic Stress Triggers PM Dislodgement of the AtTLP3 Tubby Domain
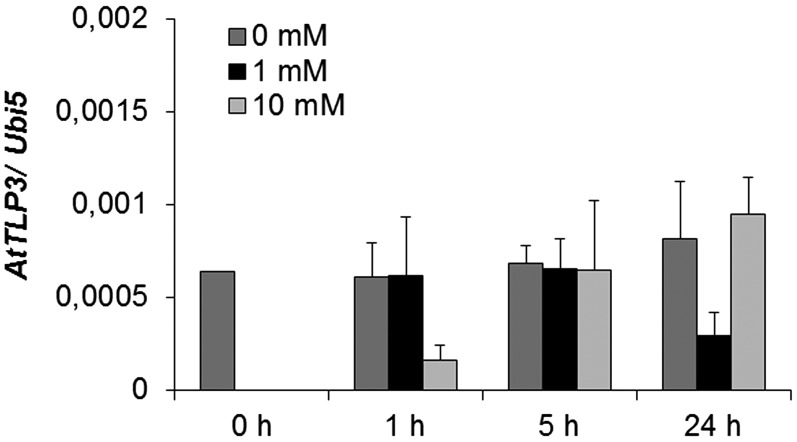
In mice, activation of G-protein-coupled receptors induces the release of Tubby and TLP3 from the PM (Santagata et al., 2001) and their subsequent translocation to the nucleus. We applied phytohormones (jasmonates, ABA, salicylic acid, indole-3-acetic acid/auxin, brassinosteroids, GA) and abiotic stress (mannitol, NaCl) to leaf cells transiently expressing the GFP-tagged Tubby domain (GFP-CT-AtTLP3 Δ1–115). None of the hormones induced the relocalization of the Tubby domain of AtTLP3 (data not shown). In contrast, treatment with either 0.3 m NaCl or 0.4 m mannitol resulted in translocation of Tubby into the cytosol and nucleoplasm within 120 min of application, as indicated by increasing colocalization with mCherry (Fig. 6, A–L). The respective mock treatment with bathing solution, which simulated apoplastic fluid conditions, did not result in the relocalization of the Tubby domain (data not shown). Next, we examined whether oxidative stress accompanying abiotic stress might be responsible for the observed translocation. Therefore, we treated transformed cells with 20 mm H2O2. This treatment initiated rapid relocalization of the Tubby domain of AtTLP3 within 5 min (Fig. 6, P–R) and complete Tubby domain detachment by 15 min (Fig. 6, S–U). Relocalization was not observed in mock-treated cells (Fig. 6, V–AA). AtTLP3 was down-regulated in Arabidopsis plants after H2O2 treatment (Fig. 7). Interestingly, whereas 1 mm H2O2 resulted in lower AtTLP3 transcript levels at 24 h after treatment, 10 mm H2O2 caused its suppression at 1 h after treatment. To determine altered oxidative stress sensitivity, we quantified ion leakage in attlp3 mutants and wild-type leaf discs in response to H2O2 treatment. This treatment did not reveal altered H2O2 sensitivity in attlp3 mutants (Supplemental Fig. S7).
Figure 6.
Osmotic stress-induced translocation of the membrane-tethered AtTLP3 Tubby domain to plastids. A, D, G, J, M, P, S, V, Y, Arabidopsis leaf cells were transformed with GFP-CT-AtTLP3 (Δ1–115). B, E, H, K, N, Q, T, W, Z, Arabidopsis leaf cells were transformed with the cytosolic and nucleoplasmic marker mCherry. C, F, I, L, O, R, U, X, AA, Merged images indicate the subcellular localization of truncated domain versions. Yellow color indicates the colocalization of green- and red-fluorescing proteins. A to F, Application of 0.3 m NaCl results in the relocalization of the C terminus of AtTLP3 [GFP-CT-AtTLP3 (Δ1–115)] from the PM (A–C) to the cytosol and nucleus within 120 min (D–F). G to L, Application of 0.4 m mannitol results in the relocalization of GFP-CT-AtTLP3 (Δ1–115) from the PM (G–I) to the cytosol and nucleus within 120 min (J–L). Exposure of cells to mannitol caused plasmolysis, thereby slightly changing the shape of cells (J–L). M to U, Application of 20 mm H2O2 results in the relocalization of GFP-CT-AtTLP3 (Δ1–115) from the PM to the cytosol and nucleus. M to O, PM localization of GFP-CT-AtTLP3 before treatment (0 min). P to R, Cytosolic and nucleoplasmic localization of GFP-CT-AtTLP3 (Δ1–115) at 5 min after application of 20 mm H2O2. S to U, Cytosolic and nucleoplasmic localization of GFP-CT-AtTLP3 (Δ1–115) at 15 min after application of 20 mm H2O2. V to AA, PM localization of GFP-CT-AtTLP3 (Δ1–115) at 0 and 30 min after mock treatment. Experiments were repeated three times with similar results. Bars = 20 µm.
Figure 7.
Expression of AtTLP3 in Arabidopsis plants treated with H2O2. Arabidopsis plants were harvested at the indicated time points after mock treatment or treatment with 1 mm or 10 mm H2O2. Expression values were calculated by the 2−ΔCt method by relating cycle thresholds of AtTLP3 to those of the housekeeping gene AtUbi5. Values are means ± se of three independent experiments.
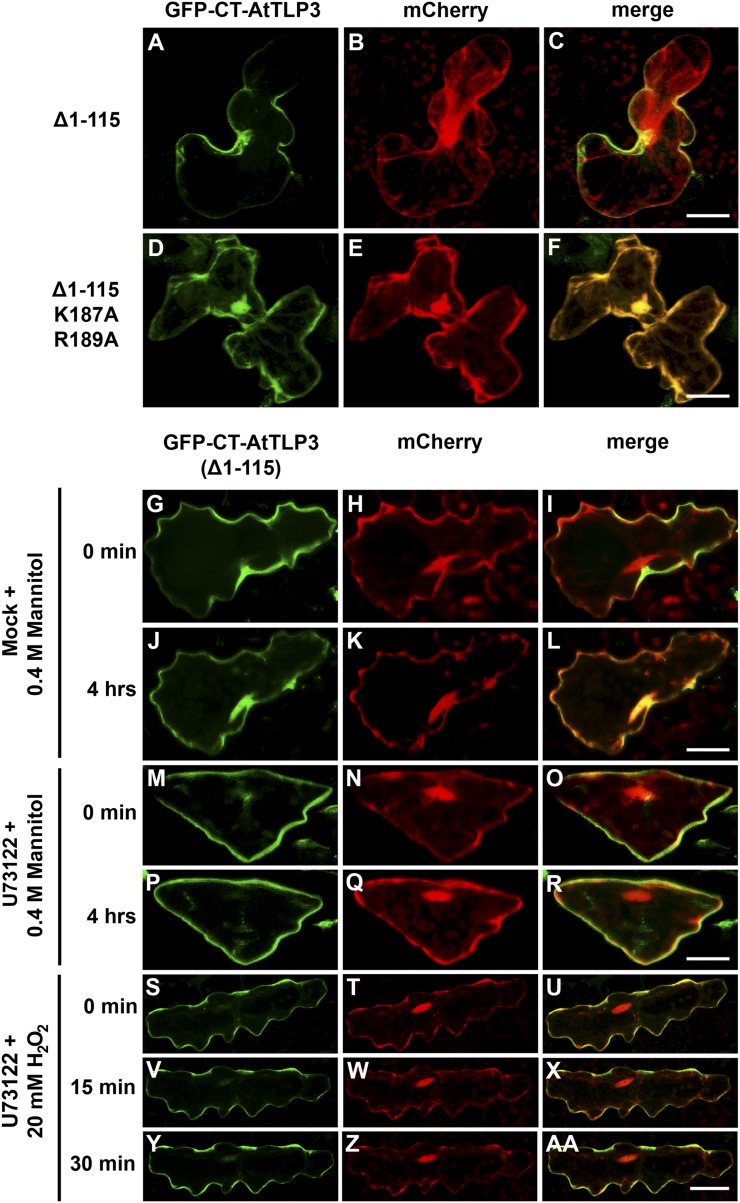
PM Tethering of the Tubby Domain Depends on Amino Acids Lys-187 and Arg-189, and Phospholipase C Activity Mediates Its Release
The structural modeling indicated the conservation of the Lys-187 and Arg-189 residues in the Tubby domain of AtTLP3 (Fig. 2, A and E). The corresponding amino acids are essentially required for PIP2 binding and PM attachment of Tubby and TLP3 in mice (Santagata et al., 2001). In order to examine whether the residues Lys-187 and Arg-189 of AtTLP3 are required for PM attachment, we transiently transformed Arabidopsis leaf cells with a GFP-CT-AtTLP3 Δ1–115 construct, in which we replaced Lys-187 and Arg-189 each by Ala. Exchange of these amino acids resulted in PM detachment of CT-AtTLP3, such that GFP-CT-AtTLP3 Δ1–115 K187A R189A colocalized with soluble mCherry (Fig. 8, A–F). In mice, phospholipase C-β mediates the dislodgement of Tubby (Santagata et al., 2001). U73122 is a well-known inhibitor of phospholipase C in plants. Consistent with mammalian systems, mannitol (Fig. 8, G–R) and H2O2 (Fig. 8, S–AA) were unable to trigger PM dislodgement of GFP-CT-AtTLP3 Δ1–115 in transformed cells if leaves were pretreated with U73122. This indicated that mannitol- and H2O2-triggered PM detachment of the Tubby domain of AtTLP3 was dependent on phospholipase C activity but was not caused by unspecific PM disintegration.
Figure 8.
Lys-187 and Arg-189 represent PIP2-binding sites in the Tubby domain of AtTLP3, and phospholipase C releases CT-AtTLP3 from the PM. A, D, G, J, M, P, S, V, Y, Arabidopsis leaf cells were transformed with GFP-CT-AtTLP3 (Δ1–115). B, E, H, K, N, Q, T, W, Z, Arabidopsis leaf cells were transformed with the cytosolic and nucleoplasmic marker mCherry. Amino acids eliminated in truncated versions used for transformation and treatments are indicated on the left. C, F, I, L, O, R, U, X, and AA, Merged images indicate the subcellular localization of truncated domain versions. Yellow color indicates the colocalization of green- and red-fluorescing proteins. A to C, A version of the C terminus of AtTLP3 fused to GFP [GFP-CT-AtTLP3 (Δ1–115)] displayed PM localization. D to F, GFP-CT-AtTLP3 (Δ1–115), in which Lys-187 and Arg-189 had been converted to Ala (A), has lost its PM-binding ability and shows cytosolic and nucleoplasmic localization. G to L, Application of 0.4 m mannitol results in the translocation of GFP-CT-AtTLP3 (Δ1–115) from the PM at 0 min (G–I) to the cytosol and nucleus within 4 h (J–L). M to R, Pretreatment with the phospholipase C inhibitor U73122 inhibits mannitol-triggered relocalization of GFP-TLP3 (Δ1–115) from the PM (M–O) to the cytosol and nucleus (P–R). S to AA, Pretreatment with the phospholipase C inhibitor U73122 inhibits H2O2-triggered relocalization of GFP-TLP3 (Δ1–115) from the PM to the cytosol and nucleus. Treated cells were observed at 0 min (S–U), 15 min (V–X), and 30 min (Y–AA) after H2O2 application. Experiments were repeated three times with similar results. Bars = 20 µm.
Heterologous Expression of AtTLP3 in Nicotiana benthamiana
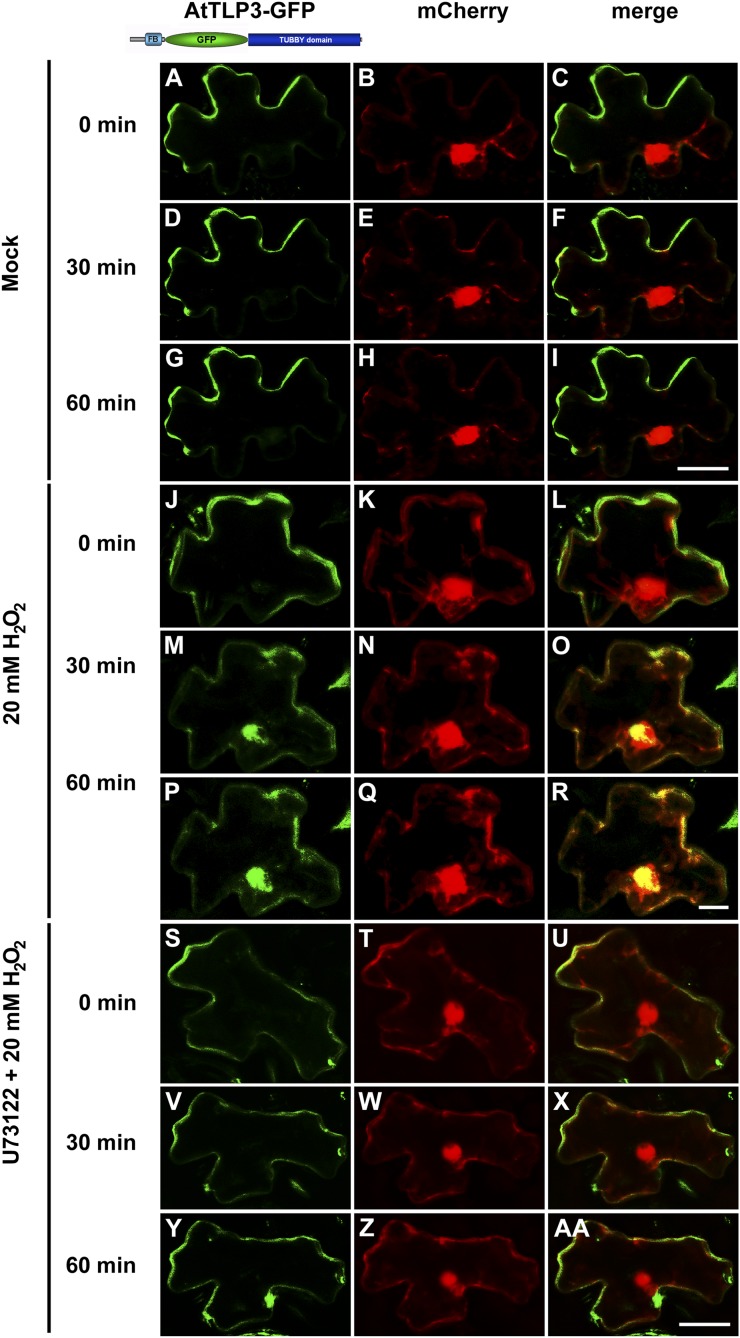
The instability of full-length AtTLP3 did not allow determination of the subcellular localization in Arabidopsis. By contrast, we detected full-length AtTLP3-GFP in biolistically transformed N. benthamiana leaf cells (Fig. 9). When GFP was cloned between the FB and Tubby domain, PM localization of full-length AtTLP3 was observed (Fig. 9, A–I). Similar to GFP-CT-AtTLP3 in Arabidopsis, 20 mm H2O2 induced PM detachment of full-length AtTLP3 (Fig. 9, J–R), and this process was blocked by the phospholipase C inhibitor U73122 (Fig. 9, S–AA). However, full-length AtTLP3 did not translocate to plastids, as did the NT-AtTLP3-GFP in N. benthamiana (Supplemental Fig. S8, A–C). GFP-CT-AtTLP3 showed PM localization (Supplemental Fig. S8, D–F), which was dependent on amino acids Lys-187 and Arg-189 (Supplemental Fig. S8, G–I).
Figure 9.
Phospholipase C releases full-length AtTLP3-GFP from the PM when heterologously expressed in N. benthamiana. A, D, G, J, M, P, S, V, Y, N. benthamiana leaf cells were transformed with full-length AtTLP3-GFP. B, E, H, K, N, Q, T, W, Z, N. benthamiana leaf cells were transformed the cytosolic and nucleoplasmic marker mCherry. C, F, I, L, O, R, U, X, AA, Merged images indicate the subcellular localization of full-length AtTLP3-GFP. Yellow color indicates the colocalization of green- and red-fluorescing proteins. A to I, Full-length AtTLP3 fused to GFP displayed PM localization over time. J to R, Application of 20 mm H2O2 results in the relocalization of full-length AtTLP3-GFP from the PM to the cytosol and nucleus. J to L, PM localization of full-length AtTLP3-GFP before treatment (0 min). M to O, Cytosolic and nucleoplasmic localization of full-length AtTLP3-GFP at 30 min after application of 20 mm H2O2. P to R, Cytosolic and nucleoplasmic localization of full-length AtTLP3-GFP at 60 min after application of 20 mm H2O2. S to AA, Pretreatment with the phospholipase C inhibitor U73122 inhibits H2O2-triggered relocalization of full-length AtTLP3-GFP from the PM to the cytosol and nucleus. Treated cells were observed at 0 min (S–U), 30 min (V–X), and 60 min (Y–AA) after H2O2 application. Experiments were repeated three times with similar results. Bars = 20 µm.
DISCUSSION
In mammals, Tubby and TLPs participate in fundamental physiological processes, as evidenced by dramatic disease syndromes in various mutants (Mukhopadhyay and Jackson, 2011). In plants, TLPs participate in immune and hormone signaling, and single gene losses display slight physiological defects (Lai et al., 2004; Cai et al., 2008). PM attachment and stimulus-dependent translocation of AtTLP3 (Figs. 6 and 8) might function as shortcut between trigger sensing and response. Therefore, the localization of AtTLPs at the PM as the cell-environment interface is plausible. The signaling processes affected by AtTLPs might have ecological and agricultural impacts. For example, various AtTLPs are required for the establishment of the mutualistic symbiosis of Arabidopsis with P. indica (Fig. 1), a fungus that supports stress resistance, development, and reproduction of its hosts (e.g. barley, maize [Zea mays], tomato [Solanum lycopersicum]; Qiang et al., 2012).
In order to uncover the function of AtTLPs, we analyzed sequences and structures of AtTLPs, in particular AtTLP3, in silico and examined the subcellular localizations of full-length and truncated protein versions. We were unable to detect the accumulation of full-length AtTLPs fused to GFP in Arabidopsis by confocal laser-scanning microscopy. This suggests a high instability of these proteins, as reported for other FB proteins (Nibau et al., 2011). Since we detected a full-length AtTLP3-GFP fusion protein in N. benthamiana, the degradation of AtTLPs is executed apparently by specific proteases (Fig. 9). Protein modeling suggested the ability of AtTLP3 to bind to PIP2 (Fig. 2, D and E). We tested one AtTLP per clade and showed PM attachment of its Tubby domain (except AtTLP8) in Arabidopsis cells (Fig. 4; Supplemental Fig. S5). In mouse Tubby, Lys-330 was required to integrate the interaction between 4- and 5-phosphates of PIP2, while Arg-332 stabilized the interaction with the 4-phosphate (Santagata et al., 2001). Indeed, corresponding Lys-187 and Arg-189 residues in AtTLP3 were necessary for PM attachment and likely PIP2 binding (Fig. 2, A, D, and E). Replacement of Lys-187 and Arg-189 by Ala resulted in nucleocytosolic localization of CT-AtTLP3 (Fig. 8, A–F). The high conservation of PIP2-binding residues in all AtTLPs (except for the nucleocytosol-localized AtTLP8), which even mediate AtTLP3 binding to PM in N. benthamiana (Supplemental Fig. S6), suggests their significance for AtTLP function.
In clear contrast to the heterogenous N terminus in mammalian TLPs, almost all AtTLPs carry an FB domain at this site. WoLF PSORT predominantly predicted plastidial localization of AtTLPs (Supplemental Table S1), although we did not detect a plastid target peptide or any other localization sequences in silico. We observed a plastidial and nucleocytosolic localization of N-terminal fragments of AtTLP2, -3, -7, and -10 (Fig. 4, H–S; Supplemental Fig. S5). The full-length AtTLP3-GFP fusion protein was not detected in plastids of N. benthamiana cells after PM dislodgement (Fig. 9, J–R). Therefore, plastidial localization might indicate an artifact due to 35S promoter-driven overexpression. However, plastidial localization occurred when the N terminus of AtTLP3 (NT-AtTLP3) was expressed in Arabidopsis cells under the control of its endogenous promoter (Fig. 4, Q–S). NT-AtTLP3 also showed plastidial localization in N. benthamiana (Supplemental Fig. S8, D–F). Most probably, in the heterologous N. benthamiana system, the intramolecular GFP fusion disturbed processes (e.g. chaperone binding) required for plastidial translocation rather than the absence of proteins required for AtTLP3 translocation. Future immunolocalization studies are required to confirm the localization of AtTLPs and to specify their function at these sites. They will further reveal whether AtTLPs are imported into plastids. The conservation of the FB domain suggests that it plays a pivotal role for AtTLP function. FB proteins represent an abundant protein group in plants that participate in the degradation of proteins. Lai et al. (2004) determined the involvement of AtTLP9 in ABA signaling. Plants overexpressing AtTLP9 displayed ABA hypersensitivity. AtTLP9 interacts with ASK1, suggesting its participation in protein degradation via an SCF complex (Lai et al., 2004). AtTLP9 also interacts with the RING-H2-type zinc finger protein XERICO, and similar to AtTLP9, the overexpression of XERICO results in ABA hypersensitivity, as indicated by reduced seed germination rates (Lai et al., 2004; Ko et al., 2006). In addition to plant development, ABA is essentially involved in abiotic stress signaling (Huang et al., 2012). The connection of ABA and AtTLP9 implies a general participation of AtTLPs in stress signaling. In support of this idea, salt (0.3 m NaCl) and drought/water deficit stress (0.4 m mannitol) triggered PM dislodgement of CT-AtTLP3 (Tubby domain; Fig. 6, A–L). In addition, CT-AtTLP3 dislodgement within 5 min after H2O2 treatment proposes a function of AtTLP3 in abiotic stress signaling (Fig. 6M–AA). Importantly, mannitol- and H2O2-triggered PM release was abolished after pretreating leaves with the phospholipase C inhibitor U73122, indicating that (1) phospholipase C activity mediated PM dislodgement of CT-AtTLP3 and (2) mannitol- and H2O2-triggered PM dislodgement of CT-AtTLP3 was not caused by the unspecific impairment of PM integrity. Furthermore, stress signaling of AtTLP3 was highly reminiscent of TLP function in vertebrates. Although we tested several stress conditions, we could not detect altered oxidative stress (Supplemental Fig. S7) or salt/mannitol sensitivity (data not shown) in attlp3 mutants. This finding suggests functional redundancy among AtTLPs in stress signaling, and future studies might include mutants lacking all members of a particular clade. The relatively specific release of AtTLP3 and maybe other AtTLPs from the PM upon stress treatments might support the future use of Tubby domains in biotechnological approaches. Ectopic fusion of Tubby domains with nuclear transcription regulators might be used for the conditional translocation of artificial transcription factors upon stress.
AtTLPs might affect signaling processes as transcriptional regulators. The C-terminal part of mouse Tubby bound to double-stranded DNA, while the N-terminal part activated transcription from a GAL4 promoter when fused to the GAL4 DNA-binding domain (Boggon et al., 1999). Nevertheless, we predicted a smaller putative DNA-binding cavity in the Tubby domain of AtTLP3 as compared with those of human and mouse TLPs (Fig. 2, F and G). In addition, the cavity of mouse Tubby possesses an abundance of positively charged amino acids required for the binding of negatively charged DNA (Boggon et al., 1999), which we did not observe in AtTLP3 (Fig. 2, F and G). Consistent with this, AtTLP9 failed to activate transcription when fused to the GAL4 DNA-binding domain (Lai et al., 2004). By contrast, OsTLP2 was shown to bind to cis-elements of the OsWRKY13 promoter (Cai et al., 2008), and the Tubby domain of AtTLP3 was translocated to the cytosol and nucleoplasm after PM dislodgement (Fig. 6, A–U). These findings suggest a functional specification of AtTLPs like that known for mammalian Tubby and TLPs, which act as transcriptional regulators or molecular adapters in vesicle trafficking (Mukhopadhyay and Jackson, 2011).
Interestingly, AtTLPs support root colonization by P. indica (Fig. 1A). Although we did not detect a differential regulation of AtTLP genes upon P. indica colonization (Supplemental Fig. S2), we observed a delayed establishment of the mutualistic Arabidopsis root-P. indica symbiosis at the biotrophic stage in plants lacking AtTLPs (Fig. 1A), suggesting a function of AtTLPs as compatibility factors in this mutualistic interaction. In addition to hormones, plants have evolved mechanisms to fine-balance and control immune signaling. For instance, similar to FB proteins, the plant U-box (PUB) proteins PUB22, -23, and -24 function as ubiquitin E3 ligases and degrade positive regulators of plant immunity (Trujillo et al., 2008). Consequently, roots of pub22/23/24 triple mutants exhibit a hyperactive immune response after recognition of pathogens and the mutualist P. indica, which results in reduced colonization (Trujillo et al., 2008; Jacobs et al., 2011). However, an immune-related function of AtTLPs is rather unlikely, since all tested attlp mutants (1) showed an unaltered colonization by biotrophic leaf (E. cruciferarum) and root (P. parasitica) pathogens (Supplemental Fig. S4) and (2) exhibited unaltered flg22 responsiveness and P. indica-induced abolishment of flg22-triggered seedling growth inhibition (Supplemental Fig. S3). In addition, attlp3 mutants did not show an elevated expression of stress and immune marker genes (Supplemental Fig. S5) of pathways that we found to restrict root colonization by P. indica (Jacobs et al., 2011).
ROS have a general function in immune signaling (Torres, 2010). In plants, the apoplast, mitochondria, and plastids are the main production sites of H2O2 (Torres, 2010). Plastidial H2O2 causes photooxidative stress and possibly promotes the hypersensitive cell death reaction (Galvez-Valdivieso and Mullineaux, 2010) under excess light. For instance, plastids accumulate H2O2 when executing mesophyll cell death in order to stop pathogen attack (Hückelhoven et al., 2000). It is conceivable that P. indica might recruit AtTLP3 to prevent ROS actions that would disturb early root colonization. Accordingly, failed ROS detoxification can impair and terminate mutualistic symbioses (Fester and Hause, 2005; Puppo et al., 2005; Torres, 2010). However, this assumption is very simplified and neglects the spatiotemporal impact of different ROS. For instance, the establishment of bacterial nodulation obviously depends on locally restricted ROS production (Jamet et al., 2007; Cárdenas et al., 2008). Moreover, ROS do not even support immunity in all instances. Cytoplasmic ROS prevent the activation of NONEXPRESSER OF PATHOGENESIS-RELATED GENES1 (NPR1), one of the main regulators of immune signaling. Cytosolic NPR1 oligomers are inactive. However, pathogen attack and salicylic acid induce NPR1 monomerization, allowing its nuclear import and the induction of defense genes. Cytoplasmic ROS prevent the monomerization of NPR1 (Peleg-Grossman et al., 2010). In addition to immune signaling, ROS participate in the regulation of physiological processes. Plastidial H2O2 functions in adaptive retrograde chloroplast-to-nucleus signaling, for which H2O2 translocation into the cytosol is thought to be essential (Galvez-Valdivieso and Mullineaux, 2010). It is tempting to speculate that cytosolic or apoplastic H2O2 elevation induces the release and relocalization of AtTLP3 to plastids as part of an adaptive stress response. Whether such a model also holds true for root cells is unknown. Currently, the function of plastids in stress signaling in root cells is almost unknown. As these examples already demonstrate the diverse and opposing functions of ROS, the question arises how ROS currents are integrated into specific signaling processes. It is the current belief that the site and amount of ROS production determine the signal output (Mittler et al., 2011). It is tempting to speculate that AtTLP3 might function as a transducer of ROS currents. Future studies have to show how AtTLPs might affect or even specify ROS signaling and whether they might act in concert with PM-localized receptors, as reported for mammals. CRK/DUF26 receptor-like kinases that were found recently to be induced by ROS (Wrzaczek et al., 2010) might represent potential interaction partners.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized with 70% (v/v) ethanol for 1 min and sodium hypochlorite (3% active chlorine) for 5 min before being washed 10 times with sterile water. If not stated otherwise, plants were grown aseptically in square petri dishes on solid one-half-strength Murashige and Skoog (MS) medium without vitamins (Duchefa) and 0.4% (w/v) Gelrite (Roth). For uniform germination, seeds were incubated at 4°C for 2 d after sowing before plants were kept in a growth chamber with a 22°C/18°C day/night cycle (8 h of light, 180 µmol m−2 s−1 photon flux density) at 60% relative humidity.
T-DNA Insertion Mutants
We aimed to select T-DNA insertion lines for all AtTLP genes except AtTLP4, which is thought to be a pseudogene (Lai et al., 2004). Where possible, we selected lines with the T-DNA insertion within the coding region of the gene to enhance the chance of successful disruption of gene function. When unavailable, lines were selected with predicted intron or promoter insertions. All mutant lines were of the Columbia ecotype (Col-0). All seeds were obtained from the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003). Homozygous plants were obtained for lines attlp2-1 (N663531), attlp2-2 (N654211), attlp3-1 (N678667), attlp3-2 (N676958), attlp5-1 (N675664), attlp7-1 (N666743), attlp8-1 (N668671), attlp9-1 (N665383), and attlp9-2 (N659871). Lines attlp9-1 and attlp9-2 are described by Lai et al. (2004). Segregating plants were obtained for lines attlp5-2 (N462147), attlp8-2 (N507790), attlp10-1 (N612155), and attlp10-2 (N550916). All lines were selfed and screened for homozygosity by PCR using T-DNA-specific primers (LBb1.3 for SALK lines, LB_GABI for GABI-Kat lines) in combination with primers flanking putative insertion sites (for primer sequences, Arabidopsis Genome Initiative [AGI] codes, SALK numbers, and insertion sites, see Supplemental Table S2).
Analysis of AtTLP Expression by Semi-Quantitative RT-PCR
AtTLP transcript abundance in Col-0 and attlp plants was determined by semi-quantitative RT-PCR. Total RNA was isolated from 3-week-old plants using TRIzol (Invitrogen), and complementary DNA (cDNA) was synthesized with the qScript cDNA synthesis kit (Quanta Biosciences) using oligo(dT) primers. The Arabidopsis Ubiquitin5 (AtUbi5) transcript was used as a loading control (for primer sequences, see Supplemental Table S2).
Inoculation with Piriformospora indica and Quantification of Fungal Colonization by qRT-PCR
P. indica isolate DSM11827 was used for all experiments. For fungal quantification, roots of 3-week-old plants were inoculated with P. indica (500,000 chlamydospores mL−1) and harvested at the indicated time points. Genomic DNA was extracted from roots with the Plant DNeasy Kit (Qiagen). Forty nanograms of DNA served as the template for qRT-PCR analyses by using 20 µL of SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) with 350 nm oligonucleotides and a 7500 FAST thermal cycler with a standard protocol (Applied Biosystems). Fungal colonization was determined by the 2−ΔCt method (Schmittgen and Livak, 2008) by subtracting the raw cycle threshold values of P. indica internal transcribed spacer from those of AtUbi5 (for primer sequences, see Supplemental Table S2). Data were analyzed by Student’s t test.
Gene Expression Analysis by qRT-PCR
Experiments were carried out according to Jacobs et al. (2011). Three-week-old Col-0 and attlp3-1 plants were inoculated with P. indica or mock treated, and root material was harvested at 0, 1, 3, and 7 dai. Total RNA was extracted using TRIzol (Invitrogen), and aliquots were used for cDNA synthesis with the qScript cDNA synthesis kit (Quanta Biosciences). Ten nanograms of cDNA was used as the template for qRT-PCR as described above for fungal quantification. The 2−ΔCt method was used to determine the differential gene expression of immune and stress marker genes. For expression of AtTLPs only, Col-0 and time points 1, 3, and 7 dai were used (for primer sequences and AGI codes, see Supplemental Table S2).
Pathogen Inoculation
Botrytis cinerea strain B05.10 was grown on HA agar (1% [w/v] malt extract, 0.4% [w/v] Glc, 0.4% [w/v] yeast extract, and 1.5% [w/v] agar, pH 5.5) as described previously (Doehlemann et al., 2006). Plants were grown in soil in a growth chamber with the same conditions as aseptically grown plants. Seedlings were transferred into individual pots 10 to 12 d after germination. Detached rosette leaves from 6- to 7-week-old plants were placed in petri dishes containing 1% (w/v) agar and inoculated with 5-µL spore suspension (2 × 105 spores mL−1 in 12 g L−1 potato dextrose broth; Duchefa). To maintain high humidity, petri dishes were covered with a transparent lid. Digital photographs were taken at different time points with a Canon EOS450D camera and analyzed using ImageJ software to measure lesion size (Abramoff et al., 2004). Phytophthora parasitica Dastur isolate 310 was isolated from Nicotiana benthamiana in Australia and maintained in the Phytophthora spp. collection at the INRA. P. parasitica growth conditions and the production of zoospores were performed as described by Galiana et al. (2005). The roots of Arabidopsis plantlets were grown in vitro and inoculated with 500 motile P. parasitica zoospores according to Attard et al. (2010). Disease symptoms were scored over 3 weeks using a wilting disease index ranked from 1 (healthy plant) to 7 (dead plant; Attard et al., 2010). The analysis was carried out on 18 to 35 inoculated plants, and all experiments were performed twice. For Erysiphe cruciferarum inoculation, Arabidopsis plants were grown at 22°C and 65% relative humidity in a 10-h photoperiod with 120 µmol m−2 s−1 light in a 2:1 soil:sand mixture (Fruhstofer Erde, type P; Quarzsand [granulation, 0.1–0.5 mm; Sakret Trockenbaustoffe Europa]). E. cruciferarum was grown on Col-0, to maintain constant aggressiveness, and on susceptible phytoalexin deficient4 (pad4) mutants, for strong conidia production. For inoculation, Arabidopsis plants were placed under an inoculation box covered with a polyamide net (0.2 mm2). Conidia were brushed off of pad4 plants through the net until a density of three to four conidia mm−2 was reached. Inoculated plants were kept in a growth chamber under the above conditions. Leaves were photographed 11 d after inoculation.
flg22-Triggered Seedling Growth Inhibition
Ten- to 12-d-old plants grown on one-half-strength MS medium plus 1% (w/v) Suc were inoculated with P. indica. At 5 dai, plants of the same size were transferred to petri dishes containing liquid one-half-strength MS medium plus 1% (w/v) Suc. For MAMP treatment, 10 µm flg22 was added. The fresh weight of mock- and flg22-treated plants was determined 10 d after treatment. The flg22 peptide was used as described (Gómez-Gómez et al., 1999; Kunze et al., 2004).
Generation of AtTLP3Prom::GUS Lines
For the generation of pCX-AtTLP3Prom::GUS, a stretch of 1,032 bp upstream of the translational start site of AtTLP3 was PCR amplified from genomic DNA (for primer sequences, see Supplemental Table S2) and ligated into XcmI-digested pCXGUS-P (Chen et al., 2009) after addition of a single 3' adenine overhang. The vector was electroporated into Agrobacterium tumefaciens strain LBA4404pSB1 (kindly provided by Japan Tobacco; Komari et al., 1996) using E. coli Pulser (Bio-Rad) according to the manufacturer’s instruction. The transformed cells were plated on yeast extract peptone agar medium containing 5 mg L−1 tetracycline, 25 mg L−1 rifampicin, and 50 mg L−1 kanamycin and subsequently incubated at 28°C in the dark for 2 d. Growing antibiotic-resistant colonies of agrobacteria were subcultured in liquid medium and then screened by PCR amplification using specific primers (for primer sequences, see Supplemental Table S2).
Plant transformation and regeneration were performed as described by the vacuum infiltration method (Bechtold et al., 1993). Briefly, a single colony of A. tumefaciens. was grown overnight at 28°C in 10 mL of YEB medium (0.1% yeast extract, 0.5% beef extract, 0.5% Suc, 0.5% casein hydrolysate, and 2 mm MgCl2) with appropriate antibiotics (5 mg L−1 tetracycline, 25 mg L−1 rifampicin, and 50 mg L−1 kanamycin). Subsequently, the culture was cultivated in 250 mL of YEB fresh medium for 6 h until a relative optical density at 600 nm of 1.8 was achieved. Agrobacteria were harvested by centrifugation and then suspended in infiltration medium consisting of one-half-strength MS salts 5% Suc, 2.3 µm 6-benzylaminopurine, and 0.01% Silwet-L77, pH 5.8, to a final optical density at 600 nm of 1.1 to 1.3. Arabidopsis Col-0 plants were infiltrated as described by Bechtold et al. (1993).
T1 seeds of transgenic Arabidopsis plants were surface sterilized as described. Seeds were germinated on one-half-strength MS medium including vitamins supplemented with 0.5% Suc, 0.4% Gelrite (Duchefa), and kanamycin (50 mg L−1) in a growth chamber under photoperiodic conditions of 16 h of light (180 μmol m−2 s−1 photon flux density) with 22°C day/18°C night temperatures. Kanamycin-resistant transformants were transferred to soil and propagated. T2 seeds of two independent transgenic lines were harvested and used for further experiments.
GUS Staining Procedure
Histochemical staining of GUS activity was performed essentially as described by Jefferson (1987). Plants were vacuum infiltrated with staining solution (0.1% [v/v] Triton X-100, 2 mm potassium ferrocyanide, 2 mm potassium ferricyanide, 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid [Duchefa], and 100 mm sodium phosphate buffer, pH 7.0) and kept at 37°C overnight. To remove chlorophyll, plants were washed three times in 70% ethanol. Images were acquired using a Leica DFC 300 FX digital camera mounted on a Leica MZ 16 F stereomicroscope.
3D Modeling
Modeling of the 3D structure of AtTLP3 was done using the project mode of the protein structure homology-modeling server Swiss model (Peitsch, 1995; Arnold et al., 2006; Kiefer et al., 2009) in combination with the program DeepView (Swiss Pdb-Viewer). Suitable template structures were searched via the Swiss model server. Resulting 3D structure templates were as follows: 1C8Z (C-terminal domain of mouse (Mus musculus) brain Tubby protein; Boggon et al., 1999); 1S31 (crystal structure analysis of the human [Homo sapiens] Tubby protein isoform a); 1I7E [mouse brain Tubby protein bound to PIP2 (Santagata et al., 2001)]; and 3C5N (human TULP1 in complex with IP3 [R.D. Busam, L. Lehtio, C.H. Arrowsmith, R. Collins, L.G. Dahlgren, A.M. Edwards, S. Flodin, A. Flores, S. Graslund, M. Hammarstrom, B.M. Hallberg, M.D. Herman, A. Johansson, I. Johansson, A. Kallas, T. Karlberg, T. Kotenyova, M. Moche, M.E. Nilsson, P. Nordlund, T. Nyman, C. Persson, J. Sagemark, L. Svensson, A.G. Thorsell, L. Tresaugues, S. Van den Berg, J. Weigelt, M. Welin, H. Berglund, and the Structural Genomics Consortium, unpublished data]). For homology modeling of AtTLP3, the crystal structures of 1C8Z as well as 1S31 were used as templates. Comparison of the amino acid sequences of the proteins used revealed identities of 38% and 35%, respectively. Visualization of the protein molecules was done using PyMOL.
GFP Fusion Constructs
A modified pAMPAT-MCS vector backbone (AY436765) containing a GFP behind the cauliflower mosaic virus 35S promoter was used to generate all constructs. For pam-35S::AtTLP-GFP and pam-35S::GFP-AtTLP constructs, cDNA fragments were cloned N or C terminally in frame with the GFP coding sequence using SalI and NotI or NotI and XmaI as terminal restriction sites, respectively. For the generation of pam-MCS-AtTLP3Prom::AtTLP3Δ116–406-GFP, the pam-MCS-35S::AtTLP3Δ116–406-GFP vector was digested with AscI and XhoI to remove the 35S promoter. A stretch of 1,032 bp upstream of the translational start site of AtTLP3 was PCR amplified from genomic DNA, adding AscI and XhoI as terminal restriction sites. After digestion of the PCR fragment with the respective enzymes, it was ligated into the opened vector (for primer sequences, see Supplemental Table S2). AtTLP3(Δ1–115) (K187A, R189A) was synthesized at Eurofins with NotI and XmaI added as terminal restriction sites and subcloned into the NotI- and XmaI-digested pam-MCS-35S::GFP vector backbone to generate the pam-35S::GFP-AtTLP3(Δ1–115) (K187A, R189A) construct.
Transient Transformation and Cytological Analyses
Plasmid DNA was introduced into Arabidopsis leaves from 4- to 5-week-old soil-grown Col-0 plants using a particle inflow gun (Schweizer et al., 1999). Leaves were placed in petri dishes containing 1% (w/v) water agar (Agar-Agar; Roth) and kept at 22°C in darkness. For N. benthamiana, leaves from 4- to 6-week-old soil-grown plants were cut in half and placed in petri dishes containing wet filter paper. Confocal images were taken 16 to 26 h after bombardment on a TCS SP2 microscope (Leica). GFP was excited with a 488-nm laser line and detected at 505 to 540 nm. The marker proteins mCherry, pm-rk, and pt-rk (Nelson et al., 2007) were excited with a 543-nm laser line and detected at 620 to 660 nm. For microscopy, leaves were placed on glass slides in a bathing solution (2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 50 mm mannitol, and 2.5 mm MES/NaOH buffer, pH 5.7; Furch et al., 2007) to simulate apoplastic conditions. Further chemicals were added as indicated. No mannitol was added to the bathing solution for experiments involving NaCl. N. benthamiana leaves were kept in water, and further chemicals were added as indicated. For experiments using U73122 (Merck), transformed leaves were placed for 3 h in either bathing solution (mock) or bathing solution containing 10 µm U73122. Then, mock-treated leaves were placed in bathing solution containing either 0.4 m mannitol or 20 mm H2O2 (Merck). Inhibitor-treated leaves were placed in the same solutions additionally containing 10 µm U73122.
H2O2 Treatment
Analysis of relative electrolyte leakage was performed as described by Lee et al. (2007) with modifications. Fifteen leaf discs (0.5 mm) of 4- to 5-week-old soil-grown plants were washed in deionized water and transferred to 50-mL reaction tubes containing 15 mL of test solutions. Tubes were placed under constant light for 48 h, and electrical conductivity was measured using a conductivity meter (Cond 315i; Wissenschaftlich-Technische Werkstätten GmbH) before (Lt) and after (L0) boiling the samples. Relative electrolyte leakage was calculated as Lt/L0 × 100%. For gene expression analysis, Col-0 plants were grown aseptically on solid one-half-strength MS medium plus 1% (w/v) Suc for 8 d. Subsequently, plants were transferred to six-well plates (Greiner Bio-One) containing liquid one-half-strength MS medium plus 1% (w/v) Suc and grown for an additional 3 d before treatment with the indicated concentrations of H2O2 (Merck). At 0, 1, 2, and 24 h after treatment, whole plants were harvested and frozen in liquid nitrogen. Generation of cDNA and qRT-PCR were performed as described above (for primer sequences, see Supplemental Table S2).
Image Processing
Images were processed for publication using Adobe Photoshop 3.0.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At2g18280, At2g47900, At1g43640, At1g53320, At1g16070, At3g06380, At1g25280, At5g26920, At2g45900, At1g18570, At3g25250, At3g62250, At5g24770, and At4g23550.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcript levels of AtTLP genes in attlp mutants.
Supplemental Figure S2. Unaltered expression of selected AtTLP genes during Arabidopsis root colonization by P. indica.
Supplemental Figure S3. flg22-induced seedling growth inhibition is unaltered in attlp mutants.
Supplemental Figure S4. Unaltered colonization of attlp mutants by E. cruciferarum, B. cinerea, and P. parasitica.
Supplemental Figure S5. Marker genes for MAMP-triggered immunity and abiotic stress do not show elevated expression in the attlp3 mutant during P. indica colonization.
Supplemental Figure S6. Separate localization of FB and Tubby domains of AtTLPs.
Supplemental Figure S7. Effects of H2O2 on relative ion leakage in leaf discs of control and mutant plants.
Supplemental Figure S8. Subcellular localization of truncated versions of AtTLP3 in N. benthamiana.
Supplemental Table S1. Prediction of the subcellular localization of AtTLPs by WoLF PSORT.
Supplemental Table S2. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank the European Arabidopsis Stock Centre for providing seeds. We also thank Dagmar Biedenkopf and Rebekka Fensch for excellent technical assistance.
Glossary
- T-DNA
transferred DNA
- PM
plasma membrane
- H2O2
hydrogen peroxide
- MAMP
microbe-associated molecular pattern
- ROS
reactive oxygen species
- PI
phosphatidylinositol
- PIP2
phosphatidylinositol 4,5-bisphosphate
- ABA
abscisic acid
- dai
d after inoculation
- qRT
quantitative real-time
- 3D
three-dimensional
- MS
Murashige and Skoog
- Col-0
ecotype Columbia
- PDB
Protein Data Bank
- cDNA
complementary DNA
References
- Abramoff MD, Magalhaes PJ, Ram SJ. (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. (2006) The SWISS-MODEL workspace: a Web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Attard A, Gourgues M, Callemeyn-Torre N, Keller H. (2010) The immediate activation of defense responses in Arabidopsis roots is not sufficient to prevent Phytophthora parasitica infection. New Phytol 187: 449–460 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci III 316: 1194–1199 [Google Scholar]
- Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L. (1999) Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 286: 2119–2125 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Cai M, Qiu D, Yuan T, Ding X, Li H, Duan L, Xu C, Li X, Wang S. (2008) Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ 31: 86–96 [DOI] [PubMed] [Google Scholar]
- Cárdenas L, Martínez A, Sánchez F, Quinto C. (2008) Fast, transient and specific intracellular ROS changes in living root hair cells responding to Nod factors (NFs). Plant J 56: 802–813 [DOI] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL. (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh S, Hückelhoven R, Schäfer P, Imani J, Sharma M, Weiss M, Waller F, Kogel KH. (2006) The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc Natl Acad Sci USA 103: 18450–18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G, Berndt P, Hahn M. (2006) Different signalling pathways involving a Galpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol Microbiol 59: 821–835 [DOI] [PubMed] [Google Scholar]
- Fester T, Hause G. (2005) Accumulation of reactive oxygen species in arbuscular mycorrhizal roots. Mycorrhiza 15: 373–379 [DOI] [PubMed] [Google Scholar]
- Furch AC, Hafke JB, Schulz A, van Bel AJ. (2007) Ca2+-mediated remote control of reversible sieve tube occlusion in Vicia faba. J Exp Bot 58: 2827–2838 [DOI] [PubMed] [Google Scholar]
- Galiana E, Rivière MP, Pagnotta S, Baudouin E, Panabières F, Gounon P, Boudier L. (2005) Plant-induced cell death in the oomycete pathogen Phytophthora parasitica. Cell Microbiol 7: 1365–1378 [DOI] [PubMed] [Google Scholar]
- Galvez-Valdivieso G, Mullineaux PM. (2010) The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol Plant 138: 430–439 [DOI] [PubMed] [Google Scholar]
- Gilissen C, Arts HH, Hoischen A, Spruijt L, Mans DA, Arts P, van Lier B, Steehouwer M, van Reeuwijk J, Kant SG, et al. (2010) Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am J Hum Genet 87: 418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T. (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: 585–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GT, Ma SL, Bai LP, Zhang L, Ma H, Jia P, Liu J, Zhong M, Guo ZF. (2012) Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep 39: 969–987 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R, Fodor J, Trujillo M, Kogel KH. (2000) Barley Mla and Rar mutants compromised in the hypersensitive cell death response against Blumeria graminis f.sp. hordei are modified in their ability to accumulate reactive oxygen intermediates at sites of fungal invasion. Planta 212: 16–24 [DOI] [PubMed] [Google Scholar]
- Jacobs S, Zechmann B, Molitor A, Trujillo M, Petutschnig E, Lipka V, Kogel KH, Schäfer P. (2011) Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol 156: 726–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet A, Mandon K, Puppo A, Hérouart D. (2007) H2O2 is required for optimal establishment of the Medicago sativa/Sinorhizobium meliloti symbiosis. J Bacteriol 189: 8741–8745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res 37: D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Han KH. (2006) Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J 47: 343–355 [DOI] [PubMed] [Google Scholar]
- Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T. (1996) Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J 10: 165–174 [DOI] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CP, Chen PH, Huang JP, Tzeng YH, Chaw SM, Shaw JF. (2012) Functional diversification of the Tubby-like protein gene families (TULPs) during eukaryotic evolution. Biocat Agric Biotechnol 1: 2–8 [Google Scholar]
- Lai CP, Lee CL, Chen PH, Wu SH, Yang CC, Shaw JF. (2004) Molecular analyses of the Arabidopsis TUBBY-like protein gene family. Plant Physiol 134: 1586–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. (2006) F-box proteins everywhere. Curr Opin Plant Biol 9: 631–638 [DOI] [PubMed] [Google Scholar]
- Lee SH, Ahsan N, Lee KW, Kim DH, Lee DG, Kwak SS, Kwon SY, Kim TH, Lee BH. (2007) Simultaneous overexpression of both CuZn superoxide dismutase and ascorbate peroxidase in transgenic tall fescue plants confers increased tolerance to a wide range of abiotic stresses. J Plant Physiol 164: 1626–1638 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Jackson PK. (2011) The tubby family proteins. Genome Biol 12: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK. (2010) TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev 24: 2180–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Nielsen E. (2011) Green light for polyphosphoinositide signals in plants. Curr Opin Plant Biol 14: 489–497 [DOI] [PubMed] [Google Scholar]
- Munnik T, Testerink C. (2009) Plant phospholipid signaling: “in a nutshell.” J Lipid Res (Suppl) 50: S260–S265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nibau C, Gibbs DJ, Bunting KA, Moody LA, Smiles EJ, Tubby JA, Bradshaw SJ, Coates JC. (2011) ARABIDILLO proteins have a novel and conserved domain structure important for the regulation of their stability. Plant Mol Biol 75: 77–92 [DOI] [PubMed] [Google Scholar]
- Peitsch MC. (1995) Protein modeling by e-mail. Nat Biotechnol 13: 658–660 [Google Scholar]
- Peleg-Grossman S, Melamed-Book N, Cohen G, Levine A. (2010) Cytoplasmic H2O2 prevents translocation of NPR1 to the nucleus and inhibits the induction of PR genes in Arabidopsis. Plant Signal Behav 5: 1401–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppo A, Groten K, Bastian F, Carzaniga R, Soussi M, Lucas MM, de Felipe MR, Harrison J, Vanacker H, Foyer CH. (2005) Legume nodule senescence: roles for redox and hormone signalling in the orchestration of the natural aging process. New Phytol 165: 683–701 [DOI] [PubMed] [Google Scholar]
- Qiang X, Weiss M, Kogel KH, Schäfer P. (2012) Piriformospora indica-a mutualistic basidiomycete with an exceptionally large plant host range. Mol Plant Pathol 13: 508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. (2001) G-protein signaling through tubby proteins. Science 292: 2041–2050 [DOI] [PubMed] [Google Scholar]
- Schäfer P, Pfiffi S, Voll LM, Zajic D, Chandler PM, Waller F, Scholz U, Pons-Kühnemann J, Sonnewald S, Sonnewald U, et al. (2009) Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica. Plant J 59: 461–474 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schweizer P, Pokorny J, Abderhalden O, Dudler R. (1999) A transient assay system for the functional assessment of defense-related genes in wheat. Mol Plant Microbe Interact 12: 647–654 [Google Scholar]
- Torres MA. (2010) ROS in biotic interactions. Physiol Plant 138: 414–429 [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. (2008) Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Uemura M, Joseph RA, Steponkus PL. (1995) Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions). Plant Physiol 109: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Salojärvi J, Kangasjärvi S, Idänheimo N, Mersmann S, Robatzek S, Karpiński S, Karpińska B, Kangasjärvi J. (2010) Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhou Y, Wang X, Gu S, Yu J, Liang G, Yan C, Xu C. (2008) Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis, rice, and poplar. Genomics 92: 246–253 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.