Abstract
Cognitive decline is a debilitating feature of most neurodegenerative diseases of the central nervous system, including Alzheimer’s disease (AD)1. The causes leading to such impairment are only poorly understood and effective treatments are slow to emerge2. Here, we show that cognitive capacities in the neurodegenerating brain are constrained by an epigenetic blockade of gene transcription that is potentially reversible. This blockade is mediated by histone deacetylase (HDAC) 2, which is increased by AD-related neurotoxic insults in vitro, in two mouse models of neurodegeneration, and in AD patients. HDAC2 associates with and reduces the histone acetylation of genes important for learning and memory, which show a concomitant decrease in expression. Importantly, reversing the buildup of HDAC2 by shRNA-mediated knockdown unlocks the repression of these genes, re-instates structural and synaptic plasticity, and abolishes neurodegeneration-associated memory impairments. These findings advocate for the development of HDAC2-selective inhibitors, and suggest that cognitive capacities following neurodegeneration are not entirely lost, but merely impaired by this epigenetic blockade.
Long-lasting forms of memories require stable gene expression changes3, which are in part orchestrated by chromatin-templated epigenetic processes4. Of the epigenetic modifications identified to date in the nervous system, histone acetylation has been unequivocally associated with facilitating learning and memory4. Acetylation diminishes the electrostatic affinity between neighboring histones and the DNA and, as a consequence, can promote a more open chromatin structure that allows for memory-related gene transcription5.
Over the past decade, several studies have reported sporadic cases of reduced histone acetylation in animal models of neurodegeneration that are characterized by cognitive decline, including models of AD4. Accordingly, pharmacological treatments aimed at increasing histone acetylation have shown promising results in reversing cognitive deficits in some of these models, predominantly by the use of non-selective HDAC inhibitors6. However, the causative agent of such memory-impairing histone acetylation changes, and, hence, the best targets for pharmacological strategies, remain unknown. One likely candidate is HDAC2, a class I HDAC that negatively regulates memory and synaptic plasticity in the healthy mouse brain7, 8.
To investigate whether HDAC2 mediates cognitive deficits associated with neurodegeneration, we measured its abundance in CK-p25 mice9, 10, which inducibly and forebrain-specifically overexpress p25, a truncated version of p35. p25 aberrantly activates cyclin-dependent kinase 5 (Cdk5), and is implicated in various neurodegenerative diseases11, including AD12. Following six weeks of p25 induction, CK-p25 mice display AD-related pathologies such as neuronal loss9, β-amyloid accumulation10, reactive astrogliosis9, and reduced synaptic density13, most prominently in the hippocampus and the cortex, two brain areas important for memory formation and storage, respectively14. Accordingly, six-week-induced CK-p25 (hereafter referred to as CK-p25) mice also display spatial and associative memory deficits13.
Using immunohistochemistry and western blot analysis (WB), we found that HDAC2 was significantly increased in neuronal nuclei in hippocampal area CA1 in CK-p25 mice compared to control littermates (Fig. 1a, d, e; see Supplementary Fig. 2a for a specificity control for the HDAC2 signal). No changes in HDAC2 were observed in hippocampal area CA3 or the dentate gyrus (Supplementary Fig. 3a, b), explaining the overall marginal increase in the entire hippocampus. Interestingly, this effect appears to be non cell-autonomous, as both p25-positive and p25-negative cells displayed elevated HDAC2. In contrast, levels of the structurally highly-related HDAC1, and of HDAC3, another class I HDAC involved in memory formation15, were not changed (Fig. 1b, c). Furthermore, HDAC2 was also increased in the prefrontal cortex of CK-p25 mice (Supplementary Fig. 4), whereas in the amygdala, a brain area not affected by neurodegeneration in the CK-p25 mice, its levels remained unchanged (Supplementary Fig. 3c). This neurodegeneration-associated increase of HDAC2 was confirmed in another mouse model of AD-related pathologies and cognitive decline, the 5xFAD mouse16, 17 (Supplementary Fig. 5).
Figure 1. Elevated HDAC2 levels epigenetically block the expression of neuroplasticity genes during neurodegeneration.
a-c, Representative immunohistochemical images depicting HDAC1-3 levels in area CA1 of CK-p25 mice and control littermates (n=3-6 slices from 3-4 mice each), scale bar, 20μm. d, Quantitative assessment of (a-c). e, Representative WB images and quantification of HDAC1-3 protein in the CK-p25 and control hippocampus (n=6-9 mice each). f-h, Quantitative PCR results of (f) HDAC2-, (g) AcH4K12-, and (h) RNA Pol II-immunoprecipitated chromatin at the promoter of neuroplasticity and housekeeping genes in the CK-p25 and control hippocampus. (i), Quantitative RT-PCR results of the same genes (f-i, n=4-8 animals each). Note that for (i) Bdnf “1”, “2” and “4” represent Bdnf exons I, II and IV, respectively. *p≤0.05; **p≤0.01; ***p≤0.001, values are mean ± s.e.m.
Next, we aimed to determine the functional consequences of elevated HDAC2. Since HDAC2 has been shown to associate with the promoter region of genes involved in memory and synaptic plasticity7, we hypothesized that it is enriched at these genes following neurodegeneration. Of the known HDAC2 targets7, we focused on those that, in several independent studies, had been demonstrated to be downregulated in the human AD brain (Supplementary Table 1). These include the immediate-early genes Arc, Bdnf exons I, II, and IV, Egr1, Homer1, and Cdk5, implicated in learning and memory, and genes related to synaptic plasticity such as the glutamate receptor subunits GluR1, GluR2, NR2A, and NR2B, as well as Nfl (neurofilament light chain), Svp (synaptophysin) and Stg (synaptotagmin). Using chromatin immunoprecipitation (ChIP, for primers see Supplementary Table 2), we found that HDAC2 is significantly enriched at these genes in the CK-p25 hippocampus, the exception being the promoter regions of the activity-dependent Bdnf exons I and II18, and the housekeeping genes β-actin, β-globin and β-tubulin (Fig. 1f). In contrast, binding of HDAC1 and HDAC3 was unaltered (Supplementary Fig. 6a, b). Interestingly, HDAC2 binding was not restricted to the promoter, as we also found HDAC2 more abundantly bound to the coding sequence of the same genes (Supplementary Fig. 7 and Supplementary Table 3 for primer sequences), in agreement with previous reports showing that HDAC2 can also bind to a gene’s coding region19.
We next assessed the acetylation of several histone (H) residues in the promoter region of these genes, for which acetylation has been shown to be important for learning, memory, and synaptic plasticity, such as H2B lysine (K) 5, H3K14, H4K5, and H4K124. ChIP analyses revealed a hypoacetylation for all residues at the neuroplasticity genes, (Fig. 1g, Supplementary Fig. 6c-e), albeit to different extents. Importantly, the acetylation of housekeeping genes was not altered. The effects of elevated HDAC2 levels appear to be restricted to histones, as we found no acetylation changes on other proteins regulated by this modification, such as tau, p53, and tubulin, nor in overall nuclear or cytoplasmic protein acetylation (Supplementary Fig. 8).
Next, to determine the functional consequences of promoter hypoacetylation, we assessed the binding of activated (i.e., phosphorylated) RNA Polymerase II (RNA Pol II), and found it to be markedly reduced (Fig. 1h). This prompted us to measure the mRNA expression of these genes by quantitative RT-PCR (primers in Supplementary Table 4). We found reduced expression for all genes with elevated HDAC2 binding and a concomitant decrease in histone acetylation and RNA Pol II binding (Fig. 1i). Of note, HDAC2 likely acts together with the transcriptional repressor complexes CoREST, NuRD, and Sin3, as we found increased association of HDAC2 with these complexes in hippocampal CK-p25 extracts by co-immunoprecipitation (Supplementary Fig. 9). Taken together, these results indicate that HDAC2 mediates a local chromatin compaction of neuroplasticity genes, which decreases their expression and may contribute to cognitive decline during neurodegeneration.
To causally examine such a scenario, we generated adeno-associated viral (AAV) vectors carrying either short-hairpin RNAs (shRNAs) directed against HDAC2 or scrambled control shRNAs (Supplementary Fig. 10a, b). The knockdown efficiency of the shRNAs in culture was about 25 - 30% (Supplementary Fig. 10c-f), ideally suited for the targeted normalization of the 20 - 50% increase of HDAC2 in the CK-p25 mice. We injected these vectors bilaterally into hippocampal area CA1 of two-week-induced CK-p25 and control mice, and assessed HDAC2 levels four weeks following viral injection (Supplementary Fig. 10g). CK-p25 animals injected with an shRNA against HDAC2 (CK-p25, shHDAC2) showed reduced HDAC2 levels compared to CK-p25 animals injected with control scrambled shRNA (CK-p25, scr), indistinguishable from control mice injected with scrambled shRNA (CON, scr) (Fig. 2a, b). Protein levels of HDAC1 and HDAC3 remained unchanged (Supplementary Fig. 11). We observed transduction efficiencies of 53 - 61% (mean ± s.e.m., 57.4 ± 2.5; n=3-4 mice per group) and comparable infection rates in both control and CK-p25 hippocampi (Supplementary Fig. 10h).
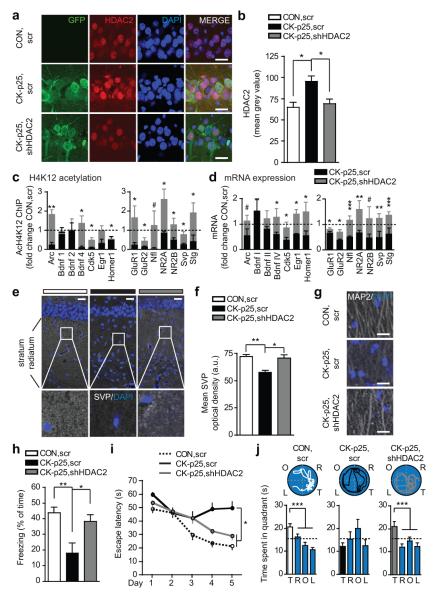
Figure 2. Reducing HDAC2 levels alleviates memory deficits.
a, Representative immunohistochemical images depicting HDAC2 in hippocampal area CA1 of CK-p25, shHDAC2, CK-p25, scr, and CON, scr animals; scale bar, 20μm. b, Quantitative assessment of (a), n=4-5 sections from 4 mice each. c, Quantitative PCR results of AcH4K12-immunoprecipitated chromatin in CK-p25, scr and CK-p25, shHDAC2 compared to CON, scr mice. d, Quantitative RT-PCR results of the same genes. (c, d, n=4-6 animals each). e, g, Representative immunohistochemical images depicting (c) Svp and (e) MAP2 immunoreactivity in the hippocampus stratum radiatum, scale bars c, 25μm, e, 20μm f, Quantitative assessment of (c), n=4 mice each. f, Freezing responses of CON, scr (n=18), CK-p25, scr (n=16) and CK-p25, shHDAC2 (n=16) mice 24 h after contextual fear conditioning. g, Escape latencies in a water maze task of CON, scr (n=19), CK-p25, scr (n=17) and CK-p25, shHDAC2 (n=19) animals. Data points are averages of two trials per day. h, Representative swim traces and time spent per quadrant during the water maze test (T, target quadrant, R=right, O=opposite, L=left of target). *p≤0.05; **p≤0.01; ***p≤0.001, values are mean ± s.e.m.
Next, we sought to determine whether reducing HDAC2 would alter the promoter histone acetylation and mRNA expression of neuroplasticity genes. We found that H4K12 acetylation was significantly enhanced on most of these genes, the majority of which also showed increased expression (Fig. 2c, d). Importantly, most of these genes showed comparable, or even higher, expression in CK-p25, shHDAC2 mice compared to CON, scr animals.
Based on these findings, we investigated whether such regained chromatin and transcriptional plasticity might translate into morphological and physiological changes in the surviving neurons. To this end, we measured synaptic density by Svp immunohistochemistry, labeling the presynaptic terminals of functional synapses, and dendritic abundance by microtubule-associated protein 2 (MAP2) immunoreactivity. We found that, whereas Svp and MAP2 were reduced in the stratum radiatum of CK-p25, scr animals (as previously described for CK-p25 mice13, 20), their abundance was markedly increased in CK-p25, shHDAC2 animals, to levels comparable to CON, scr animals (Fig. 2e-g). Interestingly, however, the number of surviving neurons, as evidenced by NeuN immunohistochemistry, was not altered by HDAC2 reduction (Supplementary Fig. 12). Then, we measured synaptic plasticity by electrophysiological recordings and observed that long-term potentiation (LTP) in CA1 neurons was undistinguishable between CK-p25, shHDAC2 and CON, scr animals, but significantly improved over CK-p25, scr animals. A similar effect was observed for basal synaptic plasticity (Supplementary Fig. 13). Together, these data indicate that, although HDAC2 normalization did not impact neuronal survival, it did reinstate morphological and synaptic plasticity in the surviving neurons.
We hypothesized that the reduction of HDAC2 would also counteract the cognitive deficits associated with neurodegeneration. For this, we assessed associative and spatial memory on a fear-conditioning and water maze task, respectively, two types hippocampus-dependent memory that are severely perturbed in CK-p25 animals13. We observed that associative memory of CK-p25, shHDAC2 animals returned to levels of CON, scr animals (Fig. 2h). Likewise, CK-p25, shHDAC2 animals showed significantly reduced escape latencies compared to CK-p25, scr animals during training in the water maze (Fig. 2i) and, 24h later, they spent significantly more time in the target quadrant, indistinguishable from the performance of CON, scr mice (Fig. 2j). Overall, swimming behavior was similar between the different groups (Supplementary Fig. 14a, b), and altering HDAC2 levels per se did not affect locomotor activity or anxiety as assessed by an open field test (Supplementary Fig. 14c-f). Together, these results suggest that elevated HDAC2 levels are causally involved in the cognitive decline associated with neurodegeneration in CK-p25 mice, but that the prevention of HDAC2 upregulation rescues memory capacities.
To gain insight into the mechanisms underlying the increase in HDAC2, we exposed primary hippocampal neurons to neurotoxic stimuli characteristic of AD-related neurodegeneration, hydrogen peroxide (H2O2) and amyloid-β (Aβ) oligomers21, 22. As revealed by immunocytochemistry and WB, treatment with either H2O2 or Aβ1-42, but not control Aβ42-1, oligomers was sufficient to increase HDAC2 (Fig. 3a-d, left panels; Supplementary Fig. 15a-d). Importantly, both neurotoxic stimuli increased Hdac2 at the mRNA level (Fig. 3e), and increased Hdac2 transcription was also evident in the CK-p25 hippocampus (Fig. 3f), suggesting the involvement of transcriptional mechanisms.
Figure 3. Neurotoxic insults increase HDAC2 via stress elements in its promoter.
a, c, Representative pictures of HDAC2 and PGR1 labeling of primary hippocampal neurons treated with (a) H2O2 and (c) Aβ-oligomers (n=20-40 neurons per group), scale bar, 10μm. b, d, Quantification of (a) and (c). e, f, Quantitative RT-PCR results showing Hdac2 expression in (e) H2O2- and Aβ-treated primary hippocampal neurons, and (f) in the CK-p25 hippocampus (n=7-9 mice each). g, Alignment of the vertebrate glucocorticoid responsive element (GRE) consensus sequence with the GRE in the proximal promoter of mouse Hdac2. h, Quantification and representative WB images of hippocampal extracts of CK-p25 versus control mice (n=3 each). i, Representative images of immunohistochemical labeling of PGR1 and HDAC2 in the CK-p25 hippocampus (n=3-6 slices from 3 mice each); scale bar, 20μm. j, Regression analysis of (i) showing a significant correlation between PGR1 and HDAC2 in CK-p25 (R2=0.686, p≤0.001), but not control mice (R2=0.019, n.s.). k, Quantification and representative WB images of Cdk5cKO and control Cdk5fl/fl forebrain extracts (n=3 each). l, Quantitative PCR results of PGR1-immunoprecipitated chromatin around the GRE in a 1.3 kb-wide Hdac2 promoter region (schematically shown above the graph; TSS, transcriptional start site) in the CK-p25 and control hippocampus (n=3-6 animals each); green lines represent fragments amplified by primer pairs. m, Luciferase activity of CAD cells transfected with the Hdac2 promoter with (orange) or without (blue) GRE (schematic of constructs shown above graph), and treated with H2O2 and Aβ1-42. n, Luciferase activity of CAD cells transfected with Hdac2-GRE in the presence of endogenous GR or of cotransfected GRS211A. In vitro results are from ≥3 independent experiments. *p≤0.05; **p≤0.01; ***p≤0.001, values are mean ± s.e.m.
This prompted us to screen the Hdac2 promoter for potential binding sites of transcriptional regulators. Using transcription factor binding databases23, we found a well-conserved recognition element for the glucocorticoid receptor 1 (GR1) in the proximal promoter region of Hdac2 (Fig. 3g). GRs are activated by phosphorylation following behavioral or cellular stress and, upon binding to the glucocorticoid responsive element (GRE) in a gene’s promoter region, GRs can act as transcriptional activators or repressors, depending, in part, on the residue phosphorylated24. Of the known phosphorylation sites, serine (S) 211 has been robustly associated with activated forms of GR125.
Based on this knowledge, we examined whether S211 phosphorylation on GR1 was increased following neurotoxic insults in vitro, and in the CK-p25 brain in vivo. Immunocytochemical labeling and WB of cultured hippocampal neurons following H2O2 and Aβ1-42 treatment revealed a significantly increased phosphorylation of GR1 on S211 (PGR1) compared to control conditions (Fig. 3a-d, right panels, Supplementary Fig. 15a-d, see Supplementary Fig. 2b for anti-PGR1S211 specificity). Furthermore, the CK-p25 hippocampus showed similarly increased PGR1 levels (Fig. 3h, i, Supplementary Fig. 16). Remarkably, we observed that the increase of PGR1 occurred concurrently with that of HDAC2 following neurotoxicity (Fig. 3a, c, i, j) and, using forebrain extracts of conditional Cdk5 knock-out (Cdk5cKO) and control mice26, we identified Cdk5 as a PGR1 kinase in vivo (Fig. 3k).
We then sought to determine whether GR1 phosphorylation increases Hdac2 transcription. We first examined the binding of PGR1 to the Hdac2 promoter by PGR1-ChIP and primer pairs (Supplementary Table 2) spanning a region from −1000bp to +300bp around the Hdac2 transcriptional start site (Fig. 3l, top). We found that PGR1 binding to the Hdac2-GRE was significantly increased in the CK-p25 hippocampus (Fig. 3l, bottom). Similar results were obtained following Aβ1-42 treatment of primary hippocampal cultures (Supplementary Fig. 15e). Second, to determine whether GR1 is directly capable of transcriptionally activating Hdac2, we cloned the Hdac2 promoter with and without the GRE into a luciferase construct and tested its activity in CAD cells, a primary neuron-like cell line. We found that the presence of the GRE alone increased the luciferase activity by approximately 3-fold but that, upon the addition of a constitutively active form of GR, GR526 (see Methods), this activity was further doubled. However, without the Hdac2-GRE, the addition of GR526 had no effect (Supplementary Fig. 17). Importantly, we found that the Hdac2-GRE also responds to neurotoxic stimuli. When CAD cells were treated with either H2O2 or Aβ1-42, luciferase activity was significantly increased, but only with the GRE present (Fig. 3m, orange bars). Furthermore, the capability of GR to activate Hdac2 critically depends upon its phosphorylation. When S211 was mutated to alanine (GRS211A), GR was no longer capable of activating Hdac2 in vitro (Fig. 3n). This dependency on S211 phosphorylation also occurs in vivo, as Cdk5cKO forebrain extracts26 had reduced PGR1 and HDAC2 levels (Fig. 3k). Taken together, these results suggest that AD-related neurotoxic stimuli lead to an increase in Hdac2 gene transcription by mechanisms involving GR activation and interaction with the Hdac2-GRE.
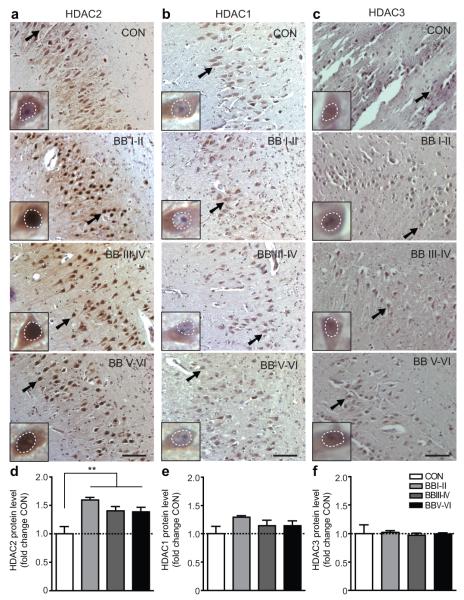
Lastly, to assess the relevance of these findings in humans, we compared HDAC2 abundance in post-mortem brain samples from patients with varying degrees of non-familial AD, the most common form of neurodegeneration-associated dementia worldwide27. The cases used here (Supplementary Table 5) are defined by the Braak and Braak (BB) stages28, which are characterized by the accumulation of hyperphosphorylated tau protein in the cortices (Supplementary Fig. 18), and by increasing neurodegeneration28 and cognitive impairment29. We found that, in all AD-related BB stages, HDAC2 was significantly elevated in hippocampal area CA1 (Fig. 4a, d) and the entorhinal cortex (Supplementary Fig. 19a, d), which are the earliest and most affected brain areas in AD2 and crucial for memory formation and storage14. HDAC2 accumulation was visible beginning at BB stage I/II, implicating it as an early event in the progress of AD. In contrast, levels of HDAC1 and HDAC3 were not altered (Fig. 4b, c, e, f; Supplementary Fig. 19b, c, e, f). Thus, elevated levels of HDAC2 may also accompany the cognitive decline of the human neurodegenerating brain.
Figure 4. HDAC2 expression is increased in Alzheimer’s disease patients.
a-c, Representative immunohistochemical images depicting nuclear HDAC1-3 levels (white dotted circles) in neurons (arrow points to magnified neuron in inset) of hippocampal area CA1 from patients with Braak and Braak (BB) stages I/II (n=4), III/IV (n=7) and V/VI (n=8) compared to healthy BB0 control brains (CON, n=7); scale bar, 100μm. d-e, Quantitative assessment of (a-c). **p≤0.01, values are mean ± s.e.m.
The findings presented in this study describe that epigenetic mechanisms substantially contribute to the cognitive decline associated with AD-related neurodegeneration. While it is well documented that neuronal loss and Aβ- or tau-induced neurotoxicity acutely disable synaptic functions, in turn leading to cognitive deficits1, 2, 30, the HDAC2-mediated epigenetic blockade of neuroplasticity-related gene expression could delineate a process by which memory functions become permanently impaired in the AD brain (Supplementary Fig. 1a). This blockade appears to be induced by GR1; thus, GRs may function as molecular mediators between neurodegeneration-associated neurotoxic stressors and cognitive impairment (Supplementary Fig. 1b).
Intriguingly, our findings may also provide a potential explanation, at least in part, as to why, in some clinical trials, cognitive impairments in AD patients persist despite successful Aβ clearance27: Once the epigenetic blockade is in place, reducing Aβ generation and deposition alone may not be sufficient to rescue against cognitive dysfunction. A more efficacious strategy may therefore lie in the combination of Aβ reduction with the inhibition of HDAC2. By extension, these findings pinpoint HDAC2 as the likely target of non-selective HDAC inhibitors that counteract cognitive decline in AD mouse models6 and, as a result, strongly advocate for the development of HDAC2-selective inhibitors. Finally, our finding that HDAC2 inhibition likely re-instates transcriptional, morphological, and synaptic plasticity in the surviving neurons of the neurodegenerating brain raises hope that such plasticity is not irrevocably lost, but merely constrained by the epigenetic blockade.
METHODS
Human material
Human material was obtained from the Massachusetts Alzheimer Disease Research Center at Massachusetts General Hospital (protocol number 2004-P-001613/4) and from the Boston Medical Center (H-24454) with informed consent by all donors. Hippocampal blocks (at the level of the lateral geniculate nucleus) were fixed in paraformaldehyde, paraffin-embedded and sectioned at 10μm thickness. Following antigen retrieval (Biogenex), brain slices were rehydrated using xylene/ethanol, blocked with 5% milk serum in TBS (pH 7.4) at room temperature and incubated at 4°C overnight in TBS containing 5% milk-serum in the following antibodies: amyloid-β, tau (Dako), HDAC1, HDAC2 (Abcam), or HDAC3 (Cell Signaling). Next, they were washed in PBS (pH 7.4), incubated in HRP-conjugated secondary antibodies (Biogenex) and visualized with DAB (Biogenex). Slides were counterstained with hematoxylin and dehydrated with ethanol/xylene. Images were quantified by an experimenter blind to BB stages using ImageJ 1.42q.
Animal models
All mouse work was approved by the Committee for Animal Care of the Division of Comparative Medicine at MIT. Adult (3-6 months old) male double transgenic CK-p25 mice9, 10, 13, 6-month old male transgenic 5xFAD mice16, and 3-4 month-old Cdk5cKO mice26 and their respective control littermates were used for the experiments unless otherwise noted; for CK-p25 mice, all behavioral experiments took place between 6-8 weeks of p25 induction, the time when cognitive deficits are first visible13. Behavioral experiments were conducted blindly and essentially as described13. Open field behavior was monitored using the VersaMax system (Accuscan) during 20min. For fear conditioning, mice were put in the conditioning chamber (TSE systems) for 3min, after which they received a one-time 2s footshock (0.8mA). Animals were then left in the box for another 30s. 24h later, the mice were put into the same box and their freezing behavior was scored during 3min. For the water maze that took place in a round tank (1.2m in diameter) filled with white opaque water, mice were first habituated to the task with the platform being visible for two trials. During habitation and the acquisition phase, mice were allowed to swim for 60s or until the reached the platform (monitored by HVS Image). Animals that did not reach the platform after 60s were gently guided towards it; all animals were allowed to remain on the platform for 15s. For testing, mice were put back into the water without the platform 24h after that last training session, from a starting position different of the last starting position during the acquisition phase, and their time spent in each quadrant was recorded (HVS Image).
Electrophysiology
To record field excitatory postsynaptic potentials, transverse hippocampal slices were prepared from CON, scr, CK-p25, scr and CK-p25, shHDAC2 mice. In brief, the brain was rapidly removed and transferred to ice-cold, oxygenated (95% O2 and 5% CO2) cutting solution containing 211mM sucrose, 3.3mM KCl, 1.3mM NaH2PO4, 0.5mM CaCl2, 10mM MgCl2, 26mM NaHCO3 and 11mM glucose. Hippocampal slices were cut with a VT1000S vibratome (Leica) and transferred for recovery to a holding chamber containing oxygenated artificial cerebrospinal fluid consisting of 124mM NaCl, 3.3mM KCl, 1.3mM NaH2PO4, 2.5mM CaCl2, 1.5mM MgCl2, 26mMNaHCO3 and 11mM glucose at 28-30°C for at least 1h before recording. CA1 field potentials evoked by Schaffer collateral stimulation were measured. After recording of a stable baseline (at least 20 min), LTP was induced by four episodes of theta burst stimulation (TBS) with 10s intervals. TBS consisted of ten bursts (each with four pulses at 100Hz) of stimuli delivered every 200ms. Recordings were performed using an AM-1800 Microelectrode amplifier (A-M systems) and a Digidata 1440A A-D converter (Axon Instruments). All data were digitized and analyzed by the use of pClamp10 software (Axon Instruments). Basal synaptic input/output relationship was obtained by plotting fEPSP slopes against stimulation intensities. All experiments were performed by an experimenter blind to treatment groups.
In vitro studies
Primary mouse hippocampal neuronal cultures (DIV 14-17) were treated with sense and antisense Aβ oligomers (1μM, Bachem) for 24h or H2O2 (50μM, Mallinckrodt Chemicals, removed after 5min, and assessed 8h later) unless otherwise noted and mRNA and proteins extracted using Qiagen’s Allprep kit. For ChIP experiments, cortical cultures (DIV10-14) were used.
Dual luciferase assays were conducted on CAD cells31 (ATCC) that were transfected using lipofectamine (Invitrogen) with 0.5μg of the proximal promoter region of HDAC2 containing the GRE consensus sequence (CAAGAAGAAAGTGGCTAC) or with the proximal promoter region without the GRE sequence subcloned into the pGL3 reporter vector (Promega) according to the manufacturer’s instructions. Cells were cotransfected with 0.05μg of the constitutively active form of GR32, GR526, or GRS211A, and treated with 1μM Aβ1-42 oligomers or with 50μM H2O2.
For site-directed mutagenesis, the cDNA for the full-length human glucocorticoid receptor (Addgene) was used and serine 211 was replaced by alanine using the QuickChange Lightning Kit (Agilent Technologies) as per the manufacturer’s instructions. All constructs were verified by sequencing (Genewiz). The following mutagenesis primers were used:
5′-GTAAAGAGACGAATGAGGCTCCTTGGAGATCAGACC-3′ (forward)
5′-GGTCTGATCTCCAAGGAGCCTCATTCGTCTCTTTAC-3′ (reverse).
Immunohistochemistry (IHC) and immunocytochemistry (ICC)
ICC and mouse IHC were performed as described7. ICC and IHCs on different experimental conditions were performed with the same antibody solution at the same time to assure identical staining conditions. A negative (i.e., no antibody) control was included simultaneously. In brief, for IHC, mice were perfused with 10% paraformaldehyde under deep anesthesia (ketamine, xylazine) and their brains sectioned at 0.35μm thickness using a vibratome (Leica). For ICC, cells were fixed using 4% paraformaldehyde. Slices/cells were permeabilized with 0.1% Triton X-100, blocked and incubated overnight with 0.1% Triton X-100/10% fetal bovine serum in PBS containing primary antibodies: HDAC1, HDAC2 (Abcam), HDAC3 (Cell Signaling), phospho-GR1 (S211) (Cell Signaling), GR (Abcam), or GFP (Aves Labs). Primary antibodies were visualized with Alexa-Fluor 488, Cy3 and Cy5 antibodies (Molecular Probes), neuronal nuclei with Hoechst 33342 (Invitrogen). Note that for staining of shRNA-injected animals, mCherry was visualized without staining, and only Cy2 and Cy5 secondary antibodies were used. Images were acquired using a confocal microscope (LSM 510, Zeiss) at identical settings at the highest intensity for each of the conditions. Images were quantified using ImageJ 1.42q by an experimenter blind to treatment groups, whenever possible. Per experimental condition, 20-40 representative cells were analyzed, and the mean signal intensity was measured. To assess the specificity of the HDAC2 immunostaining, adult HDAC2-/- mice were used as previously described7. For a specificity control of the PGR1 antibody, Calf Intestine Phosphatase (CIP) (New England Biolabs) treatment on ICC slides was performed as described previously33. CIP or H2O were applied for 40min in Buffer 3 (New England Biolabs).
Molecular analyses
Western blots, co-immunoprecipitation (Co-IP), chromatin immunoprecipitation (ChIP), and gene expression analyses were performed as described elsewhere34 with the following modifications, and expressed as fold change of the respective control conditions. For Western blots, proteins were extracted using 1x RIPA buffer containing proteinase (complete, Roche) and phosphatase inhibitors (1mM β-glycerophosphate, 10mM NaF, 0.1mM Na3VO4), transferred onto PVDF membranes (Biorad) and stripped using stripping buffer (Thermo Scientific). The following primary antibodies were used: Acetyl-K (Cell Signaling), α- and β-tubulin, β-actin (Sigma), Cdk5 (Santa Cruz), HDAC1, HDAC2 (Abcam), HDAC3, Phospho-GR1 (S211) (Cell Signaling), GR1 (Abcam), p53 (Cell Signaling), or tau (Invitrogen). Secondary antibodies were HRP-linked (GE Healthcare). Signal intensities were quantified using ImageJ 1.42q and normalized to values of β-actin, α- or β-tubulin. Phospho-GR1 was first normalized to GR1. 3-8 animals were used per condition. Cytoplasmic and nuclear fractionation was performed as described elsewhere34.
For Co-IP, hippocampal lysates were incubated with HDAC2 (Abcam) or IgG (Sigma) and the immunoprecipitated extracts probed for mSin3A, MTA2 (Abcam), CoRest (Millipore), LSD1 (Cell Signaling), or HDAC2. Signal intensities were quantified using ImageJ 1.42q, and normalized to input. 3-4 animals were used per condition.
For ChIP, tissue samples were homogenized in cell lysis buffer containing proteinase (complete, Roche) and phosphatase inhibitors (1mM β-glycerophosphate, 10mM NaF, 0.1mM Na3VO4) and chromatin was sonicated using a Branson Digital Sonifier with 10 rounds of 15s at 25% power per sample on ice to 200-400bp in length. For ChIP of primary cortical cultures, approximately 1×106 cells were crosslinked in 37% formaldehyde (Sigma), quenched with 20× glycine (Sigma), washed with PBS and cell lysis buffer containing both phosphatase and proteinase inhibitors (Roche) and sonicated in nuclear lysis buffer using a Vibra Cell Sonifier with 3× 3 pulses of 5s at 35% power (50% duty) on ice. Sheared chromatin was immunoprecipitated with antibodies against HDAC1, HDAC2 (Abcam), HDAC3 (Santa Cruz), acetyl H2BK5, acetyl H3K14 (Abcam), acetyl H4K5, acetyl H4K12 (Millipore), phospho-GR1 (S211) (Cell Signaling), or phospho-RNA Pol II (Abcam). DNA was extracted by phenol/chloroform/isoamyl alcohol (American Bioanalytical) and subjected to quantitative PCR (Biorad Thermal Cycler) using primers specific to the promoter regions of the genes assayed (see Supplementary Table 2 and 3 for primer sequences). The fluorescent signal of the amplified DNA (SYBR green, Biorad) was normalized to input. 4-8 samples were used per condition.
For gene expression analysis, mRNA was extracted (Qiagen), reverse-transcribed (Invitrogen) and quantitatively amplified on a thermal cycler (Biorad) using SYBR green (Biorad) and gene-specific primers (see Supplementary Table 4). The comparative Ct method35 was used to examine differences in gene expression. Values were normalized to expression levels of Gapdh. 4-8 samples were used per condition.
Experimental manipulations
For validation of the knockdown efficacy of HDAC2 RNAi in neurons, short hairpins targeting the open reading frame of mouse Hdac2 mRNA from the Broad Institute’s RNAi consortium shRNA library (www.broadinstitute.org/rnai/trc/lib) were packaged into lentiviral vectors36 and used to infect dissociated primary mouse embryonic (gestation day 18) cortical cultures, prepared as described7. Primary cultures were infected at DIV4. Cells were harvested ten days after infection, and protein levels were measured by western blotting. The target sequences for the two effective shRNAs are CCCAATGAGTTGCCATATAAT (HDAC2 shRNA 2-1, TRCN0000039395) and CGAGCATCAGACAAACGGATA (HDAC2 shRNA 2-4, TRCN0000039397).
After validation, scramble shRNA37 or HDAC2-shRNA constructs were subcloned under the CaMKII-U6 promoter into the pAAV entry vector fused to mCherry, tested again in mouse primary hippocampal cultures (DIV7), and high titer (1-4×1012 viral particles) adeno-associated viruses (AAV, serotype 2.5) were produced at the University of North Carolina Vector Core facility. 1μl of shRNA-containing AAV was stereotaxically injected into hippocampal area CA1 (anterior-posterior position −2.0mm, medial-lateral position ±1.6mm, dorso-ventral −1.5mm from Bregma) of both hemispheres at 0.1μl/min. Injection needles were left in place 5min after injection to assure even distribution of the virus. Injections were performed 4 weeks before behavioral testing. All infusion surgeries were performed under aseptic conditions and anesthesia (ketamine/xylazine) in accordance with the Massachusetts Institute of Technology’s Division of Comparative Medicine guidelines.
Statistics
Statistical analyses were performed using GraphPad Prism 5. One-way ANOVAs followed by Tukey post-hoc tests, or one-tailed Student’s t-tests were used unless otherwise indicated. All data are represented as mean ± s.e.m. Statistical significance was set at p≤0.05.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alison Mungenast, Stephanie Jemielity, Ram Madabushi, Froylan Calderon de Anda, and Meryl Horn for careful reading of the manuscript, A.M. for manuscript editing, Mali Eichler for mouse colony maintenance, Karlotta Fitch for sectioning the human brain samples, and M.H. for quantification of Fig. 2a. This work was partially supported by the Stanley Medical Research Institution (SJH, L-HT), NIH/NIDA (RO1DA028301, SJH) and NIH/NINDS (RO1NS078839, L-HT). JG was supported by a Bard Richmond fellowship and by the Swiss National Science Foundation, WYW by the Simons Foundation, MK by the Theodor und Ida Herzog-Egli foundation. L-HT is an investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTION This study was designed by JG and L-HT, and directed and coordinated by L-HT. JG planned and performed the in vitro, CK-p25, 5xFAD and Cdk5cKO mouse and human in vivo biochemical characterization, and the behavioral experiments. DR planned and contributed to the in vitro and CK-p25 in vivo experiments, generated the GR526 and the shRNA constructs, and contributed to the stereotaxic injections. JSG initiated and contributed to the CK-p25 and the 5xFAD biochemical characterization, and performed the HDAC2-/- experiments. WYW generated the luciferase constructs. JS performed the electrophysiological experiments. KMH, TJFN, DF, and SJH characterized the shRNA constructs. MK contributed to the qRT-PCR experiments and performed the quantification of the human data. SCS performed the site-directed mutagenesis. AS contributed to the immunohistochemistry and the qRT-/qPCR experiments. NJ contributed to the behavioral and qRT-/qPCR experiments. PFK and ID provided the human samples and contributed to the optimization of their staining. The manuscript was written by JG and L-HT, and commented on by all authors.
The authors declare no competing financial interests.
REFERENCES
- 1.Walsh DM, Selkoe DJ. Deciphering the molecular basis of memory failure in Alzheimer’s disease. Neuron. 2004;44:181–93. doi: 10.1016/j.neuron.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: the challenge of the second century. Sci Transl Med. 2011;3:77sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–8. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 4.Gräff J, Kim D, Dobbin MM, Tsai LH. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol Rev. 2011;91:603–49. doi: 10.1152/physrev.00012.2010. [DOI] [PubMed] [Google Scholar]
- 5.Brownell JE, Allis CD. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–84. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 6.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–68. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 7.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhtar MW, et al. Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J Neurosci. 2009;29:8288–97. doi: 10.1523/JNEUROSCI.0097-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH. Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron. 2003;40:471–83. doi: 10.1016/s0896-6273(03)00627-5. [DOI] [PubMed] [Google Scholar]
- 10.Cruz JC, et al. p25/cyclin-dependent kinase 5 induces production and intraneuronal accumulation of amyloid beta in vivo. J Neurosci. 2006;26:10536–41. doi: 10.1523/JNEUROSCI.3133-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz JC, Tsai LH. A Jekyll and Hyde kinase: roles for Cdk5 in brain development and disease. Curr Opin Neurobiol. 2004;14:390–4. doi: 10.1016/j.conb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Patrick GN, et al. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature. 1999;402:615–22. doi: 10.1038/45159. [DOI] [PubMed] [Google Scholar]
- 13.Fischer A, Sananbenesi F, Pang PT, Lu B, Tsai LH. Opposing roles of transient and prolonged expression of p25 in synaptic plasticity and hippocampus-dependent memory. Neuron. 2005;48:825–38. doi: 10.1016/j.neuron.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 14.Frankland PW, Bontempi B. The organization of recent and remote memories. Nature Reviews Neuroscience. 2005;6:119–130. doi: 10.1038/nrn1607. [DOI] [PubMed] [Google Scholar]
- 15.McQuown SC, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–74. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oakley H, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26:10129–40. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura R, Devi L, Ohno M. Partial reduction of BACE1 improves synaptic plasticity, recent and remote memories in Alzheimer’s disease transgenic mice. J Neurochem. 2010;113:248–61. doi: 10.1111/j.1471-4159.2010.06608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauterborn JC, et al. Differential effects of protein synthesis inhibition on the activity-dependent expression of BDNF transcripts: evidence for immediate-early gene responses from specific promoters. J Neurosci. 1996;16:7428–36. doi: 10.1523/JNEUROSCI.16-23-07428.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun JM, Chen HY, Davie JR. Differential distribution of unmodified and phosphorylated histone deacetylase 2 in chromatin. J Biol Chem. 2007;282:33227–36. doi: 10.1074/jbc.M703549200. [DOI] [PubMed] [Google Scholar]
- 20.Fischer A, Sananbenesi F, Wang XY, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–82. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 21.Markesbery WR, Carney JM. Oxidative alterations in Alzheimer’s disease. Brain Pathol. 1999;9:133–46. doi: 10.1111/j.1750-3639.1999.tb00215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yankner BA. Mechanisms of neuronal degeneration in Alzheimer’s disease. Neuron. 1996;16:921–32. doi: 10.1016/s0896-6273(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 23.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–4. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chrousos GP, Kino T. Glucocorticoid signaling in the cell. Expanding clinical implications to complex human behavioral and somatic disorders. Ann N Y Acad Sci. 2009;1179:153–66. doi: 10.1111/j.1749-6632.2009.04988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kino T, et al. Cyclin-dependent kinase 5 differentially regulates the transcriptional activity of the glucocorticoid receptor through phosphorylation: clinical implications for the nervous system response to glucocorticoids and stress. Mol Endocrinol. 2007;21:1552–68. doi: 10.1210/me.2006-0345. [DOI] [PubMed] [Google Scholar]
- 26.Guan JS, et al. Cdk5 Is Required for Memory Function and Hippocampal Plasticity via the cAMP Signaling Pathway. PLoS One. 2011;6:e25735. doi: 10.1371/journal.pone.0025735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Citron M. Alzheimer’s disease: strategies for disease modification. Nat Rev Drug Discov. 2010;9:387–98. doi: 10.1038/nrd2896. [DOI] [PubMed] [Google Scholar]
- 28.Braak H, Braak E. Diagnostic criteria for neuropathologic assessment of Alzheimer’s disease. Neurobiol Aging. 1997;18:S85–8. doi: 10.1016/s0197-4580(97)00062-6. [DOI] [PubMed] [Google Scholar]
- 29.Grober E, et al. Memory and mental status correlates of modified Braak staging. Neurobiol Aging. 1999;20:573–9. doi: 10.1016/s0197-4580(99)00063-9. [DOI] [PubMed] [Google Scholar]
- 30.Palop JJ, Mucke L. Amyloid-beta-induced neuronal dysfunction in Alzheimer’s disease: from synapses toward neural networks. Nat Neurosci. 2009;13:812–8. doi: 10.1038/nn.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Y, Wang JK, McMillian M, Chikaraishi DM. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J Neurosci. 1997;17:1217–25. doi: 10.1523/JNEUROSCI.17-04-01217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–4. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 33.Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–82. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- 34.Koshibu K, et al. Protein phosphatase 1 regulates the histone code for long-term memory. J Neurosci. 2009;29:13079–89. doi: 10.1523/JNEUROSCI.3610-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 37.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.