Abstract
Defining glioma subtypes based on objective genetic and molecular signatures may allow for a more rational, patient-specific approach to molecularly targeted therapy. However, prior studies attempting to classify glioma subtypes have given conflicting results. We aim to complement and validate the existing molecular classification system on a large number of samples from an East Asian population. A total of 225 samples from Chinese patients was selected for whole genome gene expression profiling. Consensus clustering was applied. Three major groups of gliomas were identified (referred to as G1, G2, and G3). The G1 subgroup correlates with a good clinical outcome, young age, and extremely high frequency of IDH1 mutations. Relative to the G1 subgroup, the G3 subgroup is correlated with a poorer clinical outcome, older age, and a very low rate of mutations in the IDH1 gene. Correlations of the G2 subgroup with respect to clinical outcome, age, and IDH1 mutation fall between the G1 and G3 subgroups. In addition, the G2 subtype was associated with a higher percentage of loss of 1p/19q when compared with G1 and G3 subtypes. Furthermore, our classification scheme was validated on 2 independent datasets derived from the cancer genome atlas (TCGA) and Rembrandt. With use of the TCGA classification system, proneural, neural, and mesenchymal, but not classical subtype, associated gene signatures were clearly defined. In summary, our results reveal that 3 main subtypes stably exist in Chinese patients with glioma. Our classification scheme may reflect the clinical and genetic alterations more clearly. Classical subtype–associated gene signature was not found in our dataset.
Keywords: Chinese Glioma Genome Atlas, glioma, microarray, molecular classification
Glioma is the most common type of brain tumor and is an important cause of cancer-related mortality among adults and children.1 Glioblastoma multiforme (GBM), which is the most lethal type of glioma, often demonstrates resistance to traditional cancer treatments, such as surgery, chemotherapy, and radiation, and quickly invades healthy brain tissue. Biotherapy and molecularly targeted therapy represent promising avenues for the future of effective glioma therapy.2 However, the present grading system for gliomas, which is based on histopathological diagnosis, cannot provide sufficient details for patient-specific biotherapy and molecularly targeted therapy. Furthermore, the current grading system has been associated with significant intra-observer variability. Moreover, the etiology underlying the development of the glioma subtypes is unclear. Thus, a glioma classification system based on genetic expression profiles may offer an objective means to identify subtype- or patient-specific therapeutic targets for biotherapy and molecularly targeted therapy.
Past glioma classification schemes have used gene expression data with varying success and with only moderate concordance between studies. The TCGA network describes a robust gene expression-based molecular classification of GBM into proneural, neural, classical, and mesenchymal subtypes.3 A study by Phillips et al. defined 3 subtypes of high-grade gliomas based on their molecular signatures: mesenchymal, proneural, and proliferative.4 In another study using unsupervised clustering, Li et al. identified 2 main glioma subtypes that they identified as GBM-rich (mesenchymal) and oligodendroglioma-rich (proneural).5 Only the mesenchymal and proneural subtypes were identified consistently through the various datasets. Before our current study, there existed no dataset generated from a large number of samples from an East Asian population that could be used for glioma classification.
In this study, we used 225 glioma samples from Chinese patients to complement and validate existing molecular subtyping systems. All samples were subjected to whole genome gene expression profiling. From the results, we were able to derive 3 major groups of gliomas that we call G1, G2, and G3. The G1 subgroup was characterized by good clinical outcome, young age, and a high rate of mutations in the IDH1 gene. Relative to G1, the G3 subgroup was characterized by poorer clinical outcome, older age, and a lower rate of mutations in the IDH1 gene. Correlations involving the G2 subtype with respect to clinical outcome, age, and IDH1 mutation fall between the G1 and G3 subgroups. Our classification scheme was validated on 2 independent datasets derived from the cancer genome atlas (TCGA) and Rembrandt. Using the TCGA classification system, we annotated our samples and found that the G1, G2, and G3 subgroups were enriched with proneural, neural, and mesenchymal gliomas, respectively. Of note, no classical subtype–associated gene signature was identified in our dataset.
Materials and Methods
Patients and Samples
Two hundred twenty-five samples from the Chinese Glioma Genome Atlas (CGGA) were included in this study, composed of 5 normal brain tissue samples (NB; 3 normal adult brain samples were obtained after informed consent from patients with severe traumatic brain injury who needed post-trauma surgery; 2 other normal samples were from patients who had undergone surgery for primary epilepsy), 58 astrocytomas (A), 17 oligodendrogliomas (O), 22 oligoastrocytomas (OA), 8 anaplastic astrocytomas (AA), 11 anaplastic oligodendrogliomas (AO), 15 anaplastic oligoastrocytomas (AOA), 4 secondary GBMs, and 85 primary GBMs. All of the patients underwent surgical resection from January 2005 through December 2009 and subsequently received radiation therapy and/or alkylating agent–based chemotherapy. Patients were eligible for the study if their diagnosis was established histologically by 2 neuropathologists according to the 2007 World Health Organization classification guidelines. Secondary GBM was defined according to guidelines established by Scherer. Tumor tissue samples were obtained by surgical resection before patients underwent radiation and/or chemotherapy. Only samples with >80% tumor cells were selected for analysis. This study was approved by the institutional review boards of all hospitals involved in the study, and written informed consent was obtained from all patients.
RNA Extraction and Whole Genome Gene Profiling
All tissue samples were immediately snap-frozen in liquid nitrogen after surgery. A hematoxylin and eosin–stained frozen section was prepared from each sample to assess the percentage of tumor cells before RNA extraction. Only samples with >80% tumor cells were selected for RNA extraction. Total RNA from frozen tumor samples was extracted using the mirVana miRNA Isolation kit (Ambion) according to the manufacturer's protocol. RNA concentration and quality were measured using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies).
Microarray analysis was performed on all 225 samples using the Agilent Whole Human Genome Array according to the manufacturer's instructions. The integrity of total RNA was checked using an Agilent 2100 Bioanalyzer (Agilent). cDNA and biotinylated cRNA were synthesized and hybridized to the array. Data were acquired using the Agilent G2565BA Microarray Scanner System and Agilent Feature Extraction Software (version 9.1). Probe intensities were normalized using GeneSpring GX 11.0.
Molecular Analyses
Genomic DNA was isolated from frozen tumors with the QIAamp DNA Mini Kit (Qiagen). Pyrosequencing of IDH1 mutation was supported by Genetech (Shanghai, China) and performed on a Pyro-Mark Q96 ID System (Qiagen, Valencia, Calif). The primers 5′-GCT TGT GAG TGG ATG GGT AAA AC-3′ and 5′-biotin-TTG CCA ACA TGA CTT ACT TGA TC-3′ were used for PCR amplification, and the primer 5′-TGG ATG GGT AAA ACC T-3′ was used for pyrosequencing.
Loss of the 1p and 19q chromosome arms was analyzed using denaturing high-performance liquid chromatography (DHPLC) as previously described.6
Consensus Clustering and Statistical Analysis
Median absolute deviation (MAD) was calculated using Matlab software. Probes showing highly variable expression (MAD > 1.0; probe number = 1801; gene number = 1577) were used for consensus clustering.7 Consensus clustering was performed using the hierarchical clustering method with average linkage and a distance metric equal to 1 minus the Pearson correlation coefficient. A total of 100 permutation tests were performed with a subsampling ratio of 0.8. The optimal number of glioma subgroups was determined using a consensus clustering cumulative distribution function and consensus matrices.
Kaplan-Meier survival analysis was used to estimate the survival distributions. The log-rank test was used to assess the statistical significance between stratified survival groups with use of GraphPad Prism, version 4.0 statistical software. Cox proportional hazard regression analyses were performed using SPSS, version 13.0, software for Windows (SPSS). Student's t test was used to determine significant differences. Gene ontology (GO) analysis was performed using DAVID.8 All data are presented as the mean ± standard error. A 2-sided P value of <.05 was regarded as significant. Prediction Analysis of Microarrays was used to annotate the CGCG samples with proneural, neural, classical, and mesenchymal labels.9
Results
Consensus Clustering Identified 3 Subtypes of Glioma
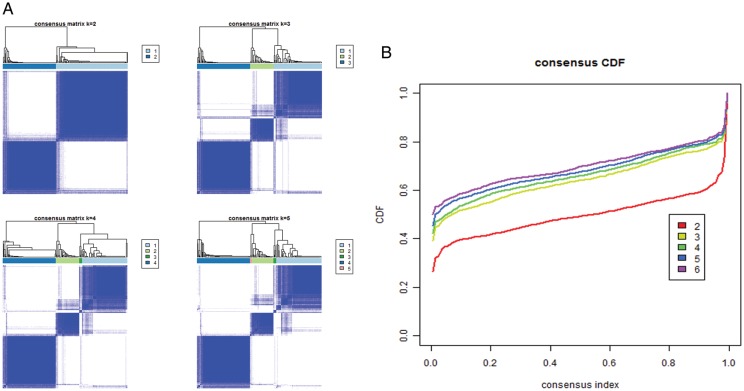
Whole genome gene expression profiles were obtained for all 225 samples using microarrays (Agilent). From the microarray data, we analyzed 1577 unique genes by selecting 1801 probes (Supplementary material, Table S1) that demonstrated highly variable expression across samples (MAD > 1.0). Consensus average linkage clustering of the 225 samples identified 3 robust clusters with clustering stability increasing between k = 2 and k = 3. However, for k > 3, clustering stability did not improve (Figs 1A, B and 2A).
Fig. 1.
Identification of 3 glioma subtypes using consensus clustering. (A) Consensus clustering matrix of 225 CGGA samples for k = 2 to k = 5. (B) Consensus clustering CDF for k = 2 to k = 6.
Fig. 2.
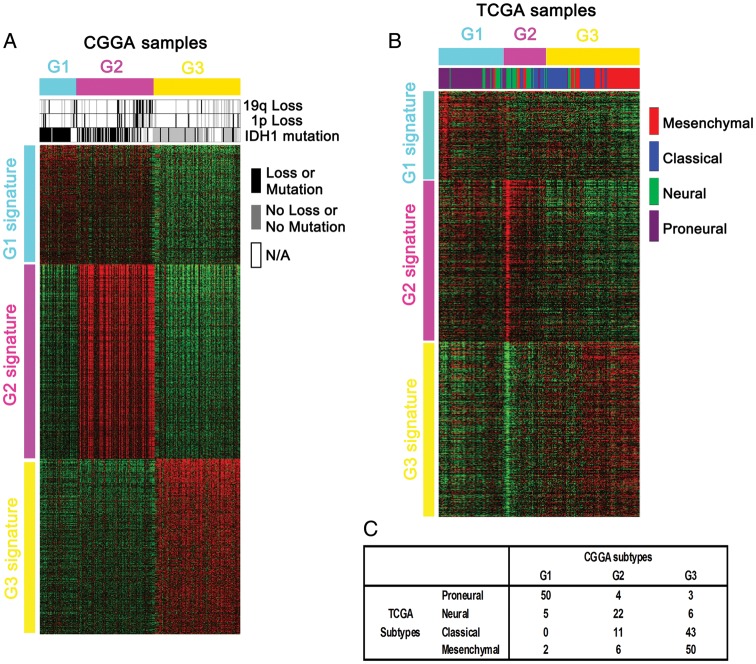
Heat maps of the 3 subtypes in the training and validation samples. (A) With use of our 1801 probe sets, samples were ordered on the basis of subtype predictions, and genes were clustered using the 225 CGGA samples. (B) Gene order from the CGGA samples was maintained in the validation data set (n = 202), which is composed of only GBMs that were molecularly classified in the TCGA. (C) Matrix of subtype grouping for the TCGA samples (n = 202) as predicted by TCGA or CGGA classification systems.
Clinical and Molecular Characteristics of the 3 Subclasses of Glioma
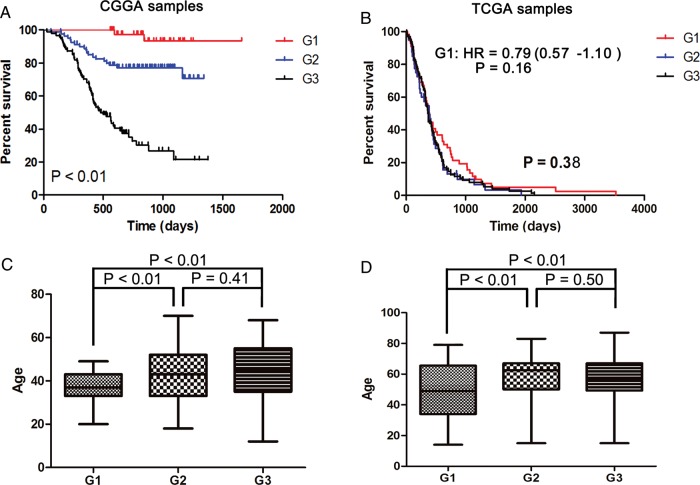
We observed that consensus clustering of 1181 of the most variable probes yielded robust differences in the clinical characteristics of the 3 glioma subclasses. Survival analysis showed that patients with the G1 glioma subtype lived significantly longer than did patients in the G2 and G3 subgroups (P < .01, log-rank) (Fig. 3A). The G2 subgroup had a better prognosis when compared with the G3 group (P < .01, log-rank) (Fig. 3A). Furthermore, Multivariable cox analysis including the new classification scheme, tumor grade, patient age, KPS score, and IDH1 mutation status was also included (Supplementary material, Table S2). The results show that the new classification scheme has an independent prognostic value when considering tumor grade, patient age, KPS score, and IDH1 mutation status. In the G1 subgroup, patients tended to be younger (P value < .01 [versus G2] and P<.01 [versus G3]) (Fig. 3C). We found no difference in the age at diagnosis in the G2 and G3 groups (P value = .41) (Fig 3C). As shown in Table 1, 100%, 60.61%, and 10.98% of samples in the G1, G2, and G3 groups, respectively, were found to carry mutations in the IDH1 gene. Loss of chromosome arm 1p was found in 30.77%, 36.67%, and 16.67% of G1, G2, and G3 samples, respectively. Loss of chromosome arm 19q was found in 23.08%, 46.67%, and 4.17% in G1, G2, and G3 samples, respectively. Our results also showed that the G3 group consisted of more GBMs than either G1 or G2. Only 3 primary GBMs and 1 secondary GBM were included in the G1 subtype, and all 4 GBMs of the G1 group harbored IDH1 mutations. We also noted that the G1 and G2 tumors tended to occur in the frontal lobe more often than did tumors of the G3 subtype.
Fig. 3.
Survival analysis and age distribution of G1, G2, and G3 subgroups in CGGA samples (A and C) and TCGA samples (B and D).
Table 1.
Clinical characteristics of the 3 subtypes in CGGA samples
| Variable | G1 subtype | G2 subtype | G3 subtype |
|---|---|---|---|
| Sex (female/male) | 21/21 | 41/46 | 37/59 |
| Age at diagnosis (year) | 36.81 ± 6.65 | 42.62 ± 11.63 | 44.12 ± 13.48 |
| IDH1 mutation (%) | 100 | 60.61 | 10.98 |
| 1p Loss (%) | 30.77 | 36.67 | 16.67 |
| 19 Loss (%) | 23.08 | 46.67 | 4.17 |
| Histopathology subtypes | |||
| Astrocytoma | 26 | 28 | 4 |
| Oligodendrocytoma | 5 | 12 | 0 |
| Oligoastrocytoma | 4 | 15 | 3 |
| Anaplastic Astrocytoma | 0 | 4 | 4 |
| Anaplastic Oligodendrocytoma | 2 | 5 | 4 |
| Anaplastic Oligoastrocytoma | 1 | 5 | 9 |
| Secondary GBM | 1 | 2 | 1 |
| Primary GBM | 3 | 11 | 71 |
| Normal brain tissues | 0 | 5 | 0 |
The new classification scheme could divide GBM samples into different prognostic groups very clearly (Supplementary material, Fig. S2A). Of them, the G1 subgroup has the best prognosis, the G2 subgroup has the poorest prognosis, and the G3 subgroup has the modest prognosis. However, in all gliomas, the G2 subgroup has the modest prognosis, whereas the G3 subgroup has the poorest prognosis (Fig. 3A). It seems a little strange, which seems unusual. Furthermore, we also compare the different prognosis among proneural, neural, classical, and mesenchymal subgroups, annotated by the TCGA classification scheme (Supplementary material, Fig. S2B). Although the overall P value was not statistically significant, we still found that the proneural subgroup has the best prognosis, the neural subtype has the poorest prognosis, and the classical and mesenchymal subgroups have the modest prognosis. In all gliomas, the neural subgroup has the modest prognosis, whereas the classical and mesenchymal subgroups have the poorest prognosis (Fig. 4B). In comparison with Supplementary material, Fig S2A and B, our classification scheme could stratify GBM samples into different prognostic subgroups more clearly. There are many overlaps between the G2 and neural samples. We may postulate that the G2 or neural subgroups may have a more rapid progression when at a different stage of development.
Fig. 4.
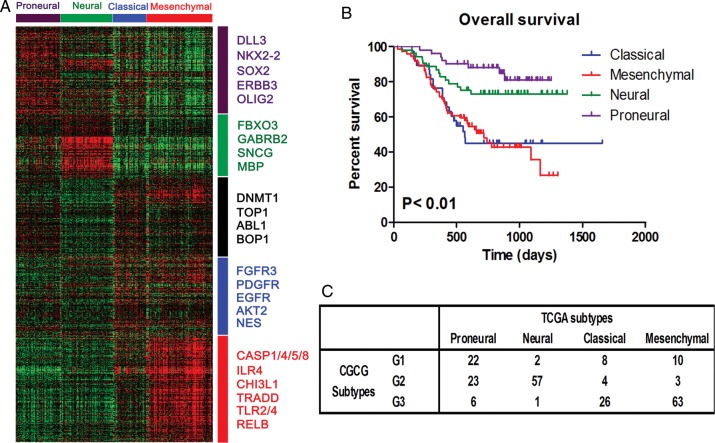
Classification of CGGA samples using the TCGA system. (A) Genes were ordered according to the predictive 840-gene list of the TCGA classification system, and the 225 CGGA samples were clustered. No classical subtype was identified in the heat map. (B) Kaplan–Meier survival analysis shows that the proneural subtype has significantly better survival for all cases (n = 217, P < .01). (C) Matrix of subtype grouping for CGGA samples (n = 225) as predicted by TCGA or CGGA classification systems.
GO analysis of the gene signatures of the 3 glioma subtypes from the 1577 genes used for classification was further performed in the CGGA samples. As shown in Supplementary material, Table S3, uniquely upregulated genes of the G1 group are associated with various processes, including neuron differentiation, cell adhesion, biological adhesion, cell-to-cell signaling, and regulation of neurogenesis. The uniquely upregulated genes in the G2 group are associated with synaptic transmission, transmission of nerve impulses, cell-cell signaling, ion transport, and regulation of system process. Uniquely upregulated genes in the G3 group are associated with response to wounding, skeletal system development, inflammatory response, immune response, and antigen processing and presentation via major histocompatibility complex class II.
Subtype Validation
After constructing our subgroups, we wanted to validate our classification scheme with an external dataset. To this end, we applied our subgrouping scheme on the gene expression data from the TCGA-containing GBMs only.3 As shown in Fig. 2A, CGGA samples were ordered on the basis of subtype predictions, and genes were clustered using the 1801 probes. Gene order from the CGGA samples was maintained in the validation dataset (n = 202; 993 genes available), which comprises all GBMs used for molecular classification of the TCGA. The G1, G2, and G3 subtypes and their respective gene signatures were clearly identified in 202 of the TCGA samples (Fig. 2B). G1 samples were enriched with the proneural subtype, whereas G2 samples were enriched with the neural subtype. G3 samples were enriched with the classical and mesenchymal subtypes (Fig. 2C). TCGA tumors of the G1 subgroup demonstrated a trend toward an increase in overall survival, although the difference was not statistically significant (P = .3767, log-rank test; hazard ratio = 0.790; 95% confidence interval, 0.569–1.097; P = .160, Cox-regression analysis). No difference in survival was found between G2 and G3 samples in the TCGA GBM samples. The age at diagnosis of patient with tumors in the G1 group in the TCGA samples was significantly younger than patients with tumors in either the G2 or the G3 groups (P = .0070 and .0030, respectively) (Fig. 3D). No difference in age at diagnosis was found between TCGA samples in groups G2 and G3. These results validate our classification system based on the CGGA samples and suggest that our subgrouping scheme could be applied to other independent samples to differentiate them based on their characteristics.
To further validate our classification scheme, we analyzed a Rembrandt dataset including all 475 samples (including all gliomas and nontumor controls). As shown in Supplementary material, Fig. S3A, our classification scheme could divide 475 samples from Rembrandt dataset into G1, G2, and G3 subgroups very clearly. Similar prognostic results are also obtained (Supplementary material, Fig. S3B). To better validate the 3 robust subtypes, we used the 2000 most differentially expressed probes in 475 samples from the Rembrandt dataset for clustering. Three main clusters (clusters 1, 2, and 3) were well defined (Supplementary material, Fig. S4). Four hundred thirty-two (91% of 475) samples of clusters 1, 2, and 3 corresponded to G1, G2, and G3 subgroups (Supplementary material, Fig. S3) respectively. The high concordance and high percentage overlap between clusters 1, 2, and 3 and G1, G2, and G3 subgroups indicate that our classification system is well validated. As in the CGCG dataset, nontumor controls in the Rembrandt dataset were all stably assigned into the G2 or cluster 2 subgroups.
Classical Subtype–Associated Gene Signature was Not Significant in CGGA Samples Using the TCGA Classification System
We applied the classification system of the TCGA to our CGGA samples. Genes were ordered using the predictive 840-gene list of the TCGA classification system, and the annotations of 225 CGGA samples were derived from the Prediction Analysis of Microarrays classifier. As shown in Fig. 4A, proneural, neural, and mesenchymal subtypes and their respective gene signatures were clearly identified in CGGA samples, but classical subtype–associated gene signature could be identified in the heat map. EGFR expression values from the expression arrays were also analyzed. As shown in Supplementary material, Fig. S1, EGFR does not show stably and consistently strong expression in the classical subtype. The differences in EGFR expression between classical and other subtypes were compared pairwise using Student's t test. This also validated the CGCG samples' lack of a robust classic gene expression signature. Kaplan-Meier survival analysis showed that the proneural subtype has significantly better survival for all cases (n = 217, P < .001) (Fig. 4B). The neural subtype also had a better prognosis when compared with the mesenchymal and classical subtypes (P < .001, log-rank) (Fig. 4B). There is no survival difference between the classical and mesenchymal subtypes. The proneural subtype was enriched with G1 samples, the neural subtype was enriched with G2 samples, and the mesenchymal and classical subtypes were enriched with G3 samples (Fig. 4C).
In addition, clinical characteristics of the proneural, neural, classical, and mesenchymal subtypes in the CGGA samples were analyzed. As shown in Table 2, 94.87%, 51.06%, 29.41%, and 19.35% of samples in the proneural, neural, classical, and mesenchymal groups, respectively, harbored IDH1 mutations. Loss of 1p was found in 31.82%, 28.57%, 25%, and 30.43% of the proneural, neural, classical and mesenchymal subtypes, respectively. Loss of 19q was seen in 40.91%, 42.86%, 0%, and 13.04% of the proneural, neural, classical, and mesenchymal samples, respectively. Many GBMs and anaplastic gliomas were grouped into the classical and mesenchymal subtypes. No difference in age at diagnosis was found among the proneural, neural, classical, and mesenchymal groups.
Table 2.
Clinical characteristics of the subtypes annotated using TCGA subtyping method in CGGA samples
| Variable | Proneural | Neural | Classical | Mesenchymal |
|---|---|---|---|---|
| Sex (female/male) | 26/25 | 26/34 | 19/19 | 28/48 |
| Age at diagnosis (year) | 41.27 ± 9.93 | 41.69 ± 11.36 | 39.61 ± 13.23 | 44.49 ± 12.95 |
| IDH1 mutation (%) | 94.87 | 51.06 | 29.41 | 19.35 |
| 1p Loss (%) | 31.82 | 28.57 | 25 | 30.43 |
| 19 Loss (%) | 40.91 | 42.86 | 0 | 13.04 |
| Histopathology subtypes | ||||
| Astrocytoma | 21 | 17 | 7 | 13 |
| Oligodendrocytoma | 7 | 8 | 2 | 0 |
| Oligoastrocytoma | 11 | 9 | 0 | 2 |
| Anaplastic Astrocytoma | 0 | 4 | 2 | 2 |
| Anaplastic Oligodendrocytoma | 4 | 3 | 3 | 1 |
| Anaplastic Oligoastrocytoma | 0 | 3 | 4 | 8 |
| Secondary GBM | 2 | 1 | 1 | 0 |
| Primary GBM | 6 | 10 | 19 | 50 |
| Normal brain tissues | 0 | 5 | 0 | 0 |
Discussion
The use of gene expression data from patient tumor samples to determine better treatment options is becoming increasingly common in clinical practice.10–12 However, inconsistencies in glioma molecular classification across various studies makes expression profiling a challenging endeavor for routine use in clinical practices outside major hospitals or commercial laboratories.13 Previous reports attempting glioma classification are all based on whole genome profiling of glioma samples derived from patients in Western countries.3–5,14 However, there exists no dataset generated from a large number of samples from an East Asian population that could be used for glioma classification. This study investigated a large number of samples from Chinese patients in an attempt to complement and/or validate existing molecular subtyping systems. In our study, a total of 225 samples were subjected to whole genome gene expression profiling. The data identified 3 major groups of gliomas.
The urgent need for an objective, molecularly based classification system for gliomas is highlighted by the high rate of divergent diagnoses, inexact prognostic capabilities, and poor therapeutic predictive properties based on the current histopathologic classification schemes.15–17 The TCGA network described a robust gene expression–based molecular classification of GBMs that divided them into proneural, neural, classical, and mesenchymal subtypes.4 Phillips et al. defined 3 subtypes (mesenchymal, proneural, and proliferative) when they molecularly profiled several high-grade glioma samples.4 Other results from Li et al. identified 2 main subtypes, which they defined as GBM-rich (mesenchymal) and oligodendroglioma-rich (proneural) with use of an unsupervised clustering method.5 With use of consensus clustering, our results have identified 3 subtypes with robust differences in clinical characteristics. The G1 subgroup was characterized by good clinical outcome, young age, low malignant behaviors, and extraordinary high IDH1 mutation. G3 groups exhibited the opposite effect. The G2 subtype is the middle class of the aforementioned 2 subtypes. Of interest, every sample in the G1 group carried the IDH1 mutation. Also of note, the G2 subgroup showed a higher percentage loss of 1p and 19q. The G3 subgroup consisted of more GBMs than did either G1 or G2. Only 3 primary GBMs and 1 secondary GBM were included in the G1 subtype, and all 4 GBMs presented with mutations in the IDH1 gene. Our classification scheme also identified a spatial difference in glioma development, with G1 and G2 tumors occurring predominantly in the frontal lobe as opposed to the G3 subtype. IDH1 mutations are early events in the development of gliomas.13,18–20 All samples in the G1 group were accompanied with IDH1 mutation and young age. On the basis of IDH1 mutation status in our molecular classification, our classification system may more accurately reflect the process of glioma development.
We next aimed to validate our classification system with the use of 2 external datasets from the TCGA dataset containing GBMs only and a Rembrandt dataset containing all gliomas.3,21 Our classification system effectively classified the 202 TCGA GBM samples into the G1, G2, and G3 subtypes. The G1 samples were enriched with the proneural subtype, whereas the G2 samples were enriched with the neural subtype. The G3 samples were enriched with the classical and mesenchymal subtypes. Furthermore, patient survival analysis and age distribution in the G1, G2, and G3 subtypes from TCGA GBM samples closely mirrored the survival and age distribution found in the CGGA samples with all grades of gliomas. Additionally, our classification scheme could divide 475 samples from Rembrandt dataset into G1, G2, and G3 subgroups with different prognoses very clearly. These results indicate that our classification system based on CGGA samples can effectively group independent glioma samples into their respective subtypes based on their different characteristics.
To analyze our dataset in greater depth, we annotated the CGGA samples with the use of the TCGA system of classification. Proneural, neural, and mesenchymal glioma subtypes and their respective gene signatures were clearly identified. However, no classical subtype–associated gene signature was identified in the heat map. Only the proneural and mesenchymal subtypes were consistently identifiable across various studies. In our study, we report that proneural, mesenchymal, and neural, but not classical, gene signatures significantly existed in the CGGA glioma dataset when the TCGA classification system was applied. Moreover, the proneural subtype was associated with significantly better survival for all cases. The neural subtype was also associated with a better prognosis when compared with the classical and mesenchymal subtype. It should be highlighted that the gene expression pattern and clinical characteristics of the classical subtype resemble those of the mesenchymal subtype. Thus, we may treat them as one mesenchymal subtype. In addition, the proneural, neural, and mesenchymal subtypes were enriched in the G1, G2, and G3 subgroups, respectively. The difference in age distribution, IDH1 mutation, and loss of 1p and 19q in the proneural, neural, and mesenchymal groups was less significant than that of the G1, G2, and G3 subtypes in CGGA samples. This indicates that our classification system may more accurately classify gliomas based on clinical and genetic characteristics. Of note, G2 or neural subgroup has the modest prognosis in all gliomas in our dataset. However, G2 or neural subgroup has the poorest prognosis when only GBMs were considered. We may postulate that G2 or neural subgroup may have a more rapid progression when at a different stage of development.
In summary, our results have identified 3 subtypes of glioma based on whole genome gene expression profiling using a large number of samples from Chinese patients with glioma. Furthermore, our results were validated on an independent dataset from GBMs in the TCGA. We annotated our samples with use of the TCGA classification system. Of note, no significant classic gene signature was identified in our dataset, potentially highlighting differences between Chinese gliomas and gliomas of other cultures. We also found that the G1, G2, and G3 subtypes were enriched with proneural, neural, and mesenchymal subgroups, respectively. Our classification scheme may discriminate more clearly between clinical and genetic alterations than when the TCGA subtyping system is applied to the CGGA dataset. This finding indicates that only 3 main subtypes clearly exist in our dataset regardless of whether the TCGA or CGGA classification system is used.
Supplementary Material
Conflict of interest statement. None declared.
Funding
This work was supported by grants from the National High Technology Research and Development Program of China (863) (No. 2012AA02A508), International Cooperation Program (No. 2012DFA30470), National Natural Science Foundation of China (No. 81201993), National Natural Science Foundation of China (No. 81101901), Jiangsu Province's Key Provincial Talents Program (No. RC2011051) and Jiangsu Province's Key Discipline of Medicine (No. XK201117).
Supplementary Material
Acknowledgments
W.Y., W.Z., and G.Y. contributed equally as first authors.
References
- 1.Yan W, Zhang W, Jiang T. Oncogene addiction in gliomas: implications for molecular targeted therapy. J Exp Clin Cancer Res. 2011;30:58. doi: 10.1186/1756-9966-30-58. doi:10.1186/1756-9966-30-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drappatz J, Norden AD, Wen PY. Therapeutic strategies for inhibiting invasion in glioblastoma. Expert Rev Neurother. 2009;9(4):519–534. doi: 10.1586/ern.09.10. doi:10.1586/ern.09.10. [DOI] [PubMed] [Google Scholar]
- 3.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. doi:10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. doi:10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Li A, Walling J, Ahn S, et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69(5):2091–2099. doi: 10.1158/0008-5472.CAN-08-2100. doi:10.1158/0008-5472.CAN-08-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.You G, Huang L, Yang P, et al. Clinical and molecular genetic factors affecting postoperative seizure control of 183 Chinese adult patients with low-grade gliomas. Eur J Neurol. 2012;19(2):298–306. doi: 10.1111/j.1468-1331.2011.03509.x. doi:10.1111/j.1468-1331.2011.03509.x. [DOI] [PubMed] [Google Scholar]
- 7.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. doi:10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. doi:10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 9.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. doi:10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konstantinopoulos PA, Spentzos D, Cannistra SA. Gene-expression profiling in epithelial ovarian cancer. Nat Clin Pract Oncol. 2008;5(10):577–587. doi: 10.1038/ncponc1178. doi:10.1038/ncponc1178. [DOI] [PubMed] [Google Scholar]
- 11.Vitucci M, Hayes DN, Miller CR. Gene expression profiling of gliomas: merging genomic and histopathological classification for personalised therapy. Br J Cancer. 2011;104(4):545–553. doi: 10.1038/sj.bjc.6606031. doi:10.1038/sj.bjc.6606031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirahata M, Iwao-Koizumi K, Saito S, et al. Gene expression-based molecular diagnostic system for malignant gliomas is superior to histological diagnosis. Clin Cancer Res. 2007;13(24):7341–7356. doi: 10.1158/1078-0432.CCR-06-2789. doi:10.1158/1078-0432.CCR-06-2789. [DOI] [PubMed] [Google Scholar]
- 13.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116(6):597–602. doi: 10.1007/s00401-008-0455-2. doi:10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 14.Gravendeel LA, Kouwenhoven MC, Gevaert O, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. doi:10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 15.Brennan C. Genomic profiles of glioma. Curr Neurol Neurosci Rep. 2011;11(3):291–297. doi: 10.1007/s11910-011-0198-7. doi:10.1007/s11910-011-0198-7. [DOI] [PubMed] [Google Scholar]
- 16.Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. 2011;59(8):1190–1199. doi: 10.1002/glia.21165. doi:10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- 17.Sulman EP, Aldape K. The use of global profiling in biomarker development for gliomas. Brain Pathol. 2011;21(1):88–95. doi: 10.1111/j.1750-3639.2010.00456.x. doi:10.1111/j.1750-3639.2010.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174(4):1149–1153. doi: 10.2353/ajpath.2009.080958. doi:10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. doi:10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 20.Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol. 2011;29(34):4482–4490. doi: 10.1200/JCO.2010.33.8715. doi:10.1200/JCO.2010.33.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madhavan S, Zenklusen JC, Kotliarov Y, et al. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167. doi: 10.1158/1541-7786.MCR-08-0435. doi:10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.