Abstract
Implementation of chemotherapy with the drug temozolomide increased the overall survival of patients with glioblastoma multiforme (GBM; WHO grade IV), in particular when the O6-methylguanine DNA methyltransferase (MGMT) promoter is epigenetically silenced. Nevertheless, the prognosis remains poor, and relapse in GBM occurs regularly. This clinical behavior seems to be due to the existence of a therapy-resistant subpopulation of cells that induce tumor regrowth. The objective of this work was to analyze the role of aldehyde dehydrogenase (ALDH) 1A1 in mediating temozolomide resistance and its value as a predictor of clinical outcome in GBM patients. Nine GBM cell lines were treated with temozolomide alone or in combination with 4-diethylaminobenzaldehyde (DEAB), an inhibitor of ALDH1A1, or with ALDH1A1 short hairpin (sh)RNA. ALDH1A1 expression and MGMT status of 70 primary GBM patients were correlated with median survival. ALDH1A1 overexpression predicted temozolomide resistance in vitro. Sensitivity of ALDH1A1 positive/MGMT-positive cells to temozolomide could be restored by inhibition of ALDH1A1 by DEAB or by knockdown with shRNA, as indicated by increased cytotoxicity, reduced clonogenicity, and accumulation in the G2/M cell-cycle phase. The prognosis of patients with a high level of ALDH1A1 expression was poor compared with that of patients with low levels (P < .0001). ALDH1A1 is a new mediator for resistance of GBM to temozolomide and a reliable predictor of clinical outcome and may serve as a potential target to improve treatment of human GBM.
Keywords: ALDH1A1, clonogenicity, G2/M arrest, glioblastoma, temozolomide resistance
Despite aggressive therapy, including surgical gross resection, radiotherapy, and chemotherapy, the prognosis of patients with glioblastoma multiforme (GBM) remains poor. Regardless of various efforts to improve postoperative therapeutic regimens, median survival is 14.6 months after diagnosis,1 and tumor relapse occurs regularly. A therapy-resistant subpopulation of cells, often described as cancer stem cells, might harbor the ability to induce tumor growth.2 Recently, aldehyde dehydrogenase (ALDH) 1A1 was presented as a novel marker for GBM cells with stem cell characteristics.3 In various solid tumors, including breast cancer, colon carcinoma, and prostate cancer,4–6 ALDH1A1 expression was found to correlate with a poor clinical prognosis. ALDH1A1 is involved in multiple biological processes, including oxidative stress response,7 cell differentiation,8 and drug resistance.9,10
Application of temozolomide (TMZ), an oral alkylating agent used as standard treatment in newly diagnosed GBM, increases the median survival of patients.11,12 TMZ transfers methyl groups to DNA, including the N7 and O6 positions of guanine, and the O3 position of adenine. The O6-methylguanine (O6-MG) adduct mispairs with thymine during replication, resulting in futile cycles of mismatch repair and double-strand breaks.13,14 Possible results of O6-MG lesions are prolonged G2/M arrest, cellular senescence, mitotic catastrophe, and induction of apoptosis.11,15 The DNA-repair protein O6-MG DNA methyltransferase (MGMT) removes O6-MG DNA lesions and thus antagonizes the cytotoxic effect of TMZ. In 2005, Hegi et al.16 demonstrated that patients with an epigenetically silenced MGMT promoter (MGMT-negative [–]) had an improved median overall survival. However, to date there is no alternative treatment option for patients with an unfavorable MGMT promoter status (MGMT-positive [+]). Combination therapy with TMZ would be an essential therapeutic regimen for patients with MGMT+ GBM.
In this study, we demonstrate that ALDH1A1 overexpression predicts resistance to TMZ in GBM cell lines. Concomitant inhibition of ALDH1A1 by 4-diethylaminobenzaldehyde (DEAB) or knockdown by short hairpin (sh)RNA resensitized MGMT+ cell lines to TMZ treatment, as indicated by increased cytotoxicity, reduced clonogenicity, and accumulation in G2/M. Moreover, retrospective immunohistochemical (IHC) analysis of ALDH1A1 expression in formalin-fixed, paraffin-embedded (FFPE) tissue sections of GBM patients revealed a significant correlation between expression of this enzyme and poor prognosis. All these findings demonstrate ALDH1A1 as a promising predictor of clinical outcome and a potential therapeutic target in the future treatment of human malignant gliomas.
Materials and Methods
Cell Culture and Neurosphere Formation
The human GBM cell lines LN18, G139, LN229, LNZ308, and U87MG and the primary cell lines T16, T30, T39, and T40 were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) under standard cell culture conditions at 37°C and 5% CO2. T98G cells were provided by Dr. Inge Tinhofer of Charité University Hospital, Berlin, Germany, and were maintained in minimum essential medium supplemented with 15% FBS. R28 GBM cells were cultured in neurobasal medium as described previously.17 In brief, R28 cells were maintained in serum-free medium supplemented with epidermal growth factor, basic fibroblast growth factor, B27 (Life Technologies), and human leukemia inhibitory factor (Millipore) for preservation of the tumors’ original molecular characteristics and for minor differentiation. Authentication of the established human cell lines LN18, LN229, and T98G was performed prior to the experiments by Cell ID Systems (Promega). For primary cell culture, freshly resected GBM specimens were obtained with patients' consent (according to the guidelines for tissue preservation of the Technische Universität München [TUM] medical faculty). To investigate neurosphere formation, cells were grown in serum-free neurobasal medium as recently described.3 Neurospheres were counted and sized using a Zeiss Axiovert 25 microscope. Spheres derived from R28 cells were analyzed using a Nikon Eclipse TS100 microscope. Regarding LN18 cells, spheres per 10 seeded cells were counted (n = 10). In the case of T98G and R28 cells, the number of spheres per microscopic field (n = 15) was analyzed.
Chemicals and Treatment
GBM cell lines were treated with different concentrations of TMZ (Sigma-Aldrich). Cytotoxicity was estimated with assay by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) according to the manufacturer's instructions (Roche Diagnostics).
To assess the amount of viable cells after treatment with TMZ and DEAB, cells were seeded at a density of 1.5–2 × 105 cells per well on a 6-well dish in duplicates. Over each of 10 days, 200 µM TMZ alone or in combination with 100 µM DEAB (Sigma-Aldrich) was added. R28 cells were treated in neurobasal medium every 48 h for 10 days. Trypan blue staining was applied to determine the number of viable cells.
Colony Formation Assay
Colony formation assay was performed to evaluate clonogenic capacity. Following treatment with 200 µM or 500 µM TMZ for the indicated time, LN18 wild-type (WT)/mock/shALDH1A1 cells were plated in duplicates, with a total of 150 or 300 viable cells per well. Following 14 days of incubation, colonies were stained and counted applying Diff-Quick set (Medion Diagnostics). In all experiments, both untreated cells and control cells treated by solvent (dimethyl sulfoxide) were analyzed.
ALDH1A1 Knockdown by shRNA
Knockdown of ALDH1A1 in LN18 and T98G cells was obtained by transduction with MISSION shRNA Lentiviral Transduction Particles (Sigma-Aldrich). Transduction and selection with puromycin (Sigma-Aldrich) were performed according to the manufacturer's instructions. A nontarget puromycin vector (mock) served as a control. Validation of the knockdown was performed at the protein level by Western blotting and at the mRNA level by relative quantitative real-time PCR (LightCycler 480, Roche). Primers used were 5′-tgttgagcgggctaagaagt-3′ and 5′-gtttggccccttctttcttc-3′ for ALDH1A1 and 5′-tactcagcgccagcatc-3′ and 5′-tgttccaatatgattccaccca-3′ for glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Stable Transfection with ALDH1A1
LNZ308 cells were stably transfected with the mammalian expression vector pCMV6-AC carrying the ALDH1A1 gene (NM_000689.3) (LNZ308 ALDH1A1; OriGene Technologies). Cells were seeded 24 h prior to transfection at a density of 1 × 105 cells per well. Transfection was carried out in Opti–MEM (Life Technologies) with 5-μg plasmid DNA and the transfection reagent lipofectamine (Life Technologies). After the indicated selection period with G418, single-cell colonies were picked and analyzed for ALDH1A1 expression at the mRNA and protein levels. LNZ308 ALDH1A1 cells were maintained in supplemented DMEM plus 0.5 mg/mL G418.
Western Blotting
Western blot analysis was performed as described.3 The primary antibodies used were anti-ALDH1A1 clone 44 (1:500) (BD Biosciences) and anti-MGMT (1:2000) (Cell Signaling Technology). Specificity of the anti-ALDH1A1 antibody to the isoform ALDH1A1 was validated by reverse transcriptase (RT)–PCR. Cell lines expressing other isoforms at the mRNA level, such as ALDH1A3, ALDH3A1, ALDH7A1, ALDH8A1, and ALDH2, showed no immunoreactivity with the applied anti-ALDH1A1 antibody (Supplementary Fig. S1). Anti–alpha-tubulin (1:10 000) and anti-GAPDH (1:50 000) (Sigma-Aldrich) served as a loading control.
Cell-Cycle Analysis
Cells were harvested and counted after the indicated treatment with TMZ, DEAB, or TMZ + DEAB:1 × 106 cells were fixed with 70% methanol on ice for at least 1 h, followed by digestion with 100 µg/mL RNAse A and staining with 50 µg/mL propidium iodide in phosphate buffered saline. The cell cycle was analyzed with a FACSCalibur flow cytometer with 10 000 events per determination. Doublet discrimination and analysis of cell-cycle distribution were performed with FlowJo analysis software (Tree Star).
Patient Characteristics and Tissue Samples
IHC analysis of ALDH1A1 expression on primary surgical specimens of 70 patients diagnosed with primary GBM (WHO grade IV18) was performed as described.3 IHC staining of isocitrate dehydrogenase 1 with R132H point mutation (clone H09; Dianova) was performed to support diagnosis of primary GBM.19 Liver tissue served as positive control for ALDH1A1 expression (Supplementary data S5 a–c). Four squares per FFPE section were counted and averaged. ALDH1A1+ cells were presented as percentage of total cell number. The specimens were collected with patients' consent (according to the TUM medical faculty guidelines for tissue preservation) at the Klinikum Rechts der Isar (RDI; n = 55) and the Berufsgenossenschaftliche Klinik Murnau (n = 15). Only GBM patients surviving at least 2 months after surgical resection and receiving adjuvant therapy were included (median survival = 16.6 months). Fifty of these patients received radiotherapy plus concomitant and adjuvant TMZ as described by Stupp et al.1 Four patients received radiotherapy and concomitant TMZ, 14 patients received only radiotherapy, and 2 patients were given only adjuvant TMZ (Table 1). Subgroup analysis of the different treatment groups was performed.
Table 1.
Characteristics of 70 primary surgical specimens of glioblastoma patients
| Total | MGMT |
ALDH1A1 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Methylation |
No methylation |
≤ 10 % |
> 10 % |
||||||
| no. | no. | % | no. | % | no. | % | no. | % | |
| Patients | 70 | 17 | 24.3 | 53 | 75.7 | 42 | 60 | 28 | 40 |
| Treatment | |||||||||
| standard* | 50 | 13 | 18.6 | 37 | 52.9 | 33 | 47.1 | 17 | 24.3 |
| others** | 20 | 4 | 5.7 | 16 | 22.9 | 9 | 12.9 | 11 | 15.7 |
| Sex | |||||||||
| men | 50 | 13 | 18.6 | 37 | 52.9 | 31 | 44.3 | 19 | 27.1 |
| women | 20 | 4 | 5.7 | 16 | 22.9 | 11 | 15.7 | 9 | 12.9 |
| Age***(years) | |||||||||
| ≤ 60 | 28 | 7 | 10 | 21 | 30 | 21 | 30 | 7 | 10 |
| 61–70 | 27 | 6 | 8.6 | 21 | 30 | 14 | 20 | 13 | 18.6 |
| > 70 | 15 | 4 | 5.7 | 11 | 15.7 | 7 | 10 | 8 | 11.4 |
*Therapeutic regimen described by Stupp et al.1
**Resection plus radiotherapy, radiotherapy and concomitant TMZ, or adjuvant TMZ.
***Age of the patients when diagnosed.
Quantification of MGMT Promoter Methylation
Quantification of MGMT promoter methylation was assessed by a specific relative quantitative real-time PCR technique called MethyQESD (methylation quantification of endonuclease-resistant DNA), as described by Bettstetter et al.20 The applied endonucleases were Hin6I (methylation-quantification digestion [MQD]) and XbaI/DraI (methylation independent calibrator digestion [CalD]). Following digestion, MGMT promoter methylation status was determined by relative quantitative real-time PCR using the LightCycler 480 (Roche). Digested DNA of U87MG cells served as a control for positive methylation, as did digested DNA of nonmethylated blood DNA for 0% methylation. Primer sequences were 5′-cccggatatgctgggacag-3′ and 5′–cccagacactcaccaagtcg-3′.
The proportion of methylated template was calculated from the differences of the cycle threshold (Ct) values from CalD and MQD (= ΔCt) according to this formula: methylation (%) = EΔCt × 100, where E is PCR efficiency. The cutoff for MGMT+ and MGMT– tumors was determined by analysis of martingale residuals.21 Tumor tissues with ≤20% MGMT promoter methylation were considered MGMT+; specimens with >20% MGMT promoter methylation were referred to as MGMT–.
Statistical Analysis
If not otherwise indicated, all experiments were performed at least 3 times. Differences between the results of experimental treatment and average neurosphere size, as well as number, were assessed by 1-way analysis of variance followed by a Bonferroni posttest with GraphPad Prism software. Differences were considered significant at values of P < .05. Patient data were analyzed with SPSS software by the Institute of Medical Statistics and Epidemiology of the RDI/TUM. Kaplan–Meier models were applied to examine survival curves. Differences between the groups were analyzed using the log-rank (Mantel–Cox) test. Multivariate analysis using a Cox regression was applied to demonstrate that ALDH1A1 expression level was an independent predictive factor. Single variables were age, sex, therapeutic modality, and MGMT status.
Results
ALDH1A1 Expression in GBM Cells Correlates with In Vitro Resistance to TMZ
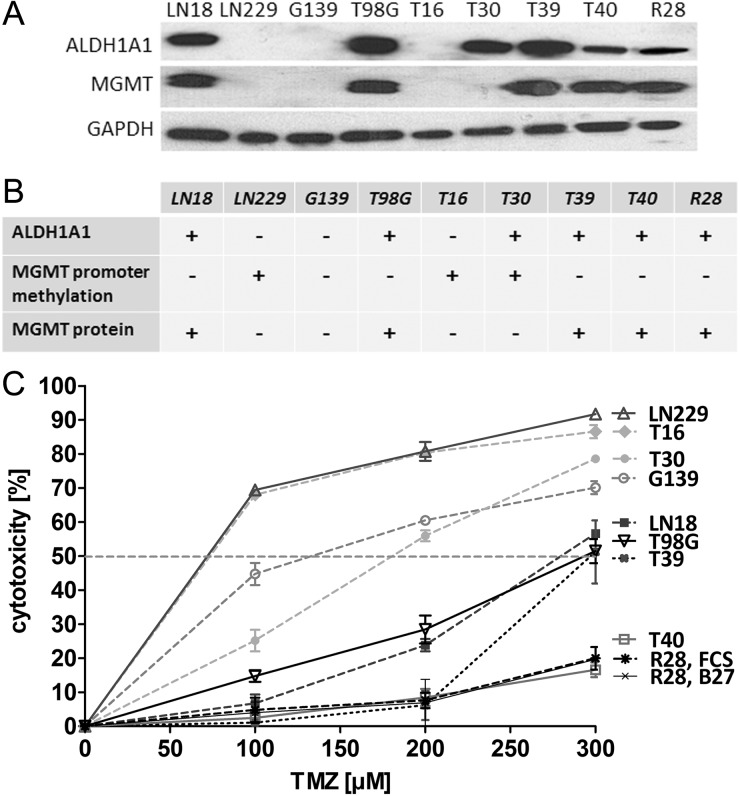
The ALDH1A1 and MGMT statuses of 9 primary and established human GBM cell lines were assessed by Western blot analysis. LN18, T98G, T30, T39, T40, and R28 cells expressed high levels of ALDH1A1 (ALDH1A1+), whereas LN229, G139, and T16 cells were negative for this enzyme (ALDH1A1–) (Fig. 1A). DNA methylation of the MGMT promoter was quantified by MethyQESD.20 Cells without MGMT protein expression (with MGMT promoter methylation) were referred to as MGMT− and cells with MGMT protein (no promoter methylation) as MGMT+. MGMT repair protein expression and no relevant promoter methylation were observed in LN18, T98G, T39, T40, and R28 cells (MGMT+). G139 cells showed no protein expression, despite an unmethylated MGMT promoter (MGMT–*). The MGMT promoter of the cell lines T16, T30, and LN229 was methylated to 60%, 30%, and 100%, respectively, and no MGMT protein was detected (MGMT–) (Fig. 1A and B).
Fig. 1.
ALDH1A1 expression, MGMT status, and response to treatment with TMZ. (A) Western blot analysis showed a high level of ALDH1A1 in LN18, T30, T39, T40, T98G, and R28 cells. The cell lines LN229, G139, and T16 were negative for ALDH1A1. MGMT protein expression was detected in LN18, T98G, T39, T40, and R28 cells. (B) No relevant MGMT promoter methylation was found in LN18, T98G, G139, T39, T40, or R28 (MGMT+). The MGMT promoter of T16, T30, and LN229 was methylated to 60%, 30%, and 100%, respectively (MGMT–).The MGMT promoter of G139 cells was completely unmethylated, but no expression of MGMT protein was detected (MGMT–*). (C) Response of GBM cell lines to treatment with TMZ for 7 days was assessed by metabolic MTT assay.
Response of GBM cell lines to TMZ was analyzed by metabolic MTT assay. ALDH1A1–/MGMT– T16 and LN229 cells were most sensitive to TMZ (half-maximal inhibitory concentration [IC50] < 100 µM). The cell line G139 (ALDH1A1–/MGMT+) responded in a comparable way to TMZ as did T30 (ALDH1A1+/MGMT–) when doses of ≥200 µM were applied. Notably, at TMZ concentrations of ≤100 µM, ALDH1A1– G139 cells were more sensitive to TMZ than were ALDH1A1+ T30 cells. ALDH1A1+/MGMT+ LN18, T98G, T39, T40, and R28 cells were resistant to TMZ with an IC50 of >200 µM (Fig. 1C).
Combination of TMZ and ALDH1A1 Inhibition Reduces the Clonogenic Capacity of ALDH1A1+/MGMT+ Cells
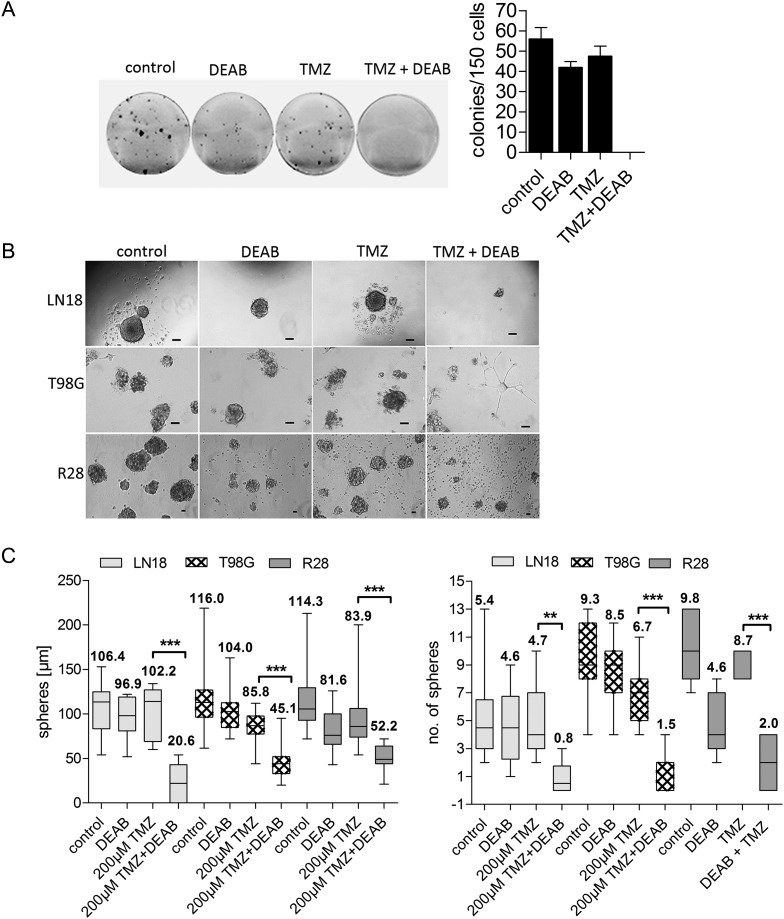
In order to analyze the anticlonogenic effect of TMZ in combination with ALDH1A1 inhibition, the TMZ-resistant cell lines LN18, T98G, and R28 (ALDH1A1+/MGMT+) were treated with either TMZ or DEAB or a combination of both for 10 days. As indicated by colony formation assay, the clonogenic capacity of LN18 cells exposed to TMZ in combination with DEAB was significantly attenuated (Fig. 2A).
Fig. 2.
Concomitant treatment with TMZ and DEAB reduced the clonogenic capacity of GBM cell lines. (A) Colony formation assay demonstrates that the clonogenic capacity of LN18 exposed to 200 µM TMZ in combination with 100 µM DEAB for 10 days was significantly attenuated. (B) TMZ in combination with ALDH1A1 inhibition impaired sphere formation. Neurospheres of LN18, T98G, and R28 cells decreased in size and number after TMZ and DEAB administration; **P < .01, ***P < .001; mean values are shown above the box plots. Scale bar = 100 µM.
In addition, neurosphere formation of LN18, T98G, and R28 was analyzed to determine clonogenicity of sphere-forming cell lines following treatment with concomitant TMZ and DEAB. Neurosphere formation was affected in ALDH1A1+/MGMT+ LN18 and T98G cell lines when the cells were pre-incubated with TMZ and DEAB; spheres were significantly decreased in size and number by combination therapy. Concomitant application of DEAB and TMZ lowered the neurosphere formation frequency from about 50% (TMZ alone) to 8% (TMZ + DEAB) and the average size to <50 µM. Similar results were obtained with T98G. The cell line R28 was permanently maintained in neurospheres. As previously described, neurosphere formation was impaired by ALDH1A1 inhibition3 and by TMZ alone.17 In line with these results, concomitant treatment with DEAB and TMZ in neurobasal medium further decreased the size and number of neurospheres (Fig. 2B).
Knockdown or Inhibition of ALDH1A1 Resensitizes ALDH1A1+/MGMT+ Cells to TMZ
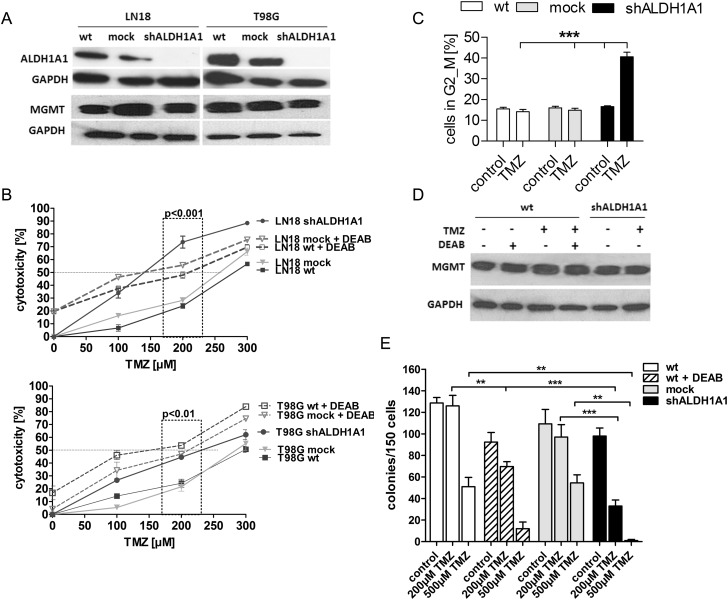
In order to prove the pivotal role of ALDH1A1 in mediating TMZ resistance and to exclude potential off-target effects of DEAB, specific shRNA was applied. Analysis by quantitative real-time PCR showed a knockdown of at least 90% at the mRNA level. Specificity of shRNA to the isoenzyme ALDH1A1 was proven by RT-PCR (Supplementary data S1A). Densitometric normalization to alpha-tubulin revealed a >90% knockdown of ALDH1A1 following shRNA transduction at the protein level compared with control cells (WT or mock) (Fig. 3A). Notably, ALDH1A1 knockdown (shALDH1A1) had no impact on MGMT repair protein expression (Fig. 3A).
Fig. 3.
Knockdown or inhibition of ALDH1A1 restored sensitivity to TMZ. (A) Downregulation of ALDH1A1 at the protein level was verified by Western blot analysis. MGMT protein expression was stable upon shRNA transduction. (B) Metabolic MTT assay revealed an increased cytotoxicity of TMZ when combined with ALDH1A1 inhibition or knockdown. LN18 and T98G shALDH1A1 cells were sensitive to TMZ compared with WT and mock transfected control cells, especially when medium doses of TMZ were applied. (C) Treatment of LN18 shALDH1A1 cells with 200 µM TMZ for 7 days led to accumulation in G2/M. (D) MGMT protein expression was not affected by treatment with TMZ, DEAB, or TMZ + DEAB for 7 days. (E) Colony formation assay showed a significant reduction of the clonogenic capacity of LN18 WT cells treated twice with TMZ + DEAB (striped bar) or LN18 shALDH1A1 cells treated twice with TMZ alone (black bar).
Strikingly, treatment of LN18 and T98G shALDH1A1 cells with TMZ alone induced a response comparable to combination therapy with DEAB in ALDH1A1+ WT and control cells (Fig. 3B). A proliferation assay confirmed the synergistic effect of TMZ and ALDH1A1 inhibition or knockdown. While high doses of TMZ (500 µM) only slightly reduced the proliferation times of LN18 WT and mock cells, numbers of LN18 cells with depleted or inhibited ALDH1A1 were significantly decreased following exposure to TMZ (Supplementary data S3). As indicated by flow cytometry, prolonged treatment with 200 µM TMZ led to G2/M arrest in LN18 shALDH1A1 cells, while the cell cycle of WT and mock cells was not affected (Fig. 3C). Western blot analysis confirmed that MGMT protein expression was not affected by treatment with TMZ, DEAB, or TMZ + DEAB (Fig. 3D). Notably, the clonogenic capacity of LN18 shALDH1A1 cells or LN18 cells treated with DEAB was significantly decreased following exposure to TMZ alone (Fig. 3E).
TMZ Induces Prolonged G2/M Cell-Cycle Arrest in ALDH1A1+ GBM Cells after Inhibition or Knockdown of ALDH1A1
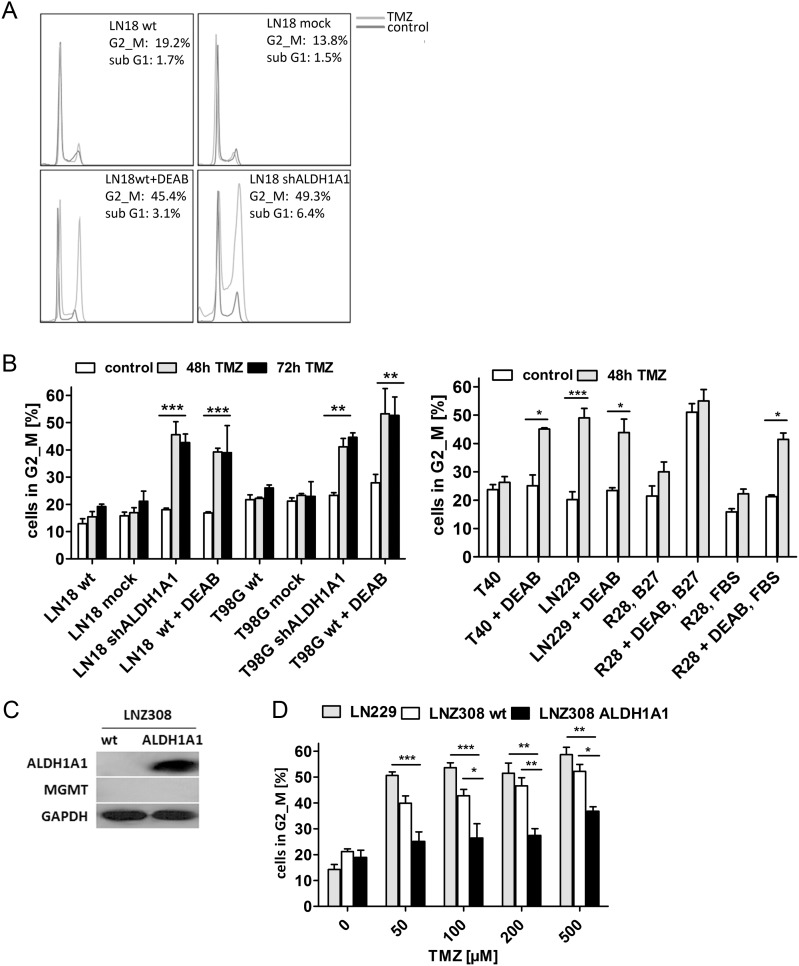
To further substantiate the hypothesis that TMZ induces a prolonged cell cycle only in ALDH1A1– or ALDH1A1 knockdown cells, LN18, T98G, T40, R28, and LN229 cells were treated with TMZ, DEAB, or a combination of both and analyzed after 48 h or 72 h by flow cytometry (Fig. 4A and B). Doses of TMZ and DEAB, showing no effect on cell-cycle distribution of MGMT+ control cells, were applied. When treated in neurobasal medium, single doses of 100 μM DEAB were used as described previously.3 Notably ALDH1A1+ cells (LN18 WT/mock, T98G WT/mock, T40, and R28) showed a cell-cycle distribution comparable to untreated control cells 48 h after TMZ application. In contrast, ALDH1A1– cells (LN229, LN18 shALDH1A1, and T98G shALDH1A1) or cells with inhibited ALDH1A1 (LN18 WT + DEAB, T98G WT + DEAB, T40 + DEAB, and R28 + DEAB) accumulated in G2/M following treatment with TMZ. ALDH1A1–/MGMT–* G139 cells responded comparably to ALDH1A1–/MGMT– LN229 cells to treatment with 200 µM TMZ (data not shown). An increase of the sub-G1 population after addition of TMZ demonstrated apoptosis of ALDH1A1– cells (Supplementary data S4). ALDH1A1– LN229 cells showed no sub-G1 peak following TMZ administration but strong cell-cycle arrest and senescence, as indicated by β–galactosidase staining (Supplementary data S2).
Fig. 4.
ALDH1A1– GBM cell lines showed prolonged G2/M arrest following treatment with TMZ. (A) Representative flow cytometry histograms of LN18 cells after treatment with 500 µM TMZ, 300 µM DEAB, or a combination of both for 48 h; y–axes: DNA content, x-axes: FL3-H. (B) Column graph shows the percentage of cells in G2/M after treatment with 500 µM (LN18, T98G) or 200 µM TMZ (LN229, T40, R28) and 300 µM DEAB for 48 h or 72 h in DMEM + FBS. R28 cells were also analyzed after treatment with 200 µM TMZ and 100 µM DEAB in neurobasal medium (R28, B27); mean values of 3 independent experiments are shown; *P < .05, **P < .01, ***P < .001. (C) Western blot analysis of LNZ308 WT and ALDH1A1 transfected (ALDH1A1) cells. No MGMT protein expression was detected. (D) Cell-cycle analysis of ALDH1A1–/MGMT– (LNZ308 WT, LN229) and ALDH1A1+/MGMT– cells (LNZ308 ALDH1A1) following exposure to 50–500 µM TMZ for 48 h.
In line with the results shown in Fig. 2B, spheres grown in neurobasal medium were strongly affected by treatment with DEAB, while cells under normal cell culture conditions did not respond to ALDH1A1 inhibition alone. When grown in DMEM + FBS, R28 cells showed only G2/M arrest after combination of TMZ and ALDH1A1 inhibition. Under a serum-free stem cell–maintaining condition, substantial accumulation in G2/M and an increased sub-G1 peak were detected after addition of DEAB alone (Fig. 4B, Supplementary data S4). Cell-cycle arrest and elevated sub-G1 population were also observed when ALDH1A1 knockdown cells were cultivated in neurobasal medium (data not shown).
High Level of ALDH1A1 Expression Is Associated with an Unfavorable Prognosis in GBM Patients
A major role of ALDH1A1 in mediating TMZ resistance was found in vitro. To explore the impact of ALDH1A1 in vivo, IHC analysis of ALDH1A1 expression was conducted in 70 surgical specimens from primary GBM patients (Table 1), with liver tissue serving as a positive control for ALDH1A1 expression (Supplementary data S5a). IHC staining of IDH1 with R132H point mutation in this cohort underlined the diagnosis as primary GBM, with only 1 positive tumor sample out of 70 cases19 (Supplementary data S5b, c). Univariate analysis according to the Cox regression model showed that clinical variables such as age and sex were not significantly associated with longer survival and that ALDH1A1 expression was an independent predictive factor. In the present investigation, clinical outcome was dependent on the therapeutic regimen and ALDH1A1 expression status (P < .0001). Patients receiving radiotherapy plus concomitant and adjuvant TMZ as described by Stupp et al1 had a better prognosis than patients who did not receive standard therapy, with median survivals of 20.5 and 8 months, respectively (P < .0001).
Multivariate analysis revealed that ALDH1A1 expression was independent of age and sex. The cutoff for ALDH1A1 high and ALDH1A1 low tumors was determined by analysis of martingale residuals.21 Tumor tissues with ≤10% positive cells were considered to be ALDH1A1 low; specimens with >10% ALDH1A1+ cells were referred to as ALDH1A1 high.
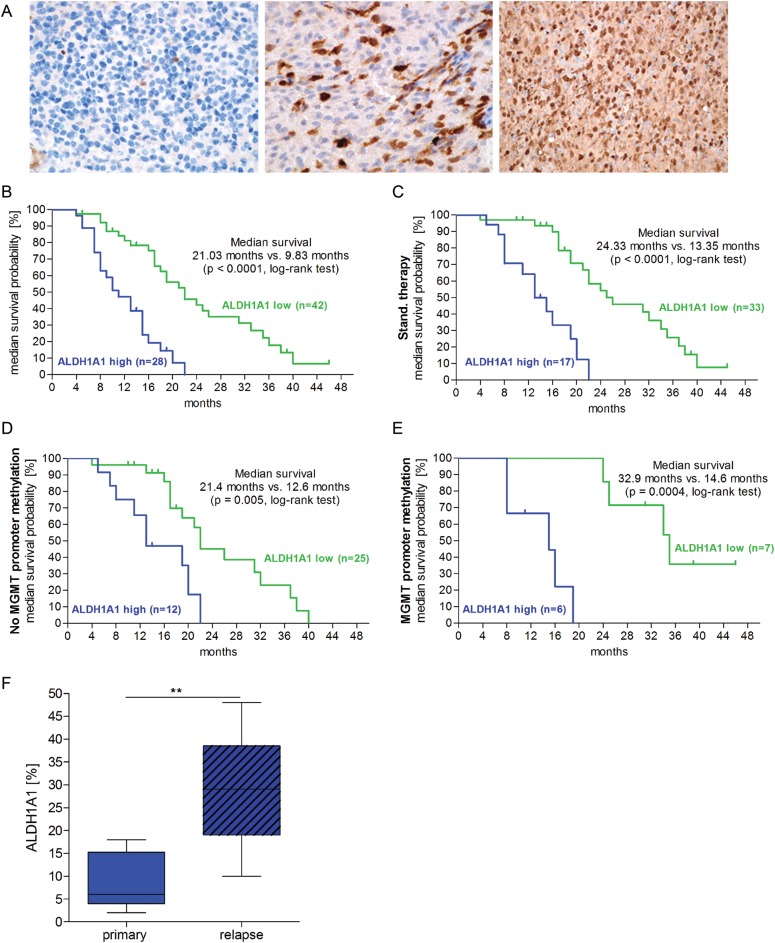
The median survival of patients with low ALDH1A1 expression (n = 42) was 21.0 months, compared with 9.8 months for patients with a high level of ALDH1A1+ cells (n = 28; P < .0001) (Fig. 5B). Comparable differences in median survival were observed when only patients receiving standard therapeutic regimen as described by Stupp et al.1 were analyzed (n = 50), with 24.3 and 13.3 months median survival, respectively (Fig. 5C). In the cases receiving no standard treatment (n = 20), the prognosis of patients with low levels of ALDH1A1 was not significantly better than that of patients with high levels, with median survivals of 8.6 and 6.7 months, respectively (P = .217). Patients receiving a standard therapeutic regimen were further classified regarding MGMT promoter status. The median survival of patients with MGMT– tumors was 24.3 months, and that of patients with MGMT+ tumors was 19.2 months (P = .21). In the cohort of MGMT+ patients, median survivals were 21.4 months and 12.6 months for ALDH1A1 low and high GBM patients, respectively (P = .005) (Fig. 5D). In the group of patients with MGMT– tumors, median survivals were 32.9 months and 14.6 months, respectively (P = .0004) (Fig. 5E).
Fig. 5.
ALDH1A1 expression was found to correlate with an adverse prognosis in human GBM. (A) The percentage of ALDH1A1+ cells in human GBM tissue was analyzed by IHC staining. FFPE sections from primary tumors with a low percentage of ALDH1A1+ cells (≤10%) (A) and with a high level of ALDH1A1 expression (>10%) (B) are shown; tissue section from relapsed GBM demonstrated a very high percentage of ALDH1A1+ cells (> 40%) (C). (B–E) Kaplan–Meier plot of patient median survival; patients with tumors with a high level of ALDH1A1 expression (blue curve) had significantly shorter median survival than did patients with low-level tumors (green curve) (P < .0001). Poor clinical outcome of patients with ALDH1A1-high tumors was found in the group of patients with and without MGMT promoter methylation (P = .005 and P = .0004). (F) The median percentage of ALDH1A1+ cells in relapsed GBM was significantly higher than in the respective primary tumors (P = .002).
In addition, the percentages of ALDH1A1+ cells of primary and relapsed tumors from 14 primary GBM patients were compared by IHC staining. Remarkably, specimens of recurrent GBM demonstrated significantly higher ALDH1A1 expression levels than the respective primary tumors. The mean ALDH1A1 level increased from 8.5% positive cells in specimens prior to combined radio/chemotherapy to 28.6% after re-resection of relapsed tumors (Fig. 5A c, F).
Discussion
In the present investigation, we introduced the enzyme ALDH1A1 as a new mediator of resistance to TMZ. High levels of ALDH1A1 were associated with resistance to TMZ in vitro, in particular in MGMT+ GBM cells. Sensitivity of ALDH1A1 and MGMT+ GBM cells to TMZ could be restored by inhibition of ALDH1A1 by DEAB or by knockdown with shRNA. Therefore, targeting ALDH1A1 is a promising therapeutic tool for an improved treatment of TMZ-resistant GBM. In addition, ALDH1A1 seems to be a feasible predictor of clinical outcome, as the prognosis of patients with high levels of ALDH1A1 expression was poor compared with that of patients with low levels.
Reduced clonogenicity and increased cytotoxicity of TMZ-resistant cells after inhibition or knockdown of ALDH1A1 indicated the important role of this enzyme in mediating TMZ resistance. ALDH1A1–/MGMT– cells were most sensitive to TMZ, whereas cell lines expressing ALDH1A1 and MGMT (ALDH1A1+/MGMT+) were highly resistant to treatment with TMZ (Fig. 1). Colony and neurosphere formation assay revealed a significantly attenuated clonogenicity of ALDH1A1+/MGMT+ cells after exposure to TMZ in combination with ALDH1A1 inhibition or depletion (Figs. 2 and 3E). Concomitant inhibition of ALDH1A1 in ALDH1A1+/MGMT+ cells (LN18, T98G) significantly decreased neurospheres in size and number, demonstrating both inhibition of proliferation (size) and reduction of clonogenicity (number) (Fig. 2B). Clinically relevant doses of TMZ are able to block neurosphere formation of MGMT– human GBM cells.22 The present data indicate that neurosphere formation of MGMT+ cells is not attenuated by pretreatment with TMZ alone, but clonogenic capacity was substantially decreased by combined ALDH1A1 inhibition. ALDH1A1+/MGMT+ R28 cells showed impaired neurosphere formation when treated in neurobasal medium with TMZ alone or DEAB. These observations are in line with previous studies demonstrating attenuated neurosphere development following exposure to DEAB or TMZ under serum-free conditions.3,17 Strikingly, combination of TMZ and DEAB led to a substantial decrease of clonogenic capacity under both normal and stem cell–maintaining conditions compared with TMZ alone and DEAB alone (Fig. 2A and B).
Further, a synergistic cytotoxic and antiproliferative effect of chemical ALDH1A1 inhibition or depletion by shRNA and TMZ was found in ALDH1A1+/MGMT+ cells (Fig. 3B, Supplementary data S3). Coexpression of ALDH1A1 and MGMT in most of the applied cell lines indicated a potential correlation between both proteins (Fig. 1A and B). However, MGMT protein expression was not affected by transduction with shRNA to ALDH1A1 or by treatment with TMZ, DEAB, or TMZ + DEAB (Fig. 3A and D). Therefore, ALDH1A1 and MGMT expression must represent different mechanisms of mediating chemoresistance in human GBM cell lines.
TMZ is known to induce DNA lesions leading to cell-cycle arrest at the G2/M boundary and to induce senescence rather than direct apoptosis in GBM.13,14,23 Analysis of cell-cycle distribution following TMZ treatment indicated that prolonged G2/M arrest was dependent on ALDH1A1 expression and MGMT status. ALDH1A1+ LN18, T98G, T40, and R28 cells exhibited no or only slight G2/M accumulation in response to TMZ, while the respective shALDH1A1 or ALDH1A1 inhibited cells showed prolonged G2/M arrest (Figs. 3C, 4A and B). In addition to cell-cycle arrest, induction of apoptosis was detected in DEAB-treated or knockdown cells following TMZ exposure (Supplementary data S4). ALDH1A1–/MGMT– LN229 cells showed prolonged G2/M arrest and became senescent following treatment with TMZ, as indicated by β-galactosidase staining (Supplementary data S2).
Prolonged G2/M arrest of ALDH1A1–/knockdown GBM cells was observed after application of both long-term medium and long-term single high doses of TMZ (Figs. 3C, 4A and B). Since no increase in cytotoxicity or G2/M fraction of ALDH1A1+/MGMT+ cells was found after irradiation with 2–5 Gy in combination with ALDH1A1 inhibition, this isoenzyme seems to play a specific role in mediating resistance to the alkylating agent TMZ (data not shown).
Recent studies showed that clinically relevant doses of 50 µM TMZ were able to eliminate MGMT– GBM cells, while IC50 of MGMT+ cells is often more than 10-fold higher.22,24 Therefore, we also analyzed the effect of TMZ on ALDH1A1+/MGMT− cells. Strikingly, LNZ308 cells stably transfected with ALDH1A1 (ALDH1A1+/MGMT−) were not affected by TMZ doses up to 200 µM, whereas ALDH1A1−/MGMT− control cells (LNZ308 WT and LN229) were arrested in G2/M cell-cycle phase after treatment with 50 µM TMZ (Fig. 4D). Thus, depletion of MGMT+ and MGMT− cells by TMZ can be strongly enhanced by inhibition of ALDH1A1.
Standardization of adjuvant therapy for newly diagnosed GBM after resection increased the median survival of patients to over 14 months.1,25 In particular, patients with an epigenetically silenced MGMT promoter (MGMT−) benefited from adjuvant and concomitant TMZ administration.16 Notably, not all patients with MGMT– GBM respond to this alkylating agent, and some are resistant to treatment with TMZ despite MGMT promoter methylation.26,27 However, to date there is no alternative treatment option for patients with an unfavorable MGMT promoter status (MGMT+). Our present in vitro data demonstrate an important role of ALDH1A1 in mediating TMZ resistance additionally to MGMT. To compare these experimental findings with clinical practice, we investigated ALDH1A1 expression retrospectively in 70 tumor specimens from patients with primary GBM. A significant link between ALDH1A1 expression and median survival probability was found in vivo. Patients with a high percentage of ALDH1A1+ cells (>10% ALDH1A1) had a poor clinical prognosis, compared with patients with low levels (≤10%), in MGMT+ and MGMT– cohorts. Our data contradict a recent study28 showing that ALDH1A1 expression correlates with better survival in GBM patients. However, the clinical data provided by Adam et al.28 demonstrate that in their setting, ALDH1A1 was not an independent factor but dependent on the type of sampling. Significantly higher levels of ALDH1A1 were found in samples analyzed after stereotactic biopsy compared with specimens from total resection. Apparently the sampling type was prognostically relevant, since patients with stereotactic biopsy had significantly lower median overall survival times compared with the cohort receiving total resection.29 Our present investigation is in line with the work by Campos et al.30; these authors showed that ALDH1A1 expression levels increase with malignancy in astrocytic gliomas. In GBM, a high level of ALDH1A1 expression was significantly associated with poor patient survival. Therefore, we suggest ALDH1A1 as a feasible predictor of clinical outcome complementary to MGMT status. A prospective study with a larger cohort of patients receiving a standard therapeutic regimen1 should be performed to verify the correlation of ALDH1A1 expression in MGMT+ and MGMT− patients with clinical prognosis and to validate the applicability of a 10% cutoff.
Despite aggressive treatment, tumor relapse occurs regularly in GBM patients. This clinical behavior might be associated with a therapy-resistant subpopulation of cells.2,26,31 Therefore, we analyzed ALDH1A1 expression in specimens of GBM patients prior to combined radio/chemotherapy and after re-resection of recurrent malignancies (Fig. 5A and F). Significantly increased ALDH1A1 levels in relapsed GBM compared with the respective primary resections underlined the hypothesis that TMZ-resistant ALDH1A1+ cells are able to escape therapy and contribute to tumor regrowth.
Lately, several approaches have attempted to target the resistant subpopulations in GBM therapy and to find appropriate biomarkers for clinical practice.32,33 Markers for stemlike brain tumor cells such as CD133, nestin, and CD15 have been suggested to negatively affect clinical outcome in glioma patients.34,35 Recently, we presented ALDH1A1 as a novel marker for a stem cell–like phenotype in GBM because the ability to form neurospheres and to differentiate into all neural lineages was found to be related to cells expressing high levels of ALDH1A1.3 Based on the present results, we hypothesize that the enzyme itself plays a major role in mediating resistance to TMZ. Expression of ALDH1A1 describes a cellular subpopulation that is resistant to therapy with the alkylating agent TMZ rather than exclusively cells with stem cell–like characteristics.
In prior studies, expression of ALDH1A1 was associated with drug resistance and cellular detoxification. It was first found in the early 1980s that ALDH1A1+ leukemic and hematopoietic stem cells are able to detoxify the alkylating agent cyclophosphamide.9,36 To date, the mechanism of ALDH1A1-mediated therapy resistance in various solid tumors is not completely understood. This enzyme is known to also bind and detoxify non-aldehydes such as flavopiridol.37 Regarding the chemical structure of TMZ and its reactive methylating agent MTIC (3-methyl-(triazen-1-yl) imidazole-4-carboxamide), direct binding of these compounds by ALDH1A1 seems rather unlikely. In general, ALDHs are able to catalyze the oxidation of a wide range of aldehydes, and a variety of data demonstrate the role of ALDHs in handling oxidative stress and in protection against DNA–damaging agents such as cyclophosphamide, mitomycin C, and Vp-16.9,38,39 Notably, recent studies have described increased production of reactive oxygen species after treatment with TMZ. Here, the ability of cells to reduce oxidative stress correlated with chemoresistance.40,41 Superoxide radicals built after TMZ application lead to peroxidation of membrane lipids. The resulting aldehydes such as malondialdehyde and 4–hydroxynonenal are highly reactive and bind to cellular proteins and DNA, giving rise to etheno-DNA adducts.42 ALDH1A1 is able to bind and detoxify these reactive aldehydes, preventing TMZ-induced DNA damage that is not repaired by MGMT.43
In conclusion, ALDH1A1 seems to play a pivotal role in mediating resistance to TMZ. Besides its value as a predictive factor, it may serve as a target for an improved therapy in human GBM. The ALDH1A1-specific inhibitor DEAB was used in various animal experiments, and its potential to resensitize GBM cells to TMZ must be further investigated in vivo.8,44 Disulfiram (DSF), a drug formerly used in the treatment of chronic alcoholism, is another inhibitor of ALDHs. Lately, the potential of DSF to reduce chemotherapy resistance in vitro and in vivo has been shown in various solid tumors.45,46 As a well-tolerated orally active compound, DSF might be a promising ALDH1A1 inhibitor in human glioma therapy. Nevertheless, all presently available ALDH inhibitors are not exclusively specific to ALDH1A147; in order to prevent side effects and to further clarify the role of ALDH1A1 in mediating chemoresistance, a specific inhibitor is required.
Supplementary Material
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 824: “Imaging for the Selection, Monitoring, and Individualization of Cancer Therapies,” Project B6).
Supplementary Material
Acknowledgments
We thank Dr. Gero Brockhoff (University of Regensburg, Germany) for technical support with cell-cycle analysis; Dr. Inge Tinhofer (Charité University Hospital, Berlin, Germany) for the cell line T98G; Dr. Natascha Schmitt (Department of Neurosurgery, BG-Unfallklinik Murnau) for providing FFPE tissue and clinical data; Dr. Tibor Schuster for statistical support (Technische Universität München, Germany); and Claire Delbridge (Technische Universität München, Germany) for discussion and critical review of the manuscript.
Conflict of interest statement. None of the authors has any financial interests or connections, direct or indirect, or other situations that might raise the question of bias in the work reported or the conclusions, implications, or opinions stated.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 3.Rasper M, Schafer A, Piontek G, et al. Aldehyde dehydrogenase 1 positive glioblastoma cells show brain tumor stem cell capacity. Neuro Oncol. 2010;12(10):1024–1033. doi: 10.1093/neuonc/noq070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69(8):3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T, Su Y, Mei Y, et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab Invest. 2010;90(2):234–244. doi: 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4(6):697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chute JP, Muramoto GG, Whitesides J, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci USA. 2006;103(31):11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilton J. Role of aldehyde dehydrogenase in cyclophosphamide-resistant L1210 leukemia. Cancer Res. 1984;44(11):5156–5160. [PubMed] [Google Scholar]
- 10.Muramoto GG, Russell JL, Safi R, et al. Inhibition of aldehyde dehydrogenase expands hematopoietic stem cells with radioprotective capacity. Stem Cells. 2010;28(3):523–534. doi: 10.1002/stem.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res. 2000;6(7):2585–2597. [PubMed] [Google Scholar]
- 12.Stupp R, Dietrich PY, Ostermann Kraljevic S, et al. Promising survival for patients with newly diagnosed glioblastoma multiforme treated with concomitant radiation plus temozolomide followed by adjuvant temozolomide. J Clin Oncol. 2002;20(5):1375–1382. doi: 10.1200/JCO.2002.20.5.1375. [DOI] [PubMed] [Google Scholar]
- 13.Karran P, Bignami M. DNA damage tolerance, mismatch repair and genome instability. Bioessays. 1994;16(11):833–839. doi: 10.1002/bies.950161110. [DOI] [PubMed] [Google Scholar]
- 14.Karran P, Macpherson P, Ceccotti S, Dogliotti E, Griffin S, Bignami M. O6-methylguanine residues elicit DNA repair synthesis by human cell extracts. J Biol Chem. 1993;268(21):15878–15886. [PubMed] [Google Scholar]
- 15.Sarkaria JN, Kitange GJ, James CD, et al. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14(10):2900–2908. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 17.Beier D, Rohrl S, Pillai DR, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. 2008;68(14):5706–5715. doi: 10.1158/0008-5472.CAN-07-6878. [DOI] [PubMed] [Google Scholar]
- 18.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 20.Bettstetter M, Dechant S, Ruemmele P, et al. MethyQESD, a robust and fast method for quantitative methylation analyses in HNPCC diagnostics using formalin-fixed and paraffin-embedded tissue samples. Lab Invest. 2008;88(12):1367–1375. doi: 10.1038/labinvest.2008.100. [DOI] [PubMed] [Google Scholar]
- 21.Therneau TM, Grambsch PM, Fleming TR. Martingale-based residuals and survival models. Biometrika. 1990;77:147–160. [Google Scholar]
- 22.Mihaliak AM, Gilbert CA, Li L, et al. Clinically relevant doses of chemotherapy agents reversibly block formation of glioblastoma neurospheres. Cancer Lett. 2010;296(2):168–177. doi: 10.1016/j.canlet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirose Y, Berger MS, Pieper RO. p53 effects both the duration of G2/M arrest and the fate of temozolomide-treated human glioblastoma cells. Cancer Res. 2001;61(5):1957–1963. [PubMed] [Google Scholar]
- 24.Beier D, Schuierer G, Beier CP, Bogdahn U. Short-term effective treatment of CNS metastasis of sarcomatoid renal cell carcinoma with temozolomide and pegylated liposomal doxorubicin: a case report. Cases J. 2008;1(1):210. doi: 10.1186/1757-1626-1-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avgeropoulos NG, Batchelor TT. New treatment strategies for malignant gliomas. Oncologist. 1999;4(3):209–224. [PubMed] [Google Scholar]
- 26.Gaspar N, Marshall L, Perryman L, et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Res. 2010;70(22):9243–9252. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hegi ME, Liu L, Herman JG, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26(25):4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 28.Adam SA, Schnell O, Poschl J, et al. ALDH1A1 is a marker of astrocytic differentiation during brain development and correlates with better survival in glioblastoma patients. [published online ahead of print March 14, 2012]. Brain Pathol. doi: 10.1111/j.1750-3639.2012.00592.x. 2012. doi:10.1111/j.1750-3639.2012.00592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlegel J, Schäfer A. ALDH1A1 is a marker of astrocytic differentiation during brain development and correlates with better survival in glioblastoma patients. Brain Pathol. doi: 10.1111/j.1750-3639.2012.00592.x. 2012; 22(5):723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campos B, Centner FS, Bermejo JL, et al. Aberrant expression of retinoic acid signaling molecules influences patient survival in astrocytic gliomas. Am J Pathol. 2011;178(5):1953–1964. doi: 10.1016/j.ajpath.2011.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frosina G. DNA repair and resistance of gliomas to chemotherapy and radiotherapy. Mol Cancer Res. 2009;7(7):989–999. doi: 10.1158/1541-7786.MCR-09-0030. [DOI] [PubMed] [Google Scholar]
- 32.Campos B, Wan F, Farhadi M, et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin Cancer Res. 2010;16(10):2715–2728. doi: 10.1158/1078-0432.CCR-09-1800. [DOI] [PubMed] [Google Scholar]
- 33.Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26(17):2839–2845. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pallini R, Ricci-Vitiani L, Montano N, et al. Expression of the stem cell marker CD133 in recurrent glioblastoma and its value for prognosis. Cancer. 2008;117(1):162–174. doi: 10.1002/cncr.25581. [DOI] [PubMed] [Google Scholar]
- 35.Zeppernick F, Ahmadi R, Campos B, et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res. 2008;14(1):123–129. doi: 10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- 36.Kastan MB, Schlaffer E, Russo JE, Colvin OM, Civin CI, Hilton J. Direct demonstration of elevated aldehyde dehydrogenase in human hematopoietic progenitor cells. Blood. 1990;75(10):1947–1950. [PubMed] [Google Scholar]
- 37.Schnier JB, Kaur G, Kaiser A, et al. Identification of cytosolic aldehyde dehydrogenase 1 from non–small cell lung carcinomas as a flavopiridol-binding protein. FEBS Lett. 1999;454(1–2):100–104. doi: 10.1016/s0014-5793(99)00773-5. [DOI] [PubMed] [Google Scholar]
- 38.Magni M, Shammah S, Schiro R, Mellado W, Dalla-Favera R, Gianni AM. Induction of cyclophosphamide-resistance by aldehyde-dehydrogenase gene transfer. Blood. 1996;87(3):1097–1103. [PubMed] [Google Scholar]
- 39.Pappa A, Brown D, Koutalos Y, DeGregori J, White C, Vasiliou V. Human aldehyde dehydrogenase 3A1 inhibits proliferation and promotes survival of human corneal epithelial cells. J Biol Chem. 2005;280(30):27998–28006. doi: 10.1074/jbc.M503698200. [DOI] [PubMed] [Google Scholar]
- 40.Oliva CR, Moellering DR, Gillespie GY, Griguer CE. Acquisition of chemoresistance in gliomas is associated with increased mitochondrial coupling and decreased ROS production. PLoS One. 2011;6(9):e24665. doi: 10.1371/journal.pone.0024665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang WB, Wang Z, Shu F, et al. Activation of AMP-activated protein kinase by temozolomide contributes to apoptosis in glioblastoma cells via p53 activation and mTORC1 inhibition. J Biol Chem. 2010;285(52):40461–40471. doi: 10.1074/jbc.M110.164046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gago-Dominguez M, Castelao JE, Pike MC, Sevanian A, Haile RW. Role of lipid peroxidation in the epidemiology and prevention of breast cancer. Cancer Epidemiol Biomark Prev. 2005;14(12):2829–2839. doi: 10.1158/1055-9965.EPI-05-0015. [DOI] [PubMed] [Google Scholar]
- 43.Lassen N, Bateman JB, Estey T, et al. Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(-/-)/Aldh1a1(-/-) knock-out mice. J Biol Chem. 2007;282(35):25668–25676. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahmoud MI, Potter JJ, Colvin OM, Hilton J, Mezey E. Effect of 4-(diethylamino)benzaldehyde on ethanol metabolism in mice. Alcohol Clin Exp Res. 1993;17(6):1223–1227. doi: 10.1111/j.1530-0277.1993.tb05233.x. [DOI] [PubMed] [Google Scholar]
- 45.Cen D, Gonzalez RI, Buckmeier JA, Kahlon RS, Tohidian NB, Meyskens FL., Jr. Disulfiram induces apoptosis in human melanoma cells: a redox-related process. Mol Cancer Ther. 2002;1(3):197–204. [PubMed] [Google Scholar]
- 46.Wang W, McLeod HL, Cassidy J. Disulfiram-mediated inhibition of NF-kappaB activity enhances cytotoxicity of 5-fluorouracil in human colorectal cancer cell lines. Internat J Cancer. 2003;104(4):504–511. doi: 10.1002/ijc.10972. [DOI] [PubMed] [Google Scholar]
- 47.Koppaka V, Thompson DC, Chen Y, et al. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64(3):520–539. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.