Abstract
Meat mutagens, including heterocyclic amines (HCAs), polycyclic aromatic hydrocarbons (PAHs) and N-nitroso compounds (NOCs), may be involved in colorectal carcinogenesis depending on their activation or detoxification by phase I and II xenobiotic metabolizing enzymes (XME). Using unconditional logistic regression to estimate odds ratios (OR) and 95% confidence intervals (CI), we examined the intake of five meat mutagens and >300 single nucleotide polymorphisms (SNPs) in 18 XME genes in relation to advanced colorectal adenoma (1205 cases and 1387 controls) and colorectal cancer (370 cases and 401 controls) within the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Dietary intake of meat mutagens was assessed using a food frequency questionnaire with a detailed meat-cooking module. An interaction was observed between 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx) intake and the NAT1 polymorphism rs6586714 in the adenoma study (P interaction = 0.001). Among individuals carrying a GG genotype, high MeIQx intake was associated with a 43% increased risk of adenoma (95% CI 1.11–1.85, P trend = 0.07), whereas the reverse was observed among carriers of the A variant (OR = 0.50, 95% CI 0.30–0.84, P trend = 0.01). In addition, we observed some suggestive (P < 0.05) modifying effects for SNPs in other XME genes (UGT1A, CYP2E1, EPHX1, AHR and GSTM3), but these were not significant after adjustment for multiple testing. This large and comprehensive study of XME genes, meat mutagens and the risk of colorectal tumours found that a NAT1 polymorphism modified the association between MeIQx intake and colorectal adenoma risk.
Introduction
Experimental evidence suggests that the carcinogenic potential of several meat-specific mutagens may be one of the underlying causal factors for the well-established epidemiological association between red and processed meat consumption and colorectal cancer risk (1). Heterocyclic amines (HCAs) are formed when meat is cooked well-done at high temperature (2,3). Grilled meat may also contain polycyclic aromatic hydrocarbons (PAHs) from smoke that coats the meat. PAHs are also present in cigarette smoke, in the environment and in other food; although concentrations in foods not prepared by grilling or smoking are minor by comparison (2,3). N-nitroso compounds (NOCs) are formed from nitrate and nitrite that are added as preservatives to processed meat (4). Despite the strong carcinogenic potential of HCAs, PAHs and NOCs observed in animal studies (5–9), evidence in humans remains inconsistent (1).
HCAs, PAHs and NOCs undergo a series of chemical reactions in the human body during which they can be activated or detoxified by phase I and phase II xenobiotic metabolizing enzymes (XMEs) (10,11). Single nucleotide polymorphisms (SNPs) in the genes encoding these XMEs may modify the ability to activate or detoxify carcinogens. Previous studies examining interactions between XME polymorphisms, meat consumption and the risk of colorectal adenomas or carcinomas have reported mixed results (12–26). In addition, most previous studies had a small number of cases or examined only a small set of SNPs from a limited number of candidate genes. Because the balance of activating and detoxifying enzymes is thought to influence carcinogen metabolism (27), comprehensive studies including numerous markers across multiple genes involved in xenobiotic metabolism are essential for studying this complex association. Furthermore, the inconsistencies in the data may partly result from the inability of most studies to estimate specific HCAs, PAHs and nitrite/nitrate due to lack of information on both cooking technique and doneness level. Although many studies ask about the consumption of well-done meat, one needs information on both cooking methods and doneness levels to get accurate intake estimates. Studies in different populations need specific databases relevant for their consumption patterns (e.g. Japan, Sweden and USA). The Computerized Heterocyclic Amines Resource for Research in Epidemiology of Disease software application was specifically developed for the US population, therefore, appropriate for our study (3).
We conducted two nested case–control studies within the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial to examine the effect of meat-related mutagens and their interaction with 513 tag SNPs from 18 selected genes involved in phase I [cytochrome P450s (CYP)] and phase II metabolism [sulfotransferases (SULT), N-acetyltransferases (NAT), UDP-glucuronosyltransferases (UGT) and glutathione S-transferases (GST)] in relation to both colorectal adenoma and colorectal cancer risk.
Materials and methods
Study population
The PLCO Cancer Screening Trial is a randomized, multicenter clinical trial investigating the efficacy of screening for prostate, lung, colorectal and ovarian cancer (28,29). Participants aged 55–74 years were recruited from 10 centres in the USA from 1993 to 2001 and randomly assigned to the screened or the control arm of the trial. The study was approved by the Institutional Review Board at the National Cancer Institute and the 10 study centres, and all participants provided written informed consent. The present investigation is restricted to the individuals in the screening arm of the trial (n = 77 469), who completed a self-administered risk factor questionnaire at baseline, filled out a food frequency questionnaire (FFQ) with detailed meat-cooking questions, and provided a blood sample.
We conducted two separate nested case–control studies with two disease outcomes: (i) advanced colorectal adenoma and (ii) colorectal cancer. To be eligible for these analyses, participants had to have no history of cancer prior to completion of the FFQ, and no prior history of self-reported colon disease (Crohn’s disease, ulcerative colitis, familial polyposis or Gardner’s syndrome). Individuals with a prior history of colorectal polyps were also excluded from the analyses of adenoma.
Advanced adenoma sample. Adenoma cases were participants found to have at least one advanced colorectal adenoma (≥1cm in size, containing villous/tubulovillous characteristics, or had severe dysplasia) of the distal colon or rectum at baseline. Controls were subjects who underwent a successful sigmoidoscopy examination (defined as insertion to at least 50cm with ≥90% of mucosa visible or suspicious lesion found) at baseline and had no evidence of a left-sided polyp. Controls were matched to cases on gender, ethnicity and for a subset, age. A total of 1243 advanced adenoma cases and 1419 controls with DNA were available for this study.
Colorectal cancer sample. Colorectal cancer cases were identified through self-report from the annual study update questionnaire, death certificates or physician reports, and confirmed by review of pathology reports and medical records. Cases were identified through 31 December 2006. Controls were subjects without a diagnosis of colorectal cancer at the time the case was diagnosed, matched on age, gender, ethnicity and year of randomization. A total of 371 cases and 405 controls with DNA were identified and genotyped in this study. Five cases from the adenoma sample later on developed colorectal cancer and were also included in the cancer sample.
Dietary assessment
Participants completed a 137-item FFQ with a detailed meat-cooking module that ascertained usual diet during the previous 12 months. Most (89%) study participants completed the FFQ prior to or on the same day as the baseline sigmoidoscopy. Using the Computerized Heterocyclic Amines Resource for Research in Epidemiology of Disease (www.charred.cancer.gov) software application (3), we generated intake estimates of three HCAs (ng/day): 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx), 2-amino-3,4, 8-trimethylimidazo[4,5-f]quinoxaline (DiMeIQx) and 2-amino-1- methyl-6-phenyl-imidazo[4,5-b]pyridine (PhIP), as well as benzo[a]pyrene (B[a]P), which is a marker of PAH exposure. We estimated nitrate and nitrite from processed meats using a nitrate/nitrite database based on laboratory measured values from 10 types of processed meat samples (bacon; sausage; hot dogs; roast beef; pork chops; ham; lunch meats, including bologna, salami and processed ham) that represented 90% of the processed meat consumed in the USA (3,17). We multiplied the frequency of consumption of each processed meat item by the portion size and by the nitrate or nitrite concentration of the respective item to estimate nitrate and nitrite intake (mg/day). Participants were excluded from these analyses if they had ≥8 missing or invalid responses of the FFQ or missing data on the meat mutagens (n = 71 and n = 18 for the adenoma and cancer samples, respectively), leaving 1386 advanced adenoma cases [171 rectal, 462 distal colon, 566 multiple adenoma (either side) and 6 with unknown location] and 1205 controls and 364 cancer cases (182 proximal colon, 102 distal colon, 79 rectal and 1 unknown location), and 394 controls for analyses.
Genotyping
A total of 513 tag SNPs were selected for 18 genes or gene regions directly involved in the metabolism of HCAs, PAHs or possibly with NOCs [aromatic hydrocarbon receptor (AHR), CYP1A1/CYP1A2, CYP1B1, CYP2A6, CYP2C9, CYP2D6, CYP2E1, epoxide hydrolase (EPHX1), GSTA1, GSTM1, GSTM3, GSTP1, GSTT1, NAT1, NAT2, NAD(P)H dehydrogenase (quinone 1) (NQO1), SULT1A1/SULT1A2 and the UGT1A locus]. Tag SNPs were selected including the region 20kb upstream and 10kb downstream of the genes, using the CEU, JPT, CHB and YRI HapMap populations and the Carlson method (30) as implemented in Tagzilla with a r 2 threshold of 0.8 and minor allele frequency ≥5%. SNPs with known or putative functional significance (i.e. non-synonymous, promoter, intron–exon splice sites) were also included whenever possible. The SNPs from the phase I and II genes for putative pathway for metabolism of HCAs, PAHs and nitrites/nitrates [Supplementary Figure 1 is available at Carcinogenesis Online (11,31,32)] were genotyped on a custom iSelect panel using Illumina’s Infinium platform.
Whole blood or buffy coat DNA was extracted with QIAamp DNA Blood Midi or Maxi Kits. For quality control purposes, replicate samples from 195 individuals (~7% of the population) were interspersed randomly within the plates. Genotyping was conducted at the National Cancer Institute Core Genotyping Facility, National Institutes of Health. We required all cases and controls to have a 90% call rate for analysis. The overall concordance rate was >99% for replicated samples. We excluded SNPs with a call rate <90%, minor allele frequency <1% in the adenoma set and 5% in the cancer set (due to lower power), or Hardy–Weinberg equilibrium P-value <1×10−6 among Caucasian controls. Of the 513 SNPs we selected, 380 SNPs remained for the adenoma analyses and 325 SNPs remained for the cancer analyses. There were a few highly correlated SNPs among the Caucasians in our study as the tag SNPs were selected for multiple populations. SNPs with an r 2 > 0.95 among Caucasians were considered to be equivalent for interaction testing.
Statistical analysis
All analyses were conducted separately for the prevalent colorectal adenoma and colorectal cancer study populations. Differences in baseline characteristics between cases and controls were assessed using chi-square tests for categorical variables, such as sex, and t-tests for continuous variables, such as age. Due to a skewed intake distribution of the meat mutagens, we transformed these variables prior to performing t-tests, using the inverse rank transformation.
Main effects: meat mutagens and SNPs
In order to confirm what has been shown previously in PLCO subgroup analyses, we examined the main effects of each mutagen and SNP on colorectal adenoma and cancer risk using unconditional multivariable logistic regression. The main effects of the meat mutagens were adjusted for all relevant covariates (listed as footnotes in the tables). Meat mutagens were categorized as low (0–39th percentile), medium (40–79th percentile) and high (≥80th percentile) intake, as the distribution of these mutagens is highly skewed with the majority of individuals consuming small amounts, and previous analyses, both within the PLCO study population and within other studies, have identified the top quintile as potentially the most important with regard to cancer risk (33,34) (e.g. PhIP intake in adenoma dataset: quintile 1: range, 0–19.5mg/day, median, 10.1mg/day; quintile 5: range, 166–3069mg/day, median, 268mg/day). The main effects of the SNPs were examined using PLINK, a whole-genome association analysis toolset (35) assuming a log-additive model for the genotype, and were adjusted for age, sex and ethnicity. Results were adjusted for multiple testing using the false discovery rate (36). We also used a Bonferroni correction for the total number of tag SNPs for each individual gene or gene region (gene-based correction).
Gene × environment (G×E) interactions
Because the experimental literature on the role of XME genes in relation to HCA, PAH and NOC metabolism is far from complete, we did not restrict our analyses to those gene–mutagen interactions shown previously, but rather used an exploratory approach when testing for interactions between all the SNPs and mutagens under study. We used a two-step test for G×E interactions as described by Murcray et al. (37), to identify SNPs involved in a G×E interaction. In the first step, we examined the association of each SNP with each mutagen in a linear regression model among cases and controls combined, adjusting for age, sex and ethnicity. Due to a skewed intake distribution, we transformed the exposure variables using the inverse normal rank transformation. Linear regressions were calculated using PLINK.
The subset of m SNPs that exceeded the significance threshold of P < 0.10 was taken forward to Step 2 for the actual case–control test for a G×E interaction. In Step 2, an unconditional logistic regression model, assuming a dominant genetic model, adjusting for age, sex and ethnicity, was calculated with a multiplicative interaction term of the SNP with the respective meat mutagen (categorical variable assigning median values), as well as the individual main effect terms. Further adjustment for all confounders selected in the mutagen main effects analyses did not change the results appreciably. Significance at this step was defined as a P-value < α/m, where α was the desired overall Type I error rate of 10%. When multiple SNPs from the same gene exceeded the P < 0.10 threshold level, a M eff was calculated to determine the effective number of independent comparisons (38).
All interactions meeting the significance threshold of P < 0.05 were further evaluated using logistic regression models to examine the association between the categorical meat mutagen intake and colorectal adenoma or cancer, stratified by genotype assuming a dominant model. We also conducted stratified analyses by ever and never smokers, rectal and non-rectal adenomas, and single versus multiple adenomas. The top interactions in the cancer set were also examined by proximal, distal and rectal cancer. Models for Step 2 and the models for the association of the meat mutagens and colorectal adenoma or cancer were calculated using STATA version 9.0.
Results
A total of 1205 cases and 1386 controls were available for the advanced colorectal adenoma analysis, and 364 cases and 394 controls were included in the colorectal cancer population (Table I). Over 90% of the study subjects were Caucasian and over 60% were males; in the adenoma study, cases were older than controls.
Main effects: meat mutagens
Consistent with a previous analysis of prevalent adenoma in the PLCO trial (34), there was an elevated risk of colorectal adenoma in the top quintile of MeIQx intake when compared with the bottom quintile (OR = 1.17, 95% CI 0.94–1.45); however, in this study population with ~2000 fewer adenoma cases, the risk did not reach statistical significance and was attenuated after adjustment for confounders (Supplementary Table I is available at Carcinogenesis Online). We observed a borderline significant positive association between nitrate and nitrite intake from meat and adenoma risk (P trend = 0.05), but this was attenuated with covariate adjustment. B[a]P, PhIP and DiMeIQx intake were not associated with adenoma risk in this population. Furthermore, none of the meat mutagens were associated with colorectal cancer. Due to subjects with missing values, the number of cases and controls in the multivariable models were lower (adenoma: 1183 cases and 1358 controls; cancer: 387 cases and 358 controls) than in the age-, ethnicity- and sex-adjusted model; however, this did not explain the observed attenuation. The ORs for MeIQx and colon cancer in particular changed considerably after adjustment for confounders; level of education and total daily energy intake contributed the most to this change.
Main effects: SNPs
We found that SNPs from the UGT1A, EPHX1 and NAT1 genes were nominally associated with both colorectal adenoma (n = 11 SNPs) and cancer risk (n = 20 SNPs) (P < 0.05) (Supplementary Table II is available at Carcinogenesis Online). In addition, one SNP from the GSTA1 locus was associated with adenoma risk, whereas one SNP at the NQO1 gene was associated with colorectal cancer. After adjusting for multiple testing for all the SNPs tested using the false discovery rate, none of these findings remained statistically significant in either the adenoma or cancer analysis. However, UGT1A rs7569014 remained associated with colorectal cancer risk at P ≤ 0.05 after the gene-based multiple testing correction.
Gene × environment interactions
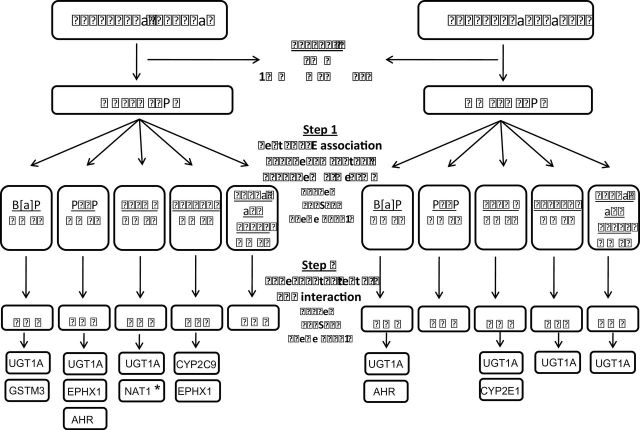
A schematic representation of the results from the two-step test for G×E interaction for both the adenoma and cancer study populations is presented in Figure 1. The subset and number of SNPs that exceeded the significant threshold of P < 0.10 in Step 1, differed by outcome and mutagen under study. SNPs that passed this initial screening step were taken forward to the actual case–control test of G×E interaction analyses in Step 2.
Fig. 1.
Schematic representation of the two-step test for G×E interaction of Murcray et al. (31). We first excluded SNPs with call rate <90%, minor allele frequency (MAF) <1% in the adenoma set and 5% in the cancer set, or Hardy–Weinberg Equilibrium (HWE) P-value <1×10−6 among Caucasian controls. In Step 1 of the test G×E interaction, G–E association were first tested in the combined case and control sample. Only SNPs with P < 0.10 in Step 1 are then tested for G×E interaction using a standard case–control analysis in Step 2. We further explored the risk pattern for all interactions that met the unadjusted significance threshold of 0.05 (see Tables 3 and 4). *Significant after adjustment for multiple testing at P < 0.05.
Advanced prevalent adenoma. The G×E interaction analyses performed in Step 2 yielded 15 interactions with P ≤ 0.10 between B[a]P, HCAs and several SNPs, including those in the GSTM3, UGT1A, AHR, EPHX, NAT1 and CYP2C9 gene regions (Table II and Figure 1). The most significant interaction was between NAT1 (rs6586714) and MeIQx (P interaction = 0.001), which was the only finding that remained significant after adjustment for multiple testing. Various SNPs from the UGT1A locus appeared to modify the association between B[a]P (3 SNPs, r > 0.79), PhIP (4 SNPs, r −0.12 to 0.89), and MeIQx intake (1 SNP) and adenoma risk (P < 0.10). Two SNPs (r = 0.39) located within the EPHX1 region (rs2671272 and rs868966) appeared to modify the effect of PhIP and DiMeIQx on colorectal adenoma risk (P ≤ 0.05).
We further explored the risk pattern for all interactions that met the unadjusted significance threshold of 0.05. Age-, ethnicity- and sex-adjusted associations between mutagen intake and colorectal adenoma stratified by genotype are presented in Table III. There was an increased risk of colorectal adenoma with increasing intake of MeIQx among individuals carrying a GG genotype for NAT1 rs6586714 (OR = 1.43, 95% CI 1.11–1.85 for high intake compared with low intake; P trend = 0.007), whereas the risk was decreased among GA and AA carriers (OR = 0.50 for high intake compared with low intake, 95% CI 0.30–0.84; P trend = 0.01; Table III). Increased intake of DiMeIQx was positively associated with adenoma risk in individuals carrying an AA genotype for EPHX1 rs868966 (OR= 1.45, 95% CI 0.98, 2.15; P trend = 0.06).
We additionally ran the G×E interaction analyses for total mutagenic activity, a measure incorporating mutagenicity of all meat-related mutagens. We found five interactions in the adenoma set, whereas one interaction (rs28969701– UGT1A locus) was observed in the cancer data. However, these SNPs came from genes similar to those interacting with the individual mutagens (CYP2C9, EPHX1, GSTM3, UGT1A locus) (data not shown). The correlation between total mutagenic activity and the individual HCAs and PAHs for adenomas was highest for MeIQx (r = 0.84), followed by PhIP (r = 0.78), DiMeIQx (r = 0.76) and B[a]P (r = 0.51).
Subgroup analyses indicated that the top interactions were similar for the subgroups of any rectal (n = 377) and any distal adenoma (n = 957), and single (n = 639) versus multiple adenomas (n = 566). However, the interaction between PhIP intake and rs2018985 was only observed with respect to multiple adenomas but not for single adenomas (data not shown). Moreover, our top interaction were similar in direction and in magnitude for ever (n = 743) and never (n = 462) smokers, except for the interaction between DiMeIQx and rs868966 that was only observed among ever smokers (data not shown).
Colorectal cancer. In the carcinoma dataset, seven interactions with P ≤ 0.10 were observed (Table II and Figure 1). We found a suggestive interaction between several SNPs in the UGT1A region and intake of B[a]P (rs6714486 and rs17868299), MeIQx (rs2011404), DiMeIQx (rs6717546) and combined nitrate and nitrite from meat (rs12466997). The association between MeIQx and colorectal cancer risk seemed to be modified by a SNP in the CYP2E1 gene (rs915908) (P interaction = 0.05). However, neither these nor other interactions for colorectal cancer were deemed significant after adjusting for multiple testing.
Age-, ethnicity- and sex-adjusted associations between mutagen intake and colorectal cancer stratified by genotype are presented in Table III (SNPs with P interaction ≤ 0.05). High consumption of B[a]P was associated with an increased risk of colorectal cancer among individuals carrying the CT genotype for UGT1A rs17868299 (OR = 6.04 for high intake compared with low intake, 95% CI 1.39–24.2; P trend = 0.06). A suggestive inverse association between MeIQx intake and colorectal cancer risk was observed among the wild-type variant of rs915908 (CYP2E1) (OR = 0.68 for high intake compared with low intake, 95% CI 0.40–1.08; P trend = 0.08).
Interactions between SNPs in the UGT1A locus and various meat mutagens were observed for both the adenoma and cancer endpoints. No strong correlation (r < 0.25) between SNPs found in the adenoma and cancer analyses were found (results not shown). However, the interaction observed between rs7571337 and MeIQx intake for adenoma could be replicated for the cancer endpoint (P interaction = 0.09), and the ORs for the SNP-stratified results were in the same direction, though attenuated. The pooled OR (95% CI) combining the adenoma and cancer results for high intake compared with low intake was 1.72 (1.03–2.39) for the TT genotype and 0.95 (0.70–1.13) for the TC+CC genotype. Nevertheless, all other interactions that were significant in the adenoma dataset were not found to be so in the cancer study (P > 0.10), and vice versa (Supplementary Table III is available at Carcinogenesis Online).
The OR and 95% CI for the main effects of the SNPs that were found to be involved in a G×E interaction (P < 0.10) are presented in Supplementary Table IV, available at Carcinogenesis Online . Two SNPs from the UGT1A locus (rs871514 and rs2018985) were marginally associated with colorectal adenoma (P = 0.06), but did not remain statistically significant after adjustment for multiple testing using either the false discovery rate or the gene-based approach.
We examined the top interactions (as described in Table III) by proximal, distal and rectal cancer subsite and observed that the interaction effect for rs6717546 remained present in all endpoints. However, the interaction between B[a]P and rs17868299 was only observed in the distal colon, and the magnitude of the SNP stratified associations for the variant allele was larger than that in the combined group of cancers (CC genotype: ORmedium = 4.36 95% CI 1.02–18.55 and ORhigh = 8.82 95% CI 1.60–48.72) (data not shown); however, since the numbers were small, this may be a chance finding.
Discussion
We observed that the effect of MeIQx intake on advanced colorectal adenoma risk differed by NAT1 genotype. Additional evidence of effect modification of the association between various meat-related HCAs, PAHs and nitrate/nitrite, and advanced colorectal adenoma and cancer by variants in UGT1A, CYP2E1, EPHX1, AHR and GSTM3 was observed; however, correction for multiple comparisons indicated that these observations may be due to chance, despite that this is the largest and most comprehensive study to date.
Our finding that NAT1 modified the effect of MeIQx on the risk of advanced colorectal adenoma is in agreement with results from animal and in vitro research. NAT1 is primarily expressed in hepatic and colonic tissue and has been extensively shown to be an important enzyme in the bioactivation of multiple HCAs via O-acetylation processes (39). NAT1 enzyme activity is characterized by strong between-population heterogeneity that probably results in inter-individual differences in acetylation rates (40). Functionality of the variant observed in our analyses (rs6586714), which is located in the intron region of NAT1, is lacking and not strongly correlated with any known functional NAT1 alleles in the iSelect panel (r 2 < 0.20). One previous analysis studied the interaction between various NAT1 alleles (*3, *4, *10, *11; categorizing participants into slow, normal and fast acetylaters) and meat intake in relation to colorectal adenoma risk but reported null results (41). Nevertheless, evidence from the few epidemiological studies that examined interactions between NAT1 and meat (15,23,24) or HCA intake (42) and colorectal cancer risk, points towards an increased risk among rapid acetylaters. However, these findings may have bias towards the null due to the generally small study samples, lack of detailed HCA exposure information and the debatable classification of NAT1 alleles. From our prior research, there is evidence that MeIQx is estimated with greater accuracy than other HCAs (43). In addition, although MeIQx is consumed at lower levels in the diet, it is a more potent mutagen than PhIP is (44). Moreover, it could be that this interaction is observed due to linkage disequilibrium with another variant in the region that has an effect on a neighbouring gene.
Although we observed some suggestive modifying effects in SNPs in other XME genes, the findings were not significant after adjusting for multiple testing. For example, multiple UGT1A polymorphisms were suggestively associated with colorectal adenoma and cancer or appeared to interact with meat mutagens and tumour risk. UGTs are phase II conjugation enzymes that are primarily expressed in the luminal cells of the gastrointestinal tract, and have been implicated in the detoxification of HCAs and B[a]P (45,46). Previous epidemiological studies suggest that alterations in UGT genes may indeed influence the elimination of meat-specific mutagens; one case–control reported an interaction between B[a]P intake and variants in UGT1A1 promoter polymorphisms at position −53 and −3156 (47), whereas a suggestive interaction between DiMeIQx and UGT1A7 (48) acetylation status was reported in a different study. However, the multiple SNPs located across the UGT1A region that were involved in interactions with specific meat mutagens in this study were found in overlapping intron regions of UGT1A1 genes (UGT1A3–A10), and mostly highly correlated, making it impossible to differentiate the associations of each statistically. Future research is needed to unravel this linkage disequilibrium and focus on individual UGT1A isoenzymes to characterize their etiological relevance with meat mutagen-induced colorectal carcinogenesis.
We found some other suggestive interactions between SNPs in the CYP2E1, AHR and EPHX1 gene, although results after multiple testing suggest that these findings may be due to chance. Although these genes are implicated in the metabolism of meat mutagens, there is little evidence in the literature for the functionality of the observed SNPs. However, rs2066853 (AHR) encodes for an amino acid change in the protein (Arg554Lys), whereas rs15864 (GSTM3) is correlated (r 2 = 0.81) with rs7483, a missense mutation in the same gene responsible for an amino acid substitution (Val224Ile) that significantly alters enzyme activity (49).
Despite the biological rationale for interactions between meat-specific mutagens and polymorphisms in phase I and II XME genes in relation to colorectal carcinogenesis, the epidemiological evidence remains inconclusive. This lack of consistency may be due to inadequate sample size, the high potential for chance findings as a result of multiple comparisons and the over-interpretation of subgroup findings in many gene–environment interaction studies. Although this study is the largest and most comprehensive to examine these associations to date, we were still underpowered to detect modest effects of single gene variants, especially for the colorectal cancers. To gain power, we applied a two-step approach (37), incorporating a preliminary screening step to identify the SNPs involved in possible gene–environment interactions, and hence protect against false positive results. Moreover, the iSelect genotyping panel was specifically designed to capture only tagging SNPs from a priori selected genes involved in carcinogen metabolism to reduce the number of tests performed.
Examination of the same set of SNPs and mutagens yielded different interactions in the adenoma and cancer analyses. Although it may be that some of our findings are false positives, the differences we observed may reflect variation in gene–environmental interactions between initiation and progression in the carcinogenesis process. There is tremendous heterogeneity among colorectal adenomas, with only a small subset of adenomas progressing to cancer. Therefore, interactions related to the formation of adenoma may not be observed for cancer if they do not also stimulate progression to cancer. In contrast, interactions associated with malignant transformation may only be observed for cancer. Nevertheless, we did observe multiple interactions between SNPs at the UGTA1 locus and both colorectal adenoma and cancer, suggesting that this locus may be important for progression and transformation. Many of the XME genes under study are part of larger multigene families. As a result there could be redundancy among these enzymes, such that if the activity of one is modified as a result of a specific genetic variation others may continue to carry out the metabolism of meat mutagens. We were not able to examine differences in etiology between proximal and distal adenomas and advanced and non-advanced adenomas; this study was restricted to advanced distal and rectal colorectal adenomas only.
We made use of a comprehensive assessment of meat-cooking methods that enabled detailed estimation of HCAs, PAHs and nitrate/nitrite, but this is nonetheless susceptible to measurement error. We did not collect information concerning all aspects of meat-cooking methods that could have influenced carcinogen production, such as microwaving prior to cooking (50), marinating the meat (51) or flipping burgers more often (52). Moreover, during meat cooking, numerous other mutagenic compounds could be formed that may be correlated with the mutagens under study, and could thus potentially explain the observed interactions (e.g. between NAT1 and MeIQx). Our article focuses on meat carcinogenesis and, therefore, we did not consider other potential dietary sources of nitrite and nitrate. Although the majority of dietary nitrite comes from processed meats (53), other food items and drinking water may contribute to intake.
Given that this study was nested within the screening arm of the PLCO Cancer Screening Trial, we examined possible interactions using colorectal adenomas as an endpoint, largely asymptomatic precursors of colorectal cancer. Moreover, selection and surveillance bias is minimal since both cases and controls had an equal opportunity to have a colorectal adenoma or cancer detected. Our colorectal cancer analyses were based on a prospective study of incident cases, eliminating the potential for recall bias. However, inherent to the nature of the study population, participants were generally more educated, less likely to smoke, more physically active and more likely to be Caucasian than the general US population, thereby limiting the generalizability of the study results. All adenoma cases and controls and the majority of cancer cases and controls underwent at least one sigmoidoscopy screening as part of the trial. Since nearly all polyps found were subsequently removed in this heavily screened population, it is possible that the colorectal cancers observed were somehow different than what would be observed in the general population, perhaps more likely to arise de novo. It is also possible that some subjects changed their diet following screening. However, this is unlikely to have affected the results of this study since diet was assessed prior to screening and cancer cases were diagnosed a median of 3.2 years later, which is shorter than the predicted time span (10–20 years) of the adenoma–carcinoma sequence.
In conclusion, we found some evidence that common variants in XME genes may modify the association between meat mutagens and colorectal neoplasia. The strongest evidence for an interaction was observed between MeIQx intake and a NAT1 polymorphism in relation to colorectal adenoma risk. Future pooled initiatives that would have larger sample sizes should further evaluate this complex interplay to better understand these relationships.
Table I.
Distribution of baseline characteristics in nested case control studies of advanced colorectal adenoma and colorectal cancer in the PLCO Cancer Screening Trial
| Adenoma | Cancer | ||||||
| Characteristic | Cases (n = 1205)a | Controls (n = 1386)a | Pb | Cases (n = 364)a | Controls (n = 394)a | Pb | |
| Age (years) | 63.1±5.2 | 62.6±5.3 | 0.02 | 67.6±6.5 | 67.5±6.3 | 0.81 | |
| Sex, n (%) | 0.83 | 0.71 | |||||
| Male | 776 (64.4) | 887 (64.0) | 216 (59.3) | 239 (60.7) | |||
| Female | 429 (35.6) | 499 (36.0) | 148 (40.7) | 155 (40.3) | |||
| Ethnicity, n (%) | 0.23 | 0.97 | |||||
| Non-Hispanic white | 1136 (94.3) | 1284 (92.6) | 311 (90.9) | 357 (90.6) | |||
| Non-Hispanic black | 29 (2.4) | 46 (3.3) | 18 (5.0) | 21 (5.3) | |||
| Other | 40 (3.3) | 56 (4.0) | 15 (4.1) | 16 (4.1) | |||
| First degree family history of colorectal cancer, n (%) | 157 (13.1) | 136 (9.9) | 0.01 | 56 (15.6) | 45 (11.6) | 0.11 | |
| Smoking status, n (%) | |||||||
| Never | 462 (38.4) | 657 (47.4) | <0.01 | 157 (43.1) | 164 (41.6) | 0.90 | |
| Former cigarette smoker | 165 (13.7) | 94 (6.8) | 167 (45.9) | 187 (47.5) | |||
| Current cigarette smoker | 577 (47.9) | 635 (45.8) | 40 (11.0) | 43 (10.9) | |||
| B[a]P (ng/day) | |||||||
| median (IQR) | 7.0 (1.3, 36.1) | 6.9 (1.2, 35.9) | 0.17 | 5.5 (1.2. 23.8) | 5.4 (1.0, 36.4) | 0.74 | |
| PhIP (ng/day) | |||||||
| Median (IQR) | 65.4 (24.6, 132.6) | 61.1 (23.3, 132.3) | 0.69 | 50.0 (21.0. 127.5) | 53.8 (22.8, 135.8) | 0.23 | |
| MeIQx (ng/day) | |||||||
| Median (IQR) | 22.7 (9.75, 43.2) | 20.5 (8.94, 41.8) | 0.20 | 18.0 (9.1. 38.7) | 18.9 (9.0, 39.6) | 0.59 | |
| DiMeIQx (ng/day) | |||||||
| Median (IQR) | 1.02 (0.23, 2.47) | 0.95 (0.27, 2.2) | 0.39 | 0.80 (0.18, 2.22) | 0.90 (0.22, 2.22) | 0.35 | |
| Combined nitrate and nitrite (ng/day) | |||||||
| Median (IQR) | 0.32 (0.13, 0.61) | 0.28 (0.12, 0.59) | 0.10 | 0.31 (0.13, 0.61) | 0.28 (0.12, 0.55) | 0.36 | |
Data are means ± standard deviations unless otherwise indicated. IQR, Interquartile range; NSAIDs, non-steroidal anti-inflammatory drugs.
aNumbers may not sum due to total due to missing values.
b P-values derived from t-test or chi-square test.
Table II.
Meat mutagen–SNP interaction and risk of colorectal adenoma and cancer for all interactions with a P ≤ 0.10
| Gene/gene region | dbSNP identifier | Main effect P valuea | Pinteractionb | ||
| Adenoma | |||||
| B[a]P | |||||
| GSTM3c | rs15864 | 0.75 | 0.07 | ||
| UGT1A locus | rs1105880 | 0.96 | 0.04 | ||
| UGT1A locus | rs12623271 | 0.44 | 0.04 | ||
| UGT1A locus | rs10168416 | 0.83 | 0.07 | ||
| PhIP | |||||
| UGT1A locus | rs871514 | 0.06 | 0.09 | ||
| UGT1A locus | rs28969701 | 0.40 | 0.08 | ||
| UGT1A locus | rs2018985 | 0.06 | 0.05 | ||
| UGT1A locus | rs10197460 | 0.37 | 0.09 | ||
| AHR | rs4236290 | 0.63 | 0.05 | ||
| EPHX1d | rs2671272 | 0.34 | 0.08 | ||
| MeIQx | |||||
| UGT1A locus | rs7571337 | 0.62 | 0.07 | ||
| NAT1 | rs6586714 | 0.64 | 0.001* | ||
| DiMeiQx | |||||
| CYP2C9 | rs9332197 | 0.47 | 0.09 | ||
| EPHX1d | rs2671272 | 0.34 | 0.05 | ||
| EPHX1e | rs868966 | 0.27 | 0.03 | ||
| Combined nitrate and nitrite | |||||
| — | — | — | — | ||
| Cancer | |||||
| B[a]P | |||||
| AHR | rs2066853 | 0.21 | 0.07 | ||
| UGT1A locus | rs6714486 | 0.50 | 0.06 | ||
| UGT1A locus | rs17868299 | 0.67 | 0.05 | ||
| PhIP | |||||
| — | — | — | — | ||
| MeIQx | |||||
| UGT1A locus | rs2011404 | 0.30 | 0.08 | ||
| CYP2E1 | rs915908 | 0.63 | 0.05 | ||
| DiMeiQx | |||||
| UGT1A locus | rs6717546 | 0.52 | 0.04 | ||
| Combined nitrate and nitrite | |||||
| UGT1A locus | rs12466997 | 0.17 | 0.08 | ||
aP value is for main effect of SNP on colorectal adenoma and cancer risk (P-values not corrected for multiple testing). Adjusted for age, sex and ethnicity and assuming a log-additive model.
bP value is for a test for interaction using the likelihood ratio test comparing models with and without the cross-product of SNP for the given gene and the median of the categories of meat mutagen intake. P-values not corrected for multiple testing. Adjusted for age, sex and ethnicity and assuming a dominant model.
cSNP located within 10kb downstream of EPS8L3.
dSNP located within 10kb downstream of TMEM63A.
eSNP located within 10kb upstream of LEFTY3.
*statistically significant at P<0.05 after adjustment for multiple testing
Table III.
Association between meat mutagen intake and colorectal adenoma and cancer risk stratified by genotypes for which mutagen–SNP interactions were P < 0.05
| Mutagen exposure | ||||||||||||||||||
| Mutagene | Gene | SNP | Geno-type | Low | Medium | High | P trend | P interacta | ||||||||||
| Ca/Co | REF | Ca/Co | ORa | (95% CI) | Ca/Co | ORa | (95% CI) | |||||||||||
| Adenoma | B[a]P | UGT1A | rs12623271 | CC | 151/184 | REF | 173/181 | 1.17 | (0.87, 1.58) | 89/123 | 0.91 | (0.75, 1.15) | 0.35 | 0.04 | ||||
| CG+GG | 315/357 | REF | 313/388 | 0.93 | (0.75, 1.15) | 163/153 | 1.25 | (0.95, 1.64) | 0.06 | |||||||||
| PhIP | UGT1A | rs2018985 | AA | 164/202 | REF | 177/199 | 1.15 | (0.85, 1.55) | 75/120 | 0.82 | (0.57, 1.19) | 0.21 | 0.05 | |||||
| AG+GG | 309/345 | REF | 314/359 | 0.97 | (0.77, 1.21) | 164/159 | 1.15 | (0.88, 0.52) | 0.26 | |||||||||
| PhIP | AHR | rs4236290 | TT | 366/411 | REF | 380/442 | 0.98 | (0.80, 1.20) | 183/229 | 0.92 | (0.72, 1.17) | 0.49 | 0.05 | |||||
| TC+CC | 105/136 | REF | 111/115 | 1.24 | (0.85, 1.80) | 57/50 | 1.45 | (0.91, 2.31) | 0.14 | |||||||||
| MeIQx | NAT1 | rs6586714 | GG | 330/413 | REF | 357/390 | 1.20 | (0.97, 1.47) | 196/182 | 1.43 | (1.11, 1.85) | 0.007 | 0.001 | |||||
| GA+AA | 90/87 | REF | 102/130 | 0.74 | (0.49, 1.10) | 38/69 | 0.50 | (0.30, 0.84) | 0.01 | |||||||||
| DiMeIQx | EPHX1 | rs2671272b | GG | 292/329 | REF | 288/352 | 0.93 | (0.74, 1.16) | 146/186 | 0.91 | (0.69, 1.20) | 0.51 | 0.05 | |||||
| GA+AA | 198/221 | REF | 173/103 | 0.93 | (0.71, 1.24) | 107/91 | 1.30 | (0.92, 1.84) | 0.12 | |||||||||
| DiMeIQx | EPHX1 | rs868966c | AA | 122/167 | REF | 142/170 | 1.12 | (0.80, 1.55) | 85/78 | 1.45 | (0.98, 2.15) | 0.06 | 0.03 | |||||
| AG+GG | 368/383 | REF | 319/388 | 0.87 | (0.70, 1.07) | 169/200 | 0.91 | (0.70, 1.17) | 0.51 | |||||||||
| Cancer | B[a]P | UGT1A | rs17868299 | CC | 136/147 | REF | 135/125 | 1.18 | (0.84, 1.66) | 54/67 | 0.92 | (0.59, 1.42) | 0.52 | 0.05 | ||||
| CT | 6/19 | REF | 17/12 | 4.67 | (1.31, 16.7) | 12/8 | 6.04 | (1.39, 24.2) | 0.06 | |||||||||
| MeIQx | CYP2E1 | rs915908 | GG | 107/107 | REF | 122/117 | 1.05 | (0.73, 1.53) | 42/63 | 0.68 | (0.40, 1.08) | 0.08 | 0.05 | |||||
| GA+AA | 41/50 | REF | 33/37 | 1.07 | (0.56, 2.05) | 18/13 | 1.69 | (0.71, 4.00) | 0.23 | |||||||||
| DiMeIQx | UGT1A | rs6717546 | GG | 68/75 | REF | 60/66 | 1.04 | (0.64, 1.70) | 35/25 | 1.53 | (0.82, 2.86) | 0.16 | 0.04 | |||||
| GA+AA | 90/89 | REF | 72/85 | 0.83 | (0.53, 1.27) | 34/50 | 0.68 | (0.39, 1.16) | 0.18 | |||||||||
Meat mutagens classified as low (0–39th percentile), medium (40th–79th percentile) and high (≥80th percentile) intake.
aP-value is for a test for interaction using the likelihood ratio test comparing models with and without the cross-product of SNP for the given gene and the median of the categories of meat mutagen intake. P-values not corrected for multiple testing. Adjusted for age, sex and ethnicity and assuming a dominant model.
b SNP located within 10kb downstream of TMEM63A.
c SNP located within 10kb upstream of LEFTY3.
Supplementary Material
Acknowledgements
This study was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics. The authors thank Drs Christine Berg and Philip Prorok, Division of Cancer Prevention, National Cancer Institute, the Screening Center investigators and staff of the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, Mr Tom Riley and Mr Matt Moore and staff, Information Management Services, Inc., and Dr Richard Hayes, New York University Medical Center. We also like to thank the Dutch Cancer Society, GROW—School for Oncology and Developmental Biology, Maastricht University, the Netherlands, and the World Cancer Research Fund (A.M.J.G.).
Conflict of Interest Statement: None declared.
Abbreviations
- AHR
aromatic hydrocarbon receptor
- B[a]P
benzo[a]pyrene
- CI
confidence intervals
- FFQ
food frequency questionnaire
- GST
glutathione S-transferases
- HCA
heterocyclic amine
- MeIQx
2-amino-3,4,8-trimethylimidazo[4,5-f]quinoxaline
- NAT
N-acetyltransferases
- NOC
N-nitroso compound
- OR
odds ratio
- PAH
polycyclic aromatic hydrocarbon
- PhIP
2-amino-1-methyl-6-phenyl-imidazo[4,5-b]pyridine
- PLCO
Prostate, Lung, Colorectal, and Ovarian
- SNP
single nucleotide polymorphisms;
- UGT
UDP-glucuronosyltransferases
- XME
xenobiotic metabolizing enzymes
Footnotes
Supplementary material
Funding
Intramural Research Program of the National Cancer Institute, National Institutes of Health.
References
- 1. World Cancer Research Fund/American Institute for Cancer Research 2007) Food, Nutrition, Physical Activity, and the Prevention of Cancer: a Global Perspective AICR: Washington DC: [Google Scholar]
- 2. Cross A.J., et al. 2004) Meat-related mutagens/carcinogens in the etiology of colorectal cancer. Environ. Mol. Mutagen 44 44 55 [DOI] [PubMed] [Google Scholar]
- 3. Sinha R., et al. 2005) Development of a food frequency questionnaire module and databases for compounds in cooked and processed meats. Mol. Nutr. Food Res. 49 648 655 [DOI] [PubMed] [Google Scholar]
- 4.Hughes R., et al. Dose-dependent effect of dietary meat on endogenous colonic N-nitrosation. Carcinogenesis. 2001) ;22:199–202. doi: 10.1093/carcin/22.1.199. [DOI] [PubMed] [Google Scholar]
- 5. Ochiai M., et al. 2002) Induction of intestinal tumors and lymphomas in C57BL/6N mice by a food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine Jpn. J. Cancer Res. 93 478 483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sugimura T. 1997) Overview of carcinogenic heterocyclic amines. Mutat. Res. 376 211 219 [DOI] [PubMed] [Google Scholar]
- 7. Ito N., et al. 1991) A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Carcinogenesis 12 1503 1506 [DOI] [PubMed] [Google Scholar]
- 8. Ohgaki H., et al. 1991) Carcinogenicities of heterocyclic amines in cooked food. Mutat. Res. 259 399 410 [DOI] [PubMed] [Google Scholar]
- 9. Bogovski P., et al. 1981) Animal species in which N-nitroso compounds induce cancer. Int. J. Cancer 27 471 474 [DOI] [PubMed] [Google Scholar]
- 10. Xue W., et al. 2005) Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review Toxicol. Appl. Pharmacol. 206 73 93 [DOI] [PubMed] [Google Scholar]
- 11. Shimada T. 2006) Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons Drug Metab. Pharmacokinet. 21 257 276 [DOI] [PubMed] [Google Scholar]
- 12. Yeh C.C., et al. 2009) Polymorphisms of cytochrome P450 1A2 and N-acetyltransferase genes, meat consumption, and risk of colorectal cancer Dis. Colon Rectum 52 104 111 [DOI] [PubMed] [Google Scholar]
- 13. Le Marchand L., et al. 2002) Well-done red meat, metabolic phenotypes and colorectal cancer in Hawaii Mutat. Res. 506–507 205 214 [DOI] [PubMed] [Google Scholar]
- 14. Kampman E., et al. 1999) Meat consumption, genetic susceptibility, and colon cancer risk: a United States multicenter case-control study. Cancer Epidemiol. Biomarkers Prev. 8 15 24 [PubMed] [Google Scholar]
- 15. Lilla C., et al. 2006) Effect of NAT1 and NAT2 genetic polymorphisms on colorectal cancer risk associated with exposure to tobacco smoke and meat consumption Cancer Epidemiol. Biomarkers Prev. 15 99 107 [DOI] [PubMed] [Google Scholar]
- 16. Chan A.T., et al. 2005) Prospective study of N-acetyltransferase-2 genotypes, meat intake, smoking and risk of colorectal cancer Int. J. Cancer 115 648 652 [DOI] [PubMed] [Google Scholar]
- 17. Ward M.H. , et al. 2007) Processed meat intake, CYP2A6 activity and risk of colorectal adenoma Carcinogenesis 28 1210 1216 [DOI] [PubMed] [Google Scholar]
- 18. Tiemersma E.W., et al. 2004) Effect of SULT1A1 and NAT2 genetic polymorphism on the association between cigarette smoking and colorectal adenomas. Int. J. Cancer 108 97 103 [DOI] [PubMed] [Google Scholar]
- 19. Shin A., et al. 2008) Meat intake, heterocyclic amine exposure, and metabolizing enzyme polymorphisms in relation to colorectal polyp risk Cancer Epidemiol. Biomarkers Prev. 17 320 329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishibe N., et al. Genetic polymorphisms in heterocyclic amine metabolism and risk of colorectal adenomas. Pharmacogenetics. 2002;12:145–150. doi: 10.1097/00008571-200203000-00008. [DOI] [PubMed] [Google Scholar]
- 21. Roberts-Thomson I.C., et al. 1996) Diet, acetylator phenotype, and risk of colorectal neoplasia Lancet 347 1372 1374 [DOI] [PubMed] [Google Scholar]
- 22. Goode E.L. , et al. 2007) Inherited variation in carcinogen-metabolizing enzymes and risk of colorectal polyps Carcinogenesis 28 328 341 [DOI] [PubMed] [Google Scholar]
- 23. Tiemersma E.W., et al. 2002) Meat consumption, cigarette smoking, and genetic susceptibility in the etiology of colorectal cancer: results from a Dutch prospective study. Cancer Causes Control 13 383 393 [DOI] [PubMed] [Google Scholar]
- 24. Chen J., et al. 1998) A prospective study of N-acetyltransferase genotype, red meat intake, and risk of colorectal cancer Cancer Res. 58 3307 3311 [PubMed] [Google Scholar]
- 25. Morita M. , et al. 2009) Genetic polymorphisms of CYP2E1 and risk of colorectal cancer: the Fukuoka Colorectal Cancer Study. Cancer Epidemiol. Biomarkers Prev. 18 235 241 [DOI] [PubMed] [Google Scholar]
- 26. Ferrucci L.M., et al. (2010) Xenobiotic metabolizing genes, meat-related exposures, and risk of advanced colorectal adenoma World Rev. Nutr. Diet 101 34 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thier R., et al. 2003) Markers of genetic susceptibility in human environmental hygiene and toxicology: the role of selected CYP, NAT and GST genes. Int. J. Hyg. Environ. Health 206 149 171 [DOI] [PubMed] [Google Scholar]
- 28. Prorok P.C. , et al. 2000) Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin. Trials 21 273S 309S [DOI] [PubMed] [Google Scholar]
- 29.Gohagan J.K., et al. The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: history, organization, and status. Control Clin. Trials. 2000;21:251S–272S. doi: 10.1016/s0197-2456(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 30. Carlson C.S., et al. 2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium Am. J. Hum. Genet. 74 106 120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Turesky R.J. 2004) The role of genetic polymorphisms in metabolism of carcinogenic heterocyclic aromatic amines. Curr. Drug Metab. 5 169 180 [DOI] [PubMed] [Google Scholar]
- 32. Mirvish S.S. 1995) Role of N-nitroso compounds (NOC) and N-nitrosation in etiology of gastric, esophageal, nasopharyngeal and bladder cancer and contribution to cancer of known exposures to NOC. Cancer Lett. 93 17 48 [DOI] [PubMed] [Google Scholar]
- 33. Cross A.J., et al. 2011) A large prospective study of meat consumption and colorectal cancer risk: an investigation of potential mechanisms underlying this association. Cancer Res. 70 2406 2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sinha R., et al. 2005) Meat, meat cooking methods and preservation, and risk for colorectal adenoma. Cancer Res. 65 8034 8041 [DOI] [PubMed] [Google Scholar]
- 35.Purcell S., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Benjamini Y., et al. 1995) Controlling the false discovery rate:a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. Methodol. 57 289 300 [Google Scholar]
- 37. Murcray C.E., et al. 2009) Gene-environment interaction in genome-wide association studies. Am. J. Epidemiol. 169 219 226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao X., et al. 2008) A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms Genet. Epidemiol. 32 361 369 [DOI] [PubMed] [Google Scholar]
- 39. Hein D.W., et al. 2000) Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms Cancer Epidemiol. Biomarkers Prev. 9 29 42 [PubMed] [Google Scholar]
- 40. Cascorbi I., et al. (1999) Arylamine N-acetyltransferase activity in man Drug Metab. Rev. 31 489 502 [DOI] [PubMed] [Google Scholar]
- 41. Tiemersma E.W., et al. 2004) Risk of colorectal adenomas in relation to meat consumption, meat preparation, and genetic susceptibility in a Dutch population Cancer Causes Control 15 225 236 [DOI] [PubMed] [Google Scholar]
- 42. Butler L.M., et al. 2008) Modification by N-acetyltransferase 1 genotype on the association between dietary heterocyclic amines and colon cancer in a multiethnic study Mutat. Res. 638 162 174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cantwell M., et al. 2004) Relative validity of a food frequency questionnaire with a meat-cooking and heterocyclic amine module Cancer Epidemiol. Biomarkers Prev. 13 293 298 [DOI] [PubMed] [Google Scholar]
- 44. Felton J.S., et al. 1991) Occurrence, identification, and bacterial mutagenicity of heterocyclic amines in cooked food. Mutat. Res. 259 205 217 [DOI] [PubMed] [Google Scholar]
- 45. Malfatti M.A., et al. 2004) Human UDP-glucuronosyltransferase 1A1 is the primary enzyme responsible for the N-glucuronidation of N-hydroxy-PhIP in vitro Chem. Res. Toxicol. 17 1137 1144 [DOI] [PubMed] [Google Scholar]
- 46.Girard H., et al. UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology. 2005;42:448–457. doi: 10.1002/hep.20770. [DOI] [PubMed] [Google Scholar]
- 47.Girard H., et al. UGT1A1 and UGT1A9 functional variants, meat intake, and colon cancer, among Caucasians and African-Americans. Mutat. Res. 2008;644:56–63. doi: 10.1016/j.mrfmmm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Butler L.M., et al. 2005) Joint effects between UDP-glucuronosyltransferase 1A7 genotype and dietary carcinogen exposure on risk of colon cancer Cancer Epidemiol. Biomarkers Prev. 14 1626 1632 [DOI] [PubMed] [Google Scholar]
- 49. Polimanti R., et al. 2011) HapMap-based study of human soluble glutathione S-transferase enzymes: the role of natural selection in shaping the single nucleotide polymorphism diversity of xenobiotic-metabolizing genes. Pharmacogenet. Genomics 21 665 672 [DOI] [PubMed] [Google Scholar]
- 50. Felton J.S., et al. 1994) Effect of microwave pretreatment on heterocyclic aromatic amine mutagens/carcinogens in fried beef patties Food Chem. Toxicol. 32 897 903 [DOI] [PubMed] [Google Scholar]
- 51. Salmon C.P., et al. 1997) Effects of marinating on heterocyclic amine carcinogen formation in grilled chicken Food Chem. Toxicol. 35 433 441 [DOI] [PubMed] [Google Scholar]
- 52. Tran N.L., et al. 2002) Experimental and simulation studies of heat flow and heterocyclic amine mutagen/carcinogen formation in pan-fried meat patties Food Chem. Toxicol. 40 673 684 [DOI] [PubMed] [Google Scholar]
- 53. Griesenbeck J.S. , et al. 2009) Development of estimates of dietary nitrates, nitrites, and nitrosamines for use with the Short Willet Food Frequency Questionnaire. Nutr. J. 8 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.