Abstract
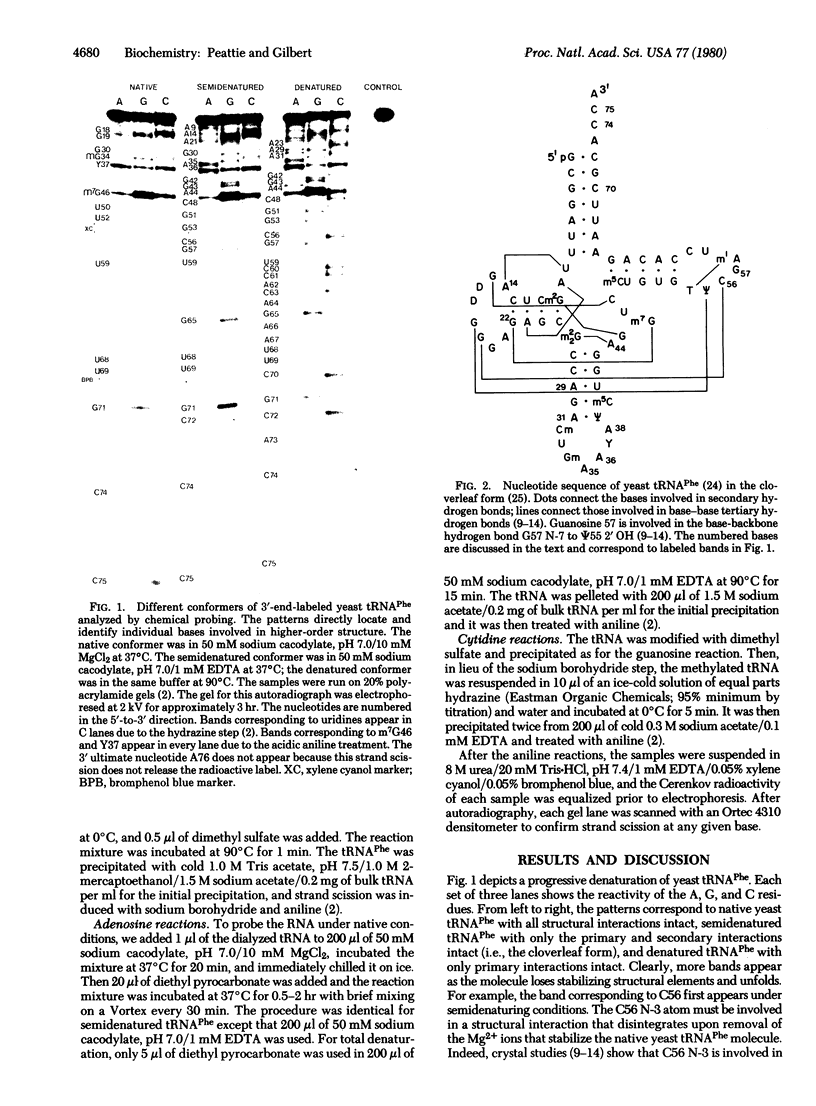
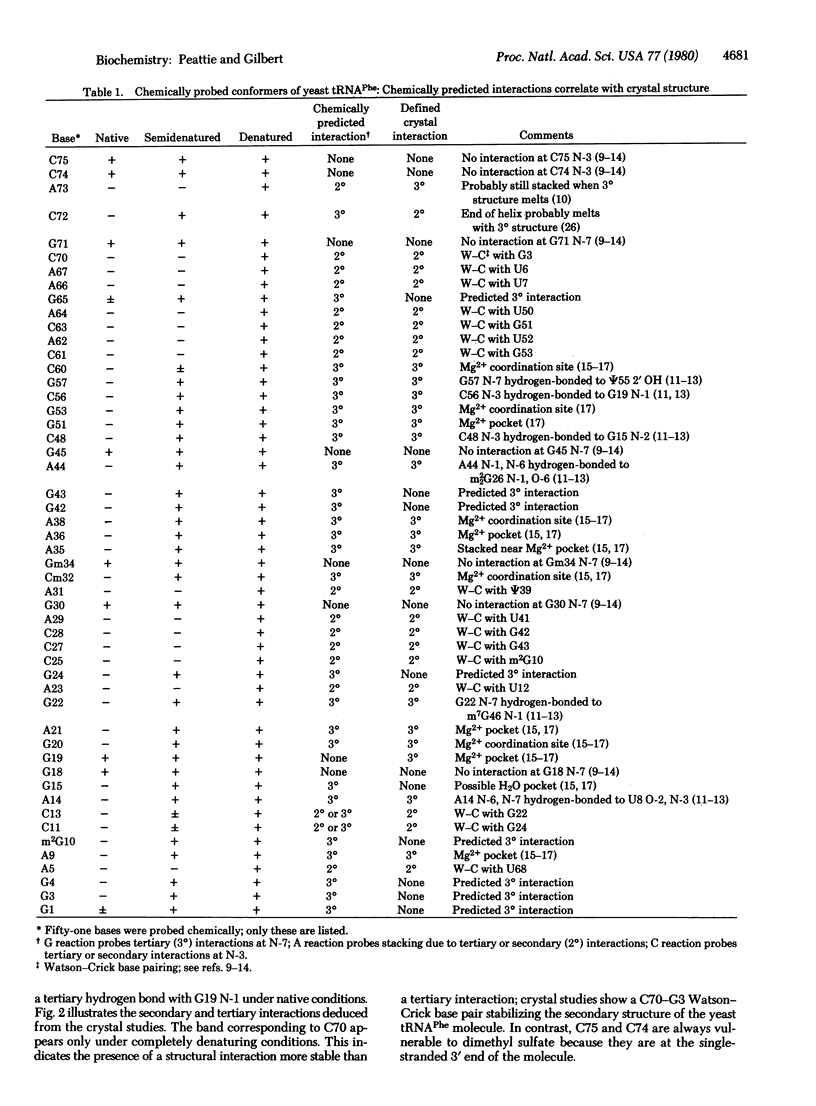
Three chemical reactions can probe the secondary and tertiary interactions of RNA molecules in solution. Dimethyl sulfate monitors the N-7 of guanosines and senses tertiary interactions there, diethyl pyrocarbonate detects stacking of adenosines, and an alternate dimethyl sulfate reaction examines the N-3 of cytidines and thus probes base pairing. The reactions work between 0 degrees C and 90 degrees C and at pH 4.5--8.5 in a variety of buffers. As an example we follow the progressive denaturation of yeast tRNAPhe terminally labeled with 32P as the tertiary and secondary structures sequentially melt out. A single autoradiograph of a terminally labeled molecule locates regions of higher-order structure and identifies the bases involved.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Fuller W., Hodgson A., Prutton I. Molecular conformations and structure transitions of RNA complementary helices and their possible biological significance. Nature. 1968 Nov 9;220(5167):561–564. doi: 10.1038/220561a0. [DOI] [PubMed] [Google Scholar]
- Brookes P., Lawley P. D. The reaction of mono- and di-functional alkylating agents with nucleic acids. Biochem J. 1961 Sep;80(3):496–503. doi: 10.1042/bj0800496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre D. S., Litvak S., Chapeville F. Purification and properties of Escherichia coli CTP (ATP)-tRNA nucleotidyltransferase. Biochim Biophys Acta. 1970 Dec 14;224(2):371–381. doi: 10.1016/0005-2787(70)90570-8. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. 1979 Sep 11;7(1):179–192. doi: 10.1093/nar/7.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresco J. R., Adams A., Ascione R., Henley D., Lindahl T. Tertiary structure in transfer ribonucleic acids. Cold Spring Harb Symp Quant Biol. 1966;31:527–537. doi: 10.1101/sqb.1966.031.01.068. [DOI] [PubMed] [Google Scholar]
- HOLLEY R. W., APGAR J., EVERETT G. A., MADISON J. T., MARQUISEE M., MERRILL S. H., PENSWICK J. R., ZAMIR A. STRUCTURE OF A RIBONUCLEIC ACID. Science. 1965 Mar 19;147(3664):1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- Hassur S. M., Whitlock H. W., Jr UV shadowing--a new and convenient method for the location of ultraviolet-absorbing species in polyacrylamide gels. Anal Biochem. 1974 May;59(1):162–164. doi: 10.1016/0003-2697(74)90020-7. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Church G. M., Kim S. H. RNA-ligant interactions. (I) Magnesium binding sites in yeast tRNAPhe. Nucleic Acids Res. 1977 Aug;4(8):2811–2820. doi: 10.1093/nar/4.8.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978 Aug 25;123(4):631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Klug A. Crystallographic refinement of yeast phenylalanine transfer RNA at 2-5A resolution. J Mol Biol. 1976 Dec 25;108(4):619–649. doi: 10.1016/s0022-2836(76)80109-x. [DOI] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Rhodes D., Brown R. S., Klug A. A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J Mol Biol. 1977 Apr 15;111(3):315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- LAWLEY P. D., BROOKES P. FURTHER STUDIES ON THE ALKYLATION OF NUCLEIC ACIDS AND THEIR CONSTITUENT NUCLEOTIDES. Biochem J. 1963 Oct;89:127–138. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard N. J., McDonald J. J., Henderson R. E., Reichmann M. E. Reaction of diethyl pyrocarbonate with nucleic acid components. Adenosine. Biochemistry. 1971 Aug 31;10(18):3335–3342. doi: 10.1021/bi00794a003. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. DNA-binding site of lac repressor probed by dimethylsulfate methylation of lac operator. J Mol Biol. 1979 Aug 25;132(4):709–728. doi: 10.1016/0022-2836(79)90384-x. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- RajBhandary U. L., Chang S. H. Studies on polynucleotides. LXXXII. Yeast phenylalanine transfer ribonucleic acid: partial digestion with ribonuclease T-1 and derivation of the total primary structure. J Biol Chem. 1968 Feb 10;243(3):598–608. [PubMed] [Google Scholar]
- Rhodes D. Initial stages of the thermal unfolding of yeast phenylalanine transfer RNA as studied by chemical modification: the effect of magnesium. Eur J Biochem. 1977 Nov 15;81(1):91–101. doi: 10.1111/j.1432-1033.1977.tb11930.x. [DOI] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Riesner D., Maass G., Thiebe R., Philippsen P., Zachau H. G. The conformational transitions in yeast tRNAPhe as studied with tRNAPhe fragments. Eur J Biochem. 1973 Jul 2;36(1):76–88. doi: 10.1111/j.1432-1033.1973.tb02887.x. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977 Dec;4(12):4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova O. B., Metelev V. G., Chichkova N. V., Smirnov V. D., Rodionova N. P., Atabekov J. G., Bogdanov A. A., Shabarova Z. A. Addressed fragmentation of RNA Molecules. FEBS Lett. 1979 Jul 1;103(1):197–199. doi: 10.1016/0014-5793(79)81280-6. [DOI] [PubMed] [Google Scholar]




