Abstract
The posttranscriptional modification of ribosomal RNA (rRNA) modulates ribosomal function and confers resistance to antibiotics targeted to the ribosome. The radical SAM (S-adenosyl-L-methionine) methyl synthases, RlmN and Cfr, both methylate A2503 within the peptidyl transferase center (PTC) of prokaryotic ribosomes, yielding 2-methyl- and 8-methyl-adenosine, respectively. The C2 and C8 positions of adenosine are unusual methylation substrates due to their electrophilicity. To accomplish this reaction, RlmN and Cfr proceed by a shared radical-mediated mechanism. However, in addition to the radical SAM CX3CX2C motif, both RlmN and Cfr contain two conserved cysteine residues required for in vivo function. These conserved cysteine residues are putatively involved in a covalent intermediate employed by RlmN and Cfr in order to achieve this challenging transformation. Currently, there is no direct evidence for this proposed covalent intermediate. We have further investigated the roles of these conserved cysteines in the mechanism of RlmN. Cysteine 118 mutants of RlmN are unable to resolve the covalent intermediate, either in vivo or in vitro, enabling us to isolate and characterize this intermediate. Additionally, tandem mass spectrometric analyses of mutant RlmN reveal a methylene-linked adenosine modification at cysteine 355. Employing deuterium-labeled SAM and RNA substrates in vitro has allowed us to further elucidate the mechanism of formation of this intermediate. Together, these experiments provide compelling evidence for the formation of a covalent intermediate species formed between RlmN and its rRNA substrate and the roles of the conserved cysteine residues in catalysis.
Introduction
The ribosome, containing both ribosomal proteins and rRNA, catalyzes the biosynthesis of proteins critical to cellular function. Within the ribosome, it is the rRNA components that play the predominant structural and functional roles. Owing to its indispensible activity, the bacterial ribosome is a frequent target of antibiotic agents.1 Unsurprisingly, compounds that interact with the ribosome and interfere with protein synthesis make close contacts with the rRNA located in functionally important regions of the ribosome.2–5 Consequently, modifications to the rRNA in these regions can abrogate binding of antibiotics, leading to restoration of ribosomal activity and antibiotic resistance. One of the most common rRNA modifications leading to antibiotic resistance is the SAM-dependent installation of a methyl group on a specific nucleobase.6 The recently-discovered, clinically significant resistance enzyme, Cfr, catalyzes the formation of an 8-methyladenosine residue at position 2503 (E. coli numbering) of 23S rRNA,7 which resides within the PTC of the bacterial ribosome. This modification confers resistance to five classes of PTC-targeted antibiotics,8 including the synthetic oxazolidinone linezolid, an agent commonly reserved as a “last line of defense” against resistant pathogens. RlmN, a homolog of Cfr, modifies the same adenosine base as Cfr (A2503),9 resulting in the formation of 2-methyladenosine.10 RlmN has recently been demonstrated to methylate adenosine residues at position 37 of several E. coli tRNAs, indicating a broader substrate specificity and a larger role in fine-tuning interactions critical to translation.11 While the biological role of Cfr’s activity may be more striking,8 the positioning of the constitutive RlmN modification at the intersection of the PTC and the exit tunnel, as well as the impact of the modified base on ribosomal interactions with the nascent polypeptide, makes this modification no less critical to understand.12,13
These two homologous enzymes are members of the radical SAM superfamily and are presumed to employ a shared, radical-based mechanism to modify the target base in rRNA.10,14,15 Both Cfr and RlmN contain the conserved CX3CX2C cysteine-rich motif, which ligates the [4Fe-4S] iron-sulfur cluster employed by all members of the radical SAM superfamily.16 Members of this class of enzymes have been implicated in a broad range of pathways and reactions,17,18 catalyzing amine shifts,19–23 nucleotide reduction,24–26 sulfur insertions,27–32 DNA repair,33,34 methylthiolation,35–40 oxidative decarboxylation,41–43 peptide modifications, 44–47 and complex rearrangements48 among many others. Within this superfamily, RlmN and Cfr are the only known examples of methyl synthases, and both assemble a methyl group on their products from a hydrogen atom initially present on the substrate and a methylene fragment ultimately derived from SAM.14,15
These methyl synthases share significant homology, including two conserved cysteine residues that are not associated with the iron-sulfur cluster: cysteines 105 and 338 in Cfr and cysteines 118 and 355 in RlmN. These residues are proposed to play important roles in catalysis. Prior in vivo work indicates that both of these cysteines are crucial to the ability of Cfr to elicit antibiotic resistance.49 Further, primer extension assays demonstrated that rRNA isolated from C105A mutant strains exhibits a strong stop at A2503, consistent with a modification other than a methyl group—which yields a weaker stop.49
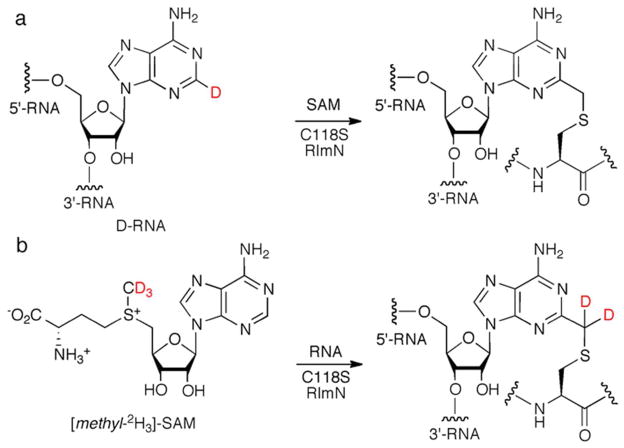
In conjunction with the foregoing in vivo data, elegant single turnover experiments14 and an X-ray crystallographic structure of RlmN50 point to a pathway initiated by the pre-methylation of cysteine 355 (RlmN numbering), by a first molecule of SAM bound at the [4Fe-4S] cluster, followed by the binding of a second molecule of SAM. With the injection of an electron, the 5′-deoxyadenosyl (5′-dA) radical formed from the second equivalent of SAM activates the thiomethyl group at cysteine 355 by abstracting a hydrogen atom. The resulting thiomethylene radical adds into the adenine ring of A2503. Loss of an electron from this initial adduct (species IV in Scheme 1), possibly to the now oxidized [4Fe-4S] cluster,51 leads to the formation of a covalent RlmN-RNA thioether species. This adduct is subsequently resolved by the action of cysteine 118 in conjunction with acid/base catalysis (Scheme 1).14
Scheme 1.
Proposed mechanism of RlmN-mediated methylation of RNA.
While this proposed mechanism is consistent with the observed formation of the thiomethyl species on cysteine 355, no conclusive evidence of the putative intermediate species has been provided. To address this uncertainty, we sought to characterize the mechanistically critical RNA-protein adduct (VI in Scheme 1). Herein, we describe a combination of in vivo and in vitro approaches to characterize the adduct formed between RlmN and RNA as well as its mechanism of formation.
Experimental Procedures
General
Anaerobic manipulations were carried out in a glovebox (MBraun) under a 99.997% nitrogen atmosphere containing less than 2 ppm oxygen. All reagents were analytical grade or the highest grade commercially available and used as supplied unless noted otherwise. Iron quantitation,10,52 preparation of truncated rRNA substrates,10 RNA digestion,10 HPLC separation and LC-MS analysis of nucleotides15 were performed as previously noted. The RlmN C118S and C118G mutants were generated from the C118A mutant plasmid under conditions analogous to those previously employed, using the oligonucleotide primers noted in the supporting information (Table S1) and the DNA sequence confirmed (Elim Biopharmaceuticals, Inc.).
Expression, Purification, and Reconstitution of Wild-type and Mutant RlmN
Wild-type RlmN and C355A RlmN were expressed, purified and reconstituted as previously.10,14 C118 mutants were prepared by modified versions of those previous protocols. Holo-C118A/G/S RlmN was overexpressed and purified by His6 affinity chromatography with Talon resin. Anion exchange chromatography, employing either HiTrap Q10 or MonoQ columns (GE) was subsequently used to separate protein covalently modified by RNA in vivo from RNA-unmodified protein. These proteins were reconstituted and further purified by size exclusion chromatography prior to use. Apo-C118S RlmN was overexpressed and purified as described previously for wild-type apoprotein.53 This protein did not form an RNA-RlmN adduct in vivo (see below), so the anion exchange step noted above was omitted, and the remainder of the protocol was performed as described. RNA-RlmN adduct samples to be analyzed by RNA-digest, gel-shift or MS methods were subjected to RNase treatment with a mixture of RNases A and T1 (Thermo) according to the manufacturer’s protocol. The in-gel RNase digestion protocol is similar and is described in detail in the supporting information.
2-Methyladenosine Production by Wild-type, C118 and C355 Mutant RlmN
In order to assess the ability of mutant proteins to install methyl groups on RNA substrates, the amount of 2-methyladenosine from hydrolysates of RNA reacted with mutant enzymes was compared to that from wild-type. As previous work indicated that flavodoxin (FldA)54 and its cognate reductase (Fpr)55 were a suitable reducing system for RlmN and Cfr activity assays,14 these proteins were employed in our assays. 250 μL reactions were performed under the following conditions: 200 mM HEPES pH 7.5, 500 mM KCl, 10 mM MgCl2, 2 mM DTT, 200 μM SAM, 20 μM RNA (rRNA truncation 2496–2582) and 25 μM RlmN WT or mutant enzyme. The reducing system was composed of 20 μM FldA, 2 μM Fpr and 2 mM NADPH, and reactions were allowed to proceed for one hour at 37 °C. The RNA was recovered from the reactions, digested and separated as previously. Fractions corresponding to the elution time of authentic 2-methyladenosine were lyophilized and further analyzed by LC-MS to detect the presence of a species at m/z=282 corresponding to intact 2-methyladenosine as previously. To determine the threshold of detection of 2-methyladenosine from wild-type enzyme, the 2-methyladenosine from WT reactions corresponding to between 25 and 500 nmol of recovered RNA were analyzed. Subsequently, samples that would contain any 2-methyladenosine produced in reactions of C355A, C118A or C118S and corresponding to between 600 and 2000 nmol of RNA were analyzed in an identical fashion. If no 2-methyladenosine can be detected in a mutant sample representing ten times the RNA of the threshold amount of WT RNA, the mutant was presumed to have less than 10% of WT activity, allowing for threshold activities of mutants to be established.
Generation of Endogenous RlmN-FLAG
A C-terminal FLAG sequence was fused to the Genomic version of rlmN with a FLAG sequence were generated on the chromosome of E. coli using the techniques and constructs first described by Datsenko and Wanner.56 The C-terminal FLAG sequence was incorporated into the primer pair (Table S1) used to PCR amplify kan from pKD4.56 This PCR product was transformed into E. coli strain BW25113 carrying the λ-red system on pKD4656 and successful incorporation onto the chromosome was selected through kanamycin resistance. The kanamycin resistance cassette was removed using the FLP expression plasmid pCP20 and the tagged locus was verified by sequencing. These strains were used in subsequent immunoprecipitation experiments.
Immunoprecipitation of Endogenous RlmN-FLAG
An overnight culture of E. coli BW25113 in which a C-terminal FLAG sequence was fused to the genomic copy of rlmN was diluted 1:100 into fresh LB medium, grown at 37 °C with agitation at 200 rpm, and aliquots were removed at 30-minute intervals. Cell pellets were frozen at −80 °C and all processed after collection of the final timepoint. Cells were lysed using BugBuster protein extraction reagent according to the manufacturer’s instructions, and protein concentrations were determined by the method of Bradford.57 Ten μg of total protein from each time-point was run on a polyacrylamide gel and the protein was subsequently electroblotted to a PVDF membrane. Proteins bearing a FLAG-tag were detected with an anti-FLAG HRP conjugated antibody and visualized by chemiluminescence.
Four liters of LB were inoculated 1:100 from an overnight culture of E. coli expressing the FLAG-tagged endogenous RlmN. The cells were harvested after 90 minutes. The resulting cell pellet was re-suspended in 5 mL TBST buffer containing 1 mM EDTA and 100 μM PMSF and lysed by sonication. The resulting lysate was treated with anti-FLAG M2 affinity gel as per the manufacturer’s instructions to isolate FLAG fusion proteins. The resulting FLAG fusions were separated by SDS-PAGE and the band exhibiting the expected MW of RlmN-FLAG (~45 kDa) was excised from the gel. The protein was subjected to in-gel trypsin digestion and the tryptic peptides were characterized by LC-MS/MS as described in the supporting information.
Generation and PAGE Analysis of In Vitro Generated RNA-RlmN C118 Mutant Adduct
Reactions to form an RNA-protein adduct from T7-transcribed RNA and purified C118 mutant enzyme were performed with two types of RlmN samples. The first were Fe-S cluster-containing C118 mutant proteins purified away from the in vivo RNA-RlmN adduct, exhibiting chromatographic behavior similar to wild-type and either methylated or unmodified at C355. The second were apo (Fe-S cluster free) versions of C118 mutant RlmN expressed in minimal media under iron-deficient conditions in the presence of a strong chelating agent. Apo-RlmN had previously been shown to lack methylation at C355,53 and we further examined whether C118 RlmN mutants expressed under these conditions formed adducts in vivo or could form adducts in vitro. As such, they were purified and reconstituted as previously indicated. In order to assess the ability of these unmodified mutant proteins to form RNA-RlmN adducts in vitro, small scale (50–100 μL) reactions were performed under the following conditions: 200 mM HEPES pH 7.5, 500 mM KCl, 10 mM MgCl2, 1mM DTT, 100 μM SAM (200 μM with apo, reconstituted samples to compensate for the lack of any C355 methylation), 25 μM RlmN mutant enzyme and 20 μM RNA encompassing residues 2496–2582 of 23S rRNA. The reducing system was composed of 20 μM flavodoxin, 2 μM ferrodoxin:NADPH oxidoreductase and 2 mM NADPH. All assays were initiated by the addition of RlmN. Aliquots of the reaction mixtures were separated by PAGE in gels containing 5% polyacrylamide, 7 M urea and 0.1% SDS; RNA species were visualized by ethidium bromide staining and imaged using a Typhoon imaging system.
Mass spectrometry
Tryptic digests were analyzed on an LTQ-Orbitrap XL or VELOS hybrid tandem mass spectrometer (Thermo Scientific) coupled with a nanoACQUITY HPLC system (Waters) in LC/MS/MS mode. Details of the HPLC separation are provided in the supporting information. Mass measurements (MS survey) were performed in the Orbitrap with high resolution and mass accuracy; collision-induced dissociation experiments (CID) were performed in the linear ion trap on computer-selected, multiply charged precursor ions properly adjusting the CID conditions to the m/z and z values of the precursor ions.
Data processing
Peaklists were generated from the raw datafiles using an in-house peak-picking software, PAVA. With the peaklist, a strict database search was performed using the search engine, Protein Prospector (prospector.ucsf.edu) against the SwissProt database, with no species specificity, permitting only tryptic cleavages (1 missed cleavage). Cysteine carbamidomethylation was a fixed modification; methionine oxidation, N-terminal glutamine cyclization and protein acetylation as variable modifications were considered. Mass accuracy was 20 ppm for the precursor ions, and 0.8 Da for the fragments. Once it was established that RlmN was indeed present and at a reasonable purity, a subsequent search was performed targeting unspecified modifications occurring on the protein in general or specifically on cysteine residues.58 Finally, the CID data supporting the assignments were carefully inspected, and the mass accuracy of the precursor ions was manually calculated. This latter step was necessary because the software only indicates potential mass modification at certain residues, without providing an elemental composition.
Results
Mutant Activity Compared to WT
Prior in vivo experiments with Cfr and its cysteine mutants have implicated cysteine 105 and cysteine 338 as necessary to catalyze the 8-methyl modification of A2503.49 Therefore we anticipated that corresponding mutations to RlmN cysteines 118 and 355, would result in enzymes with virtually no ability to form 2-methyladenosine on RNA substrates. Using known quantities of RNA recovered from in vitro WT reactions as a frame of reference, we were able to establish a detection limit for the 2-methyladenosine product to which the products of mutant reactions could be compared. Whereas the 2-methyladenosine from 25 nmol of RNA from a WT reaction was still detectable (Figure S1), no signal was evident from between 600 and 2000 nmol of RNA from mutant reactions under identical conditions. From these results, it was determined that C355A, C118A and C118S exhibit ≤ 1.3, 3.1, and 4.2 % of WT activity, respectively. These low activities agree well with the putatively indispensible roles of the conserved cysteine residues implicated in prior in vivo experiments, as well as the proposed chemical mechanism (Scheme 1).
Methylation of Endogenous RlmN
Previously, over-expressed RlmN has been shown to be methylated at cysteine 355 by both X-ray crystallographic analysis50 and protease digestion followed by LC-MS/MS.14 To evaluate the methylation state of the protein during active rRNA modification and ribosome assembly, the genomic copy of rlmN was altered to encode a C-terminal FLAG sequence to ensure that all experiments were performed under conditions where RlmN was expressed at its normal cellular level. Time-dependent expression of RlmN-FLAG was evaluated by Western blotting for the RlmN-FLAG in lysates at various timepoints (Figure S2). RlmN-FLAG is maximally expressed approximately 90 minutes after dilution of a saturated culture 1:100 into fresh LB medium, corresponding to an OD600 of approximately 0.25. This timing is consistent with the growth rate-dependent control observed for ribosome biosynthesis,59 suggesting that RlmN produced under these conditions is the catalytically relevant resting state of the enzyme. RlmN-FLAG was immunoprecipitated from the 90-minute timepoint and the isolated protein was characterized by tryptic digestion, LC-MS/MS similar to prior experiments.14 This analysis revealed the peptide encompassing residues 348 to 365 yields an m/z (2+) = 909.4149 species consistent with methylated peptide (calculated value = 909.4203: mass difference of 6 ppm). Fragmentation of this precursor ion yielded b/y series ions that confirmed the site of the methyl modification as cysteine 355,60 indicating modification of the enzyme under physiological conditions (Figure S3).
Characterization of RNA-RlmN C118 Mutant Adduct Formation In Vivo
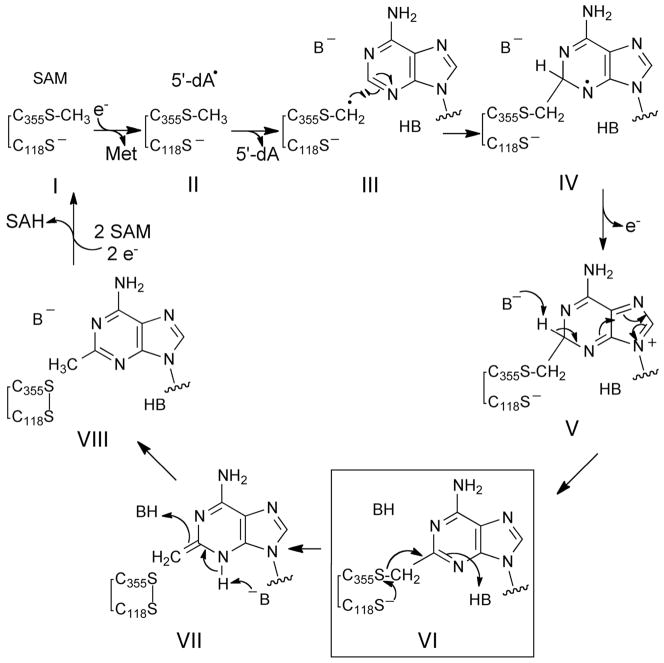
Prior in vivo work with C105A Cfr demonstrated a strong stop at A2503 of the isolated rRNA in primer extension assays, indicating a significant perturbation of the rRNA at this position, although the precise nature of this change was not assessed.49 These observations are consistent with the attachment of a large substituent to A2503 and possibly the formation of a covalent adduct between Cfr and rRNA at A2503 in vivo. C118A RlmN was also shown to exhibit a shift in its λmax from 278 to 265 nm in addition to the release of modified RNA nucleotides upon RNase and phosphatase treatment; however, no direct evidence of covalent attachment or the putative points of attachment between C118A RlmN and RNA were provided.14 To further investigate the possibility that a covalent adduct between RNA and RlmN cysteine 118 mutant enzymes forms in vivo, the C118A, C118G and C118S mutants of RlmN were over-expressed and purified by Talon chromatography. The various mutants were examined to determine if the nature of the residue at position 118 might influence the relative amounts of RNA-adducted and unmodified protein. When the isolated His-tagged proteins were analyzed by SDS-PAGE, the samples exhibited two distinct bands, one at the native molecular weight of RlmN, 43 kDa, and the second at approximately 65 kDa (Figure 1a&b). From this analysis, it appeared that the C118S RlmN formed the greatest proportion of adduct in vivo, so it was employed in subsequent in vitro experiments (see below).
Figure 1.
SDS-PAGE analysis of RlmN C118 mutant proteins. Protein markers are in lane 1 of all gels. a) C118A RlmN purified by immobilized metal ion affinity chromatography (lane 2). b) C118G RlmN (lane 2) and C118S RlmN (lane 3) purified by immobilized metal ion affinity chromatography. c) Untreated C118A RlmN-RNA adduct purified by anion exchange (lane 2). Sample from lane 2, after treatment with RNases A&T1 (lane 3). d) C118S RlmN grown under metal deficient conditions and purified by immobilized metal ion affinity chromatography (lane 2). In this sample, no RNA adduct could be detected.
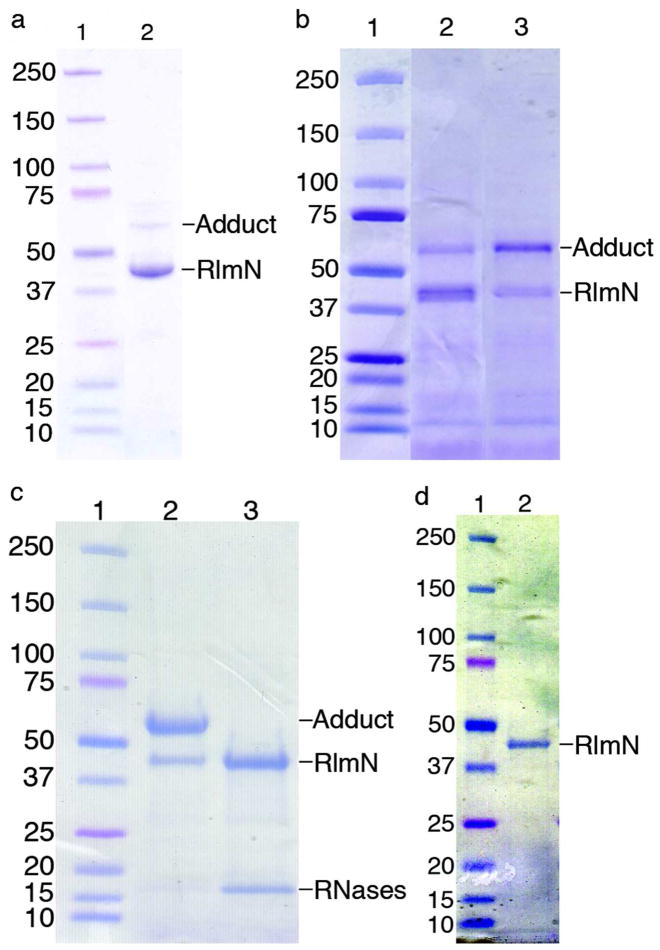
In the case of C118A RlmN, when this heterogeneous material was purified by anion exchange chromatography, the two species were resolved, with the lower MW species eluting at a much lower salt concentration than the higher MW species (Figure S4). This observation is indicative of the higher MW species being much more anionic in character at pH 7.5–8. This higher MW species also exhibits a much greater absorbance at 260 nm than the lower MW species. Together, these results are consistent with the addition of the polyanionic phosphate backbone of an RNA molecule to the protein. Further, when the putative RNA-RlmN adduct purified by anion exchange was treated with a mixture of RNases A and T1, the resulting protein exhibited native-like MW (Figure 1c). This native-like, RNase-treated material was analyzed by tryptic digestion, LC-MS/MS, revealing the peptide containing cysteine 355 is modified by a 147 Da species (Figure 2). In this instance, the fragment ions were measured in the Orbitrap with high resolution and mass accuracy, and the main sequence ions unambiguously indicate that the modification resides on C355. The modified peptide was chromatographically resolved from the unmodified or S-methylated peptide, and the precursor ion bearing the + 147 Da modification was subjected to further fragmentation. Analysis of the main sequence ions generated from this modified precursor localized the modification to cysteine 355 (Figure 2). The observed mass of 147 Da is consistent with a methylene-linked adenine adduct analogous to the covalent intermediate species in the proposed mechanism (VI in Scheme 1). Though these samples were not specifically treated to cleave the glycosidic bonds between the RNA nucleobases and ribose, it appears that the linkage is not stable under the experimental conditions. Neutral loss of ribose from ribonucleosides in MS experiments61 could easily account for our observation of an adenine adduct instead of adenosine. Interestingly, when C118S RlmN was prepared as the apoprotein (lacking the Fe-S cluster), the material exhibited only one band at the native MW of 43 kDa (Figure 1d). This observation is consistent with the previously noted requirement of an intact Fe-S cluster for S-methylation,53 as well as the requirement of the cluster for subsequent radical SAM chemistry and adduct formation.
Figure 2.
Ion trap CID data of 348GDDIDAAC(CH2adenine)GQLAGDVIDR375 formed from RlmN C118A protein in vivo and subsequently RNase treated. The precursor mass was m/z 650.9631(3+), within ~1 ppm of the calculated mass of 650.9623 (3+ ion of RlmN 348–375 peptide +147). The table at right lists all main sequence ions detected within 20 ppm of the calculated value. In comparison to the methyl derivative (Figure S3), the higher charge state of the precursor ion yielding the best CID data as well as the presence of doubly charged N- and C-terminal fragments clearly indicate the incorporation of a basic modification. The site of modification within the peptide and the deduced structure of the modification are highlighted in yellow.
RNA-RlmN C118 Mutant Adduct Formation In Vitro
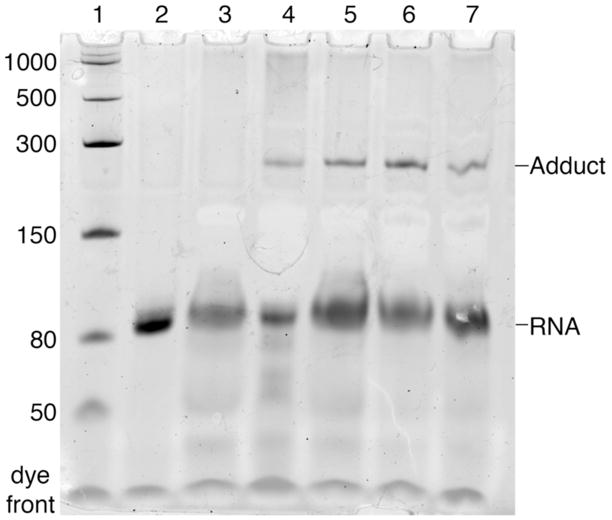
To investigate whether a covalent adduct between RlmN cysteine 118 mutants and RNA also forms in vitro, the recombinant mutant proteins C118A and C118S were incubated with an RNA fragment encompassing residues 2496–2582 of 23S rRNA and the flavodoxin reducing system. In light of the previously observed protein gel-shift of the in vivo RNA-modified material (Figure 1a,b&c), a gel-shift assay monitoring RNA was employed to monitor covalent modification of the RNA fragments in vitro. When RNA from these reactions is separated by PAGE and visualized with ethidium bromide, complete reaction mixtures containing C118A or C118S demonstrate two distinct bands (Figure 3, lanes 4 and 5, respectively). The first species is unreacted RNA (87 nt, ~28 kDa); the second species appears at a molecular weight consistent with an RNA-RlmN adduct, expected MW ~71 kDa (mass equivalent to 215 nt). The formation of the adduct species is dependent on the presence of a complete reducing system, as when NADPH is omitted, no gel-shifted species appears (Figure 3, lane 3). This finding is consistent with the role of NADPH as the ultimate electron source in the FldA/Fpr (flavodoxin and reductase) system that leads to the reductive cleavage of SAM, resulting in the formation of the critical 5′-dA radical that initiates the subsequent radical-based reactions. When the adduct species was excised from the gel and subjected to in-gel RNase A&T1 and trypsin digestion, subsequent LC-MS/MS experiments analogous to those above for the in vivo adduct revealed an identical 147 Da modification on cysteine 355 of C118S mutant protein exposed to rRNA substrate (Figure S5), again consistent with the proposed methylene-linked adenosine intermediate (VI in Scheme 1). These in vitro experiments with defined RNA substrate demonstrate that a covalent adduct is formed between RlmN cysteine 118 mutant enzyme and rRNA substrate.
Figure 3.
In vitro gel-shift assay of RlmN C118 mutants. RlmN mutant proteins were incubated with RNA fragments as indicated, and RNA was separated by denaturing PAGE and visualized with ethidium bromide. Lane 1 contains RNA markers of the indicated size, in nt. Lane 2 contains only the 87 nt RNA fragment used in assays. Lane 3 is a negative control lacking NADPH. Lane 4 is the reaction with C118A RlmN, SAM and RNA. Lane 5 is the reaction with C118S RlmN, SAM and RNA. Lane 6 is the reaction with C118S RlmN, SAM and D-RNA. Lane 7 is the reaction with C118S RlmN, [methyl-2H3]-SAM and RNA.
To further interrogate the mechanism of formation of this intermediate arrested in mutant enzymes, the incorporation of deuterium into the protein-RNA adduct was examined. RlmN C118S mutant protein was expressed in minimal media in the presence of a strong chelator, resulting in protein lacking both the iron-sulfur cluster and methylation at cysteine 355.53 The Fe-S cluster of this apoprotein was then chemically reconstituted, resulting in [4Fe-4S]-containing RlmN C118S enzyme with a free thiol at cysteine 355. This protein was then incubated with the same RNA fragment as above and [methyl-2H3]-SAM. In a separate experiment, C118S RlmN (either free thiol or S-methyl at C355) was incubated with unlabeled SAM and D-RNA (i.e. substrate in which all adenosine residues have been replaced with [2-2H]-adenosine; similar deuterium substituted rRNA has previously been demonstrated as a substrate for RlmN15). The resulting RNA-RlmN C118S adducts (Figure 3, lanes 6 and 7) were digested with RNases and trypsin as above. The presence and location of deuterium in the modified cysteine 355 peptides was assessed by LC-MS/MS and compared to the spectra of the same species formed from completely unlabeled reactants. In agreement with the proposed formation of the covalent adduct via addition of the thiomethylene radical into the substrate adenine (Scheme 1), the species generated from [methyl-2H3]-SAM and unlabeled RNA incorporates two deuterium atoms into the group modifying cysteine 355, consistent with a CD2-methylene linkage between the cysteine 355 sulfur and the adenine ring (Figure 4). Reactions with unlabeled SAM and D-RNA yielded MS and MS/MS data (Figure S6) indistinguishable from those generated by reactions with completely unlabeled precursors (Figure S5), consistent with the loss of the amidine proton from the RNA-protein adduct to an adjacent, solvent shielded, general base. This result agrees with the previously observed retention of deuterium label in product 2-methyladenosine,15 as the shielded proton is returned to product only subsequent to resolution of the adduct. Such retention in the product of a proton abstracted from substrate has previously been observed.62,63
Figure 4.
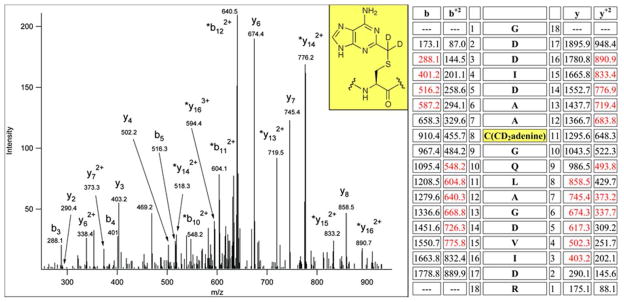
Ion trap CID data of 348GDDIDAAC(CD2adenine)GQLAGDVIDR375 formed from RlmN C118S protein in vitro in the presence of [methyl-2H3]-SAM. The precursor mass was m/z 651.6353(3+), within 3 ppm of the calculated mass of 651.6332 (3+ ion of RlmN 348–375 peptide +149). Fragments that showed the 2 Da shift due to deuterium incorporation are labeled with asterisks. Main sequence ions detected are shown in red in the table. The site of modification within the peptide and the deduced structure of the modification are highlighted in yellow.
Discussion
The radical SAM methyl synthases, RlmN and Cfr, utilize a novel, methyl group assembly mechanism to achieve methylation of the 2- and 8- amidine carbons of A2503 within 23S rRNA, thereby exerting significant influence on the biological function of the modified ribosomes.8,13 Multiple mechanistic scenarios have been suggested to account for the experimental findings.14,15 In this work, we provide strong evidence for the mechanism invoking methylation of a cysteine residue in the protein and subsequent formation of a covalent RNA-protein adduct as originally proposed by Grove et al.14 Importantly, our mutagenesis-enabled trapping of the covalent intermediate provides the first direct evidence of any of the proposed reactive intermediates occurring subsequent to the initial electron insertion.
Crucial to this mechanism are cysteine residues 118 and 355 in E. coli RlmN, which are conserved among RlmN and Cfr homologs across species.49 We have investigated the roles of both cysteines in this study. For cysteine 355, we examined methylation status under endogenous conditions. We showed that protein produced from the genomic copy of rlmN under the native promoter at the time of maximal protein expression and ribosome biosynthesis exhibits methylation at C355. The fact that this modification is observed during optimal growth strongly implies that this modification is involved in the in vivo rRNA methylation catalyzed by RlmN, confirming its catalytic relevance. Interestingly, methylation of a reactive cysteine in the active site of the DNA methyltransferase Dnmt3a has recently been observed and attributed to either spurious methylation of a nucleophilic cysteine residue or a regulatory mechanism inactivating excess enzyme in the absence of substrate.64 While either of these possibilities are conceivable for RlmN, the above argument suggests that, for RlmN, this methylation is directly involved in catalysis.
For cysteine 118, the resolving cysteine, we examined C118A and C118S mutant RlmN to obtain insight into the covalent RlmN-RNA species formed both in vivo and in vitro. Previous studies showed that purified C118A RlmN exhibited an absorbance maximum at 265 nm vs. 278 nm for WT,14 and that ribonuclease and phosphatase treatment of these samples released nucleotides including pseudouridine.14 Our observation of the significant increase in anionic character of this species, as revealed by ion exchange chromatography, as well as the gel shift upon ribonuclease treatment together strongly argue that this species is a covalent adduct between RNA and RlmN. Significantly, LC-MS/MS experiments localize the site of RNA modification to cysteine 355 on RlmN. This cysteine exhibits a +147 modification consistent with a methylene-linked adenosine covalent intermediate in the RlmN mechanism.
The in vivo formation of a protein-RNA adduct with cysteine 118 mutant enzymes can be recapitulated in vitro, with the formation dependent on the putative in vivo reducing system, flavodoxin and ferrodoxin:NADPH oxidoreductase. Further, the incorporation of two deuterium atoms into the in vitro adduct from [methyl-2H3]-SAM, as determined by LC-MS/MS, supports the addition of the cysteine 355-derived thiomethylene radical into the substrate adenosine in RNA. The lack of deuterium in the intermediate formed from D-RNA is consistent with the one electron oxidation of the initial adduct (IV in Scheme 1) and subsequent deprotonation, leading to re-aromatization of adenine and the formation of the methylene-linked RNA-cysteine 355 adduct. The observed incorporation of deuterium from labeled substrates into the covalent adduct is indicated in Scheme 2. Our results demonstrate the obligatory role of cysteine 118 in product formation, specifically by acting subsequent to the formation of the covalent intermediate. These observed critical roles for both cysteines 118 and 355 agree well with the prior in vivo observations that Cfr C105A and C338A mutants are not resistant to florfenicol or tiamulin, and short polynucleotides, encompassing A2503, isolated from the rRNA of these mutant strains exhibit no methylation by MALDI-MS analysis.49
Scheme 2.
Deuterium incorporation from substrates into covalent adduct between C118 mutants of RlmN and RNA.
We also note that the adduct formed by cysteine 118 mutants both in vivo and in vitro remains relatively stable in the presence of high concentrations of thiol reducing agents. The in vivo adducts were purified in the presence of 10 mM β-mercaptoethanol or 5 mM DTT depending on the purification step. Despite this exposure, the covalent RNA adduct is not resolved during purification. The in vitro adducts were formed in the presence of 1 mM DTT, indicating that low concentrations of this reagent do not perturb this intermediate. Further, when the in vivo adduct is prepared for SDS-PAGE, it is exposed to ~350 mM β-mercaptoethanol under denaturing conditions, yet the RNA-modified band is still evident. Cumulatively, these observations indicate that a sulfur nucleophile is necessary, though not sufficient, to resolve the covalent RNA-protein adduct. This may be due to conformational constraints on the approach of the nucleophile, possibly concomitant with protonation of the intermediate, which is unlikely to be readily accomplished with externally supplied thiol.
Conclusion
In summary, the findings presented here clearly support the existence and the intermediacy of the covalent adduct between RlmN and rRNA substrates and highlight the crucial roles for the two conserved cysteines unassociated with the [4Fe-4S] cluster. These results lend experimental evidence to the proposed mechanism14 that combines two canonical roles of the common metabolite S-adenosylmethionine: methylation and radical activation. The unique methylations catalyzed by RlmN and Cfr at aromatic amidine positions of a critically positioned nucleotide enable the modification of complex RNA substrates with significant biological impact.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. A. L. Burlingame for support. Dr. Kevan Shokat and Dr. Peter Walter graciously provided use of their Typhoon imaging systems. This work was supported by NIAID (R01AI095393-01) and NSF (CAREER 1056143) to D.G.F., and by NIH NIGMS grants R01 GM085697 and GM057755 to C.A.G. K.F.M and D.A.M. are members of the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A. L. Burlingame, Director) supported by the Biomedical Technology Research Centers program of the NIH National Institute of General Medical Sciences, NIH NIGMS 8P41GM103481.
Abbreviations
- rRNA
ribosomal RNA
- SAM
S-adenosyl-l-methionine
- PTC
peptidyltransferase center
- 5′-dA
5′-deoxyadenosyl/5′-deoxyadenosine
- HPLC
high-performance liquid chromatography
- LC-MS
liquid chromatography-mass spectrometry
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- DTT
dithiothreitol
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- WT
wild-type
- PCR
polymerase chain reaction
- TBST
Tris-buffered saline containing Tween-20
- EDTA
ethylenediaminetetraacetic acid
- PMSF
phenylmethanesulfonyl fluoride
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- CID
collision-induced dissociation
- LB
Luria-Bertani
- MW
molecular weight
- Fe-S
iron-sulfur
- tRNA
transfer RNA
- HRP
horseradish peroxidase
- FLAG
DYKDDDDK octapeptide
Footnotes
Supporting Information. Experimental procedures for the generation of FldA and Fpr expression plasmids, purification of FldA and Fpr and in-gel RNase and tryptic digests of RlmN; sequences of oligonucleotide primers used to generate FldA and Fpr PCR products, C118S RlmN and endogenous RlmN-FLAG (Table S1); LC-MS analysis of 2-methyladenosine from WT and mutant RlmN reactions (Figure S1); Western blot analysis of endogenous RlmN-FLAG (Figure S2); CID spectra of endogenous RlmN-FLAG C355-me peptide (Figure S3); Anion exchange chromatograph of C118A mutant RlmN (Figure S4); CID spectra of the in vitro generated C355 methyleneadenine-modified peptide from C118S RlmN (Figure S5); CID spectra of the in vitro generated C355 methyleneadenine-modified peptide from C118S RlmN produced using D-RNA substrate (Figure S6). This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Poehlsgaard J, Douthwaite S. Nat Rev Micro. 2005;3:870. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 2.Schlünzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Nature. 2001;413:814. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 3.Dunkle JA, Xiong L, Mankin AS, Cate JHD. Proc Natl Acad Sci USA. 2010;107:17152. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulkley D, Innis CA, Blaha G, Steitz TA. Proc Natl Acad Sci USA. 2010;107:17158. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tenson T, Mankin A. Mol Microbiol. 2006;59:1664. doi: 10.1111/j.1365-2958.2006.05063.x. [DOI] [PubMed] [Google Scholar]
- 6.Douthwaite S, Fourmy D, Yoshizawa S. In: Topics in Current Genetics. Grosjean H, editor. Vol. 12. 2005. p. 285. [Google Scholar]
- 7.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. Mol Microbiol. 2005;57:1064. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 8.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. Antimicrob Agents Chemother. 2006;50:2500. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toh SM, Xiong L, Bae T, Mankin AS. RNA. 2008;14:98. doi: 10.1261/rna.814408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan F, LaMarre JM, Röhrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. J Am Chem Soc. 2010;132:3953. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benítez-Páez A, Villarroya M, Armengod ME. RNA. 2012 doi: 10.1261/rna.033266.112. [DOI] [Google Scholar]
- 12.LaMarre JM, Howden BP, Mankin AS. Antimicrob Agents Chemother. 2011;55:2989. doi: 10.1128/AAC.00183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. EMBO J. 2010;29:3108. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. Science. 2011;332:604. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 15.Yan F, Fujimori DG. Proc Natl Acad Sci USA. 2011;108:3930. doi: 10.1073/pnas.1017781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Res. 2001;29:1097. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frey PA, Magnusson OT. Chem Rev. 2003;103:2129. doi: 10.1021/cr020422m. [DOI] [PubMed] [Google Scholar]
- 18.Booker SJ. Curr Opin Chem Biol. 2009;13:58. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moss M, Frey PA. J Biol Chem. 1987;262:14859. [PubMed] [Google Scholar]
- 20.Ballinger MD, Frey PA, Reed GH. Biochemistry. 1992;31:10782. doi: 10.1021/bi00159a020. [DOI] [PubMed] [Google Scholar]
- 21.Baraniak J, Moss ML, Frey PA. J Biol Chem. 1989;264:1357. [PubMed] [Google Scholar]
- 22.Chang CH, Ballinger MD, Reed GH, Frey PA. Biochemistry. 1996;35:11081. doi: 10.1021/bi960850k. [DOI] [PubMed] [Google Scholar]
- 23.Ballinger MD, Reed GH, Frey PA. Biochemistry. 2002;31:949. doi: 10.1021/bi00119a001. [DOI] [PubMed] [Google Scholar]
- 24.Eliasson R, Fontecave M, Jörnvall H, Krook M, Pontis E, Reichard P. Proc Natl Acad Sci USA. 1990;87:3314. doi: 10.1073/pnas.87.9.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eliasson R, Pontis E, Fontecave M, Gerez C, Harder J, Jörnvall H, Krook M, Reichard P. J Biol Chem. 1992;267:25541. [PubMed] [Google Scholar]
- 26.King DS, Reichard P. Biochem Biophys Res Commun. 1995;206:731. doi: 10.1006/bbrc.1995.1103. [DOI] [PubMed] [Google Scholar]
- 27.Ugulava NB, Gibney BR, Jarrett JT. Biochemistry. 2001;40:8343. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, Tu L, Booker SJ. Biochemistry. 2004;43:6378. doi: 10.1021/bi049528x. [DOI] [PubMed] [Google Scholar]
- 29.Ugulava NB, Sacanell CJ, Jarrett JT. Biochemistry. 2001;40:8352. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tse Sum Bui B, Benda R, Schünemann V, Florentin D, Trautwein AX, Marquet A. Biochemistry. 2003;42:8791. doi: 10.1021/bi034426c. [DOI] [PubMed] [Google Scholar]
- 31.Cicchillo RM, Lee KH, Baleanu-Gogonea C, Nesbitt NM, Krebs C, Booker SJ. Biochemistry. 2004;43:11770. doi: 10.1021/bi0488505. [DOI] [PubMed] [Google Scholar]
- 32.Tse Sum Bui B, Lotierzo M, Escalettes F, Florentin D, Marquet A. Biochemistry. 2004;43:16432. doi: 10.1021/bi048040t. [DOI] [PubMed] [Google Scholar]
- 33.Yang L, Lin G, Liu D, Dria KJ, Telser J, Li L. J Am Chem Soc. 2011;133:10434. doi: 10.1021/ja110196d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheek J, Broderick JB. J Am Chem Soc. 2002;124:2860. doi: 10.1021/ja017784g. [DOI] [PubMed] [Google Scholar]
- 35.Pierrel F, Douki T, Fontecave M, Atta M. J Biol Chem. 2004;279:47555. doi: 10.1074/jbc.M408562200. [DOI] [PubMed] [Google Scholar]
- 36.Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ. Proc Natl Acad Sci USA. 2008;105:1826. doi: 10.1073/pnas.0708608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee KH, Saleh L, Anton BP, Madinger CL, Benner JS, Iwig DF, Roberts RJ, Krebs C, Booker SJ. Biochemistry. 2009;48:10162. doi: 10.1021/bi900939w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arragain S, Handelman SK, Forouhar F, Wei FY, Tomizawa K, Hunt JF, Douki T, Fontecave M, Mulliez E, Atta M. J Biol Chem. 2010;285:28425. doi: 10.1074/jbc.M110.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arragain S, Garcia-Serres R, Blondin G, Douki T, Clemancey M, Latour JM, Forouhar F, Neely H, Montelione GT, Hunt JF, Mulliez E, Fontecave M, Atta M. J Biol Chem. 2010;285:5792. doi: 10.1074/jbc.M109.065516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernández HL, Pierrel F, Elleingand E, García-Serres R, Huynh BH, Johnson MK, Fontecave M, Atta M. Biochemistry. 2007;46:5140. doi: 10.1021/bi7000449. [DOI] [PubMed] [Google Scholar]
- 41.Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. EMBO J. 2003;22:6214. doi: 10.1093/emboj/cdg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Layer G, Kervio E, Morlock G, Heinz DW, Jahn D, Retey J, Schubert WD. Biol Chem. 2005;386:971. doi: 10.1515/BC.2005.113. [DOI] [PubMed] [Google Scholar]
- 43.Layer G, Pierik AJ, Trost M, Rigby SE, Leech HK, Grage K, Breckau D, Astner I, Jänsch L, Heathcote P, Warren MJ, Heinz DW, Jahn D. J Biol Chem. 2006;281:15727. doi: 10.1074/jbc.M512628200. [DOI] [PubMed] [Google Scholar]
- 44.Fang Q, Peng J, Dierks T. J Biol Chem. 2004;279:14570. doi: 10.1074/jbc.M313855200. [DOI] [PubMed] [Google Scholar]
- 45.Grove TL, Lee KH, St Clair J, Krebs C, Booker SJ. Biochemistry. 2008;47:7523. doi: 10.1021/bi8004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, Wang E, Freed J, Krebs C, Ealick SE, Lin H. Nature. 2010;465:891. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flühe L, Knappe TA, Gattner MJ, Schafer A, Burghaus O, Linne U, Marahiel MA. Nat Chem Biol. 2012;8:350. doi: 10.1038/nchembio.798. [DOI] [PubMed] [Google Scholar]
- 48.Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. Nat Chem Biol. 2008;4:758. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaminska KH, Purta E, Hansen LH, Bujnicki JM, Vester B, Long KS. Nucleic Acids Res. 2010;38:1652. doi: 10.1093/nar/gkp1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC. Science. 2011;332:1089. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruszczycky MW, Choi SH, Liu HW. J Am Chem Soc. 2010;132:2359. doi: 10.1021/ja909451a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fish WW. Methods Enzymol. 1988;158:357. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 53.Grove TL, Radle MI, Krebs C, Booker SJ. J Am Chem Soc. 2011;133:19586. doi: 10.1021/ja207327v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osborne C, Chen LM, Matthews RG. J Bacteriol. 1991;173:1729. doi: 10.1128/jb.173.5.1729-1737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bianchi V, Reichard P, Eliasson R, Pontis E, Krook M, Jornvall H, Haggard-Ljungquist E. J Bacteriol. 1993;175:1590. doi: 10.1128/jb.175.6.1590-1595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datsenko KA, Wanner BL. Proc Natl Acad Sci USA. 2000;97:6640. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bradford MM. Anal Biochem. 1976;72:248. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 58.Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, Burlingame AL. Mol Cell Proteomics. 2008;7:2386. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. Annu Rev Microbiol. 1996;50:645. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- 60.Biemann K. Methods Enzymol. 1990;193:886. doi: 10.1016/0076-6879(90)93460-3. [DOI] [PubMed] [Google Scholar]
- 61.Dudley E, Tuytten R, Bond A, Lemiere F, Brenton AG, Esmans EL, Newton RP. Rapid Commun Mass Spectrom. 2005;19:3075. doi: 10.1002/rcm.2151. [DOI] [PubMed] [Google Scholar]
- 62.Rose IA. Biochim Biophys Acta. 1957;25:214. doi: 10.1016/0006-3002(57)90453-5. [DOI] [PubMed] [Google Scholar]
- 63.Hall SS, Doweyko AM, Jordan F. J Am Chem Soc. 1976;98:7460. doi: 10.1021/ja00439a077. [DOI] [PubMed] [Google Scholar]
- 64.Siddique AN, Jurkowska RZ, Jurkowski TP, Jeltsch A. FEBS J. 2011;278:2055. doi: 10.1111/j.1742-4658.2011.08121.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.