SUMMARY
The effectiveness of RNA interference (RNAi) in many organisms is potentiated through the signal-amplifying activity of a targeted RNA directed RNA polymerase (RdRP) system that can convert a small population of exogenously-encountered dsRNA fragments into an abundant internal pool of small interfering RNA (siRNA). As for any biological amplification system, we expect an underlying architecture that will limit the ability of a randomly encountered trigger to produce an uncontrolled and self-escalating response. Investigating such limits in C. elegans, we find that feed-forward amplification is limited by a critical biosynthetic and structural distinction at the RNA level between (i) triggers that can produce amplification and (ii) siRNA products of the amplification reaction. By assuring that initial (primary) siRNAs can act as triggers but not templates for activation, and that the resulting (secondary) siRNAs can enforce gene silencing on additional targets without unbridled trigger amplification, the system achieves substantial but fundamentally limited signal amplification.
INTRODUCTION
Canonical RNAi is a biochemical pathway triggered by foreign dsRNA that ultimately results in the destruction of endogenous target RNA of corresponding sequence (reviewed in Boisvert and Simard, 2008). The core RNAi factors that initiate this process center on Dicer (DCR-1 in C. elegans), along with its dsRNA-binding partner (RDE-4 in C. elegans) which start the pathway by processing the dsRNA trigger into short interfering RNAs (siRNAs) (Knight and Bass, 2001; Grishok et al., 2001; Tabara et al., 2002). The double-stranded primary siRNAs have structures characteristic of RNase III type cleavages; 2nt 3’ overhangs, 3’ hydroxyls (3'-OH) and 5’ monophosphates (5’-monoP) (Elbashir et al., 2001a, b, c). Primary siRNAs are then transferred to Argonaute class RNA-binding proteins, with cleavage of one strand leaving a single-stranded guide RNA which can engage complementary sequences in a target RNA pool. Argonautes possess three main domains; the PAZ domain which binds the 3’ terminus of the siRNA, the MID domain which binds the 5’ terminus of the siRNA, and the PIWI domain which folds into an RNase H-like structure (Song et al., 2004). In a number of systems, most notably in mammals and Drosophila, the destruction of the target RNA has been shown to be mediated by a cleavage activity of the Argonaute RNase H domain, guided by the bound siRNA (reviewed in Nowotny and Yang, 2009). Although the simplicity of this canonical RNAi paradigm provides some indications of its potential biological effect, it appears from the stoichiometry in several systems that a simple one-siRNA-one-target relationship would be insufficient for the degree of gene silencing that is observed.
How does the RNAi pathway ensure robust silencing of target RNAs? One solution relies on the mechanistic ability of individual RNA-Induced Silencing Complex (RISC) assemblies to serially target multiple mRNAs (Hutvagner and Zamore, 2002). In some organisms such as fungi, plants, and C. elegans, the potentially-multiturnover core RNAi mechanism is supplemented through the action of RdRP which expand the initial siRNA pool with the generation of secondary siRNAs (Cogoni and Macino, 1999; Mourrain et al., 2000; Sijen et al., 2001). (Although Drosophila and mammals do not appear to possess canonical RdRPs, the latter may use other polymerases to perform RNAi-mediating RdRP function; e.g., Lipardi and Paterson, 2009; Maida et al., 2009).
The C. elegans genome encodes four putative RdRPs; RRF-1, -2, -3, and EGO-1. While rrf-1 and ego-1 were initially shown to be required for RNAi in the soma and germline, respectively, rrf-3 appears to mediate endogenous gene silencing functions and rrf-2’s function(s) has yet to be discovered (Sijen et al., 2001; Simmer et al., 2002; Smardon et al., 2000; Gent et al., 2009; Pavelec et al., 2009; Vasale et al., 2010). Current models posit at least two different small RNA pools with key roles in RNAi: a "primary" pool produced upon action of Dicer on the initial dsRNA trigger, with additional small RNA produced through targeted activity of the RdRPs. Previous work has illustrated a potentially important structural distinction between primary siRNAs and RdRP products, in that the latter often retain the 5’ triphosphate (5’-PPP) from the initiating nucleotide (Pak and Fire, 2007; Sijen et al., 2007).
Biological effects of 20–30nt RNAs in eukaryotes frequently involve their incorporation into RISCs that include a member of the Argonaute protein family whose sequence-directed activity is programmed by a single small RNA effector. The C. elegans genome encodes 27 putative Argonautes, a diversity supporting the varied biological activities and molecular roles of several small RNA families in this organism, including microRNAs, piRNAs, and several types of endogenous siRNAs (Grishok et al., 2001; Yigit et al., 2006; Guang et al., 2008; reviewed in Fischer, 2010). Small RNA distributions during an RNAi response in C. elegans further exemplify the potential for mechanistic plurality inherent in a diversity of Argonautes, with 5’-monoP (primary) siRNAs having been shown to bind the Argonaute RDE-1, while 5’-PPP siRNAs (RdRP products) bind the "WAGO" group of worm-specific Argonautes (Yigit et al., 2006).
Current models posit the expansion of small RNA pools during RNAi as resulting from an ability of individual siRNA RISC complexes interacting with a target message to recruit RdRP and thereby generate a target-limited population of RdRP products. This model, involving four types of RNA species (initial dsRNA trigger, target mRNA, primary siRNAs, and RdRP products), certainly allows for amplification and propagation of RNAi responses as long as populations of trigger and target RNAs are present.
Given the efficacy of RdRP activity, why don't RNAi processes amplify indefinitely (Sijen et al., 2001; Bergstrom et al., 2003; Pak and Fire, 2007; Sijen et al., 2007)? Unbridled secondary siRNA generation could potentially create havoc, generating diverse sequences which could potentially direct the destruction of unrelated RNAs. Experimental analysis of RNAi in C. elegans offers the flexibility of altering the structure and delivery of trigger RNA as well as the genetic background of the recipient animals in characterizing the interference response. Combining these two capabilities with high throughput sequencing to characterize populations of RNAs associated with RNAi, we have investigated the mechanistic basis for limitations to amplification of RNAi in C. elegans.
RESULTS
Properties of primary and secondary siRNA pools during RNAi
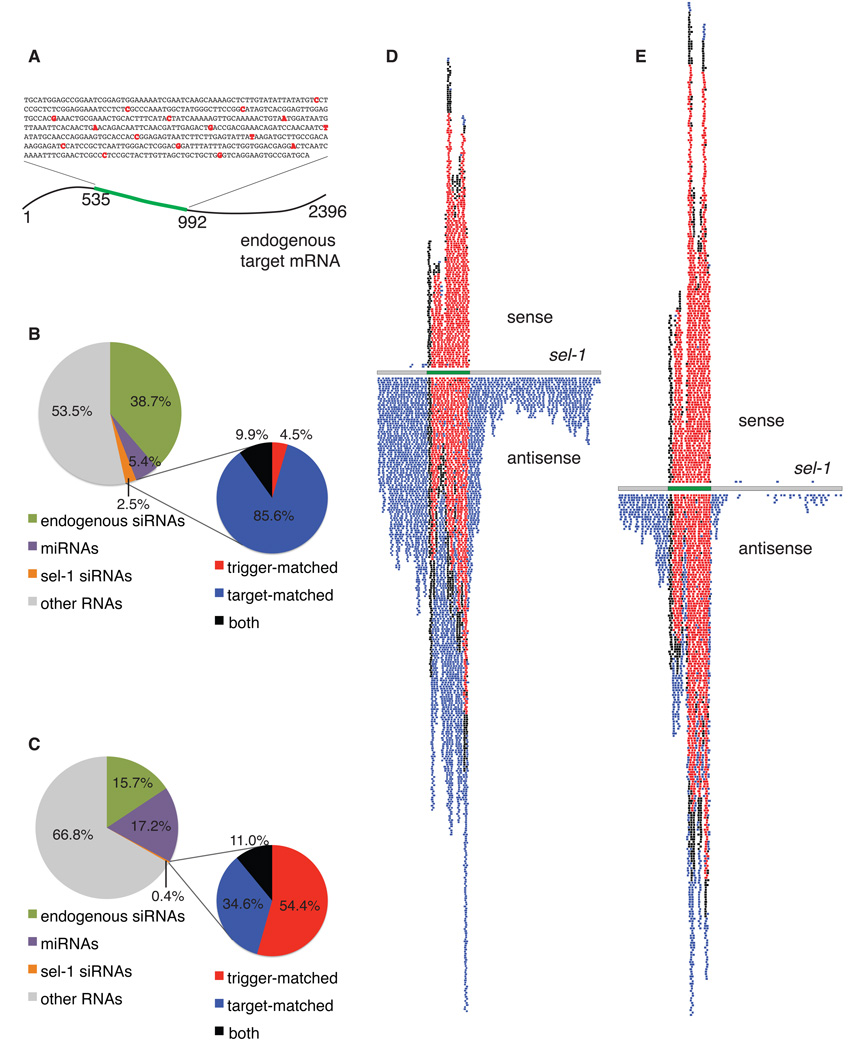
In C. elegans, RNAi proceeds in two phases; the primary and secondary siRNA responses. To distinguish between these phases, we established an assay wherein RNAi against the sel-1 gene was initiated by feeding (Timmons et al., 2001) with a dsRNA trigger that contained a series of mismatches from the wild type sequence at 25 nt intervals (Figure 1A). This allows a sequence-based distinction between trigger-derived RNA sequences and sequences derived from copying of target RNA.
Figure 1.
Characterization of primary and secondary siRNA pools using a mismatched trigger. (A) Mismatched trigger assay. (B) Proportions of distinct small RNA populations from 5’-P-independent small RNA capture from N2 (wild type) animals fed mismatched sel-1 dsRNA. (C) Proportions of distinct small RNA populations from 5’-monoP-enriched small RNA capture from N2 (wild type) animals fed mismatched sel-1 dsRNA. (D) Plot of sel-1 siRNAs from 5’-P-independent capture from N2 animals fed mismatched sel-1 dsRNA. (E) Plot of sel-1 siRNAs from 5’-monoP-enriched capture from N2 animals fed mismatched sel-1 dsRNA. (D, E) grey: sel-1 mRNA, green: region of sel-1 mRNA encompassed in mismatched trigger, black: siRNA matching both target and trigger RNAs, blue: siRNA matching only target RNA, red: siRNA matching only trigger RNA. Lightened shading indicates multiple incidence. (See also Figure S1.)
Sequencing was used to detect sel-1 siRNAs and determine their identity with the original target and trigger sequences. We employed two small RNA (sRNA) capture protocols for high-throughput sequencing using the Illumina platform. To examine the broadest spectrum of siRNAs, we used a 5’-P-independent protocol, which does not distinguish between different 5’ phosphoforms (Gent et al., 2010). A second protocol that depends on ligation to a 5’-monoP should enrich for 5’-monoP species such as primary DCR-1 products (Lau et al., 2001). As previously observed (Pak and Fire, 2007), RNAs retaining a 5'-PPP terminus (such as direct RdRP products) would be depleted in the 5'-monoP-enriched pool. All of the protocols used for library production rely on a dephosphorylated 3'-OH terminus.
Using the 5’-P-independent protocol, we sequenced 6,420,803 captured RNAs, of which 159,424 (2.5%) corresponded to sel-1 sequence (Table 1, Figure 1B). As a control, we examined 25,109,432 sequences compiled from multiple sets of animals that had no exposure to sel-1 dsRNA (Gent et al., 2010; Maniar and Fire, 2011; Wu et al., 2011); from this control group, only 223 (<0.001%) sel-1 siRNAs were found, suggesting that the observed exogenously-triggered sel-1 siRNAs in the RNAi experiments are indeed specific to the RNAi response.
Table 1.
Small RNAs from various strains captured using the 5'-P-independent capture protocol
| Strain | N2 | rde-4(ne299) | rrf-1(pk1417) glp-4(bn2) | rde-1(ne300) | MAGO | rde-1(ne300); Σrde-1(+) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total barcoded, linkered reads | 6,420,803 | 3,134,550 | 2,421,299 | 3,178,834 | 2,057,199* | 14,964,152 | |||||||
| WS190 cDNAs | sense | 61,207 | 48,661 | 66,112 | 46,995 | 42,060 | 197,055 | ||||||
| antisense | 2,482,796 | 1,065,915 | 62,707 | 1,092,025 | 383,063* | 5,077,387 | |||||||
| microRNAs | 345,640 | 445,467 | 1,627,753 | 626,642 | 338,297 | 1,286,322 | |||||||
| sel-1 siRNAs/miRNAs: | sel-1 siRNAs# | % of N2$ | sel-1 siRNAs | % of N2$ | sel-1 siRNAs | % of N2$ | sel-1 siRNAs | % of N2$ | sel-1 siRNAs | % of N2$ | sel-1 siRNAs | % of N2$ | |
| match target only | 0.395 | [100%] | 0.001 | 0.2% | 0.005 | 1.2% | 0.004 | 1.0% | 0.019* | 4.8% | 0.178 | 45.0% | |
| Antisense | match trigger only | 0.011 | [100%] | 0.003 | 24.7% | 0.008 | 69.3% | 0.008 | 69.0% | 0.003 | 25.3% | 0.009 | 77.9% |
| match both | 0.043 | [100%] | 0.001 | 1.4% | 0.002 | 4.1% | 0.000 | 0.9% | 0.001 | 1.3% | 0.027 | 61.1% | |
| total | 0.449 | [100%] | 0.004 | 0.9% | 0.014 | 3.1% | 0.012 | 2.7% | 0.022 | 5.0% | 0.213 | 47.3% | |
| match target only | 0.000 | [100%] | 0.000 | 16.3% | 0.000 | 60.3% | 0.000 | 31.9% | 0.000 | 26.9% | 0.000 | 104.7% | |
| Sense | match trigger only | 0.010 | [100%] | 0.003 | 28.7% | 0.007 | 76.7% | 0.003 | 26.0% | 0.003 | 29.1% | 0.005 | 55.5% |
| match both | 0.002 | [100%] | 0.000 | 17.0% | 0.001 | 51.6% | 0.001 | 23.7% | 0.001 | 27.0% | 0.001 | 65.7% | |
| total | 0.012 | [100%] | 0.003 | 26.6% | 0.009 | 72.2% | 0.003 | 25.7% | 0.003 | 28.7% | 0.007 | 57.5% | |
| Total | 0.461 | [100%] | 0.007 | 1.6% | 0.023 | 4.9% | 0.152 | 32.9% | 0.026 | 5.6% | 0.220 | 47.6% | |
An additional unique 29 nt sel-1 siRNA found in 15,261 incidences in the MAGO sample has not been further characterized (see Experimental Procedures).
In order to control for variability in sRNA capture between samples, sel-1 siRNA counts were normalized to miRNA counts within the same sample.
miRNA-normalized sel-1 siRNA counts are expressed as a fraction of miRNA-normalized counts of the equivalent siRNA class in N2 animals.
i) Trigger-matched siRNAs
siRNAs which match the mismatched sel-1 dsRNA trigger accounted for only 5% of total sel-1 siRNAs in the responding animals. We found that the sense:antisense ratio of trigger-matched siRNAs was close to 1 (0.88 and 0.93 respectively in the 5’-monoP-enriched and 5’-P-independent pools). This contrasts with the sense:antisense ratios for the remaining (target-matched) sel-1 siRNAs (0.00014 and 0.00284, respectively) and argues that the trigger-matched population is indeed a defined siRNA class (Tables 1 and 2). A priori, at least three classes of RNA molecules may comprise this trigger-matched pool: (i) fragments of RNA generated in the bacteria fed to the animals (potentially with diverse 5' structures), (ii) primary siRNAs generated directly through DCR-1-mediated processing of dsRNA trigger molecules (with 5'-monoP termini), and (iii) products of RdRP copying of the trigger RNA in C. elegans (possessing 5'-PPP termini). Although we can not exclude the presence of any of the aforementioned classes of RNAs, our data are consistent with a substantial fraction of the trigger-matched siRNA population carrying a 5'-monoP. In particular, enriching for 5’-monoP-sRNAs produced a 12-fold enrichment for trigger-matched siRNAs among total sel-1 siRNAs (p=0.0189) (compare N2 trigger-matched siRNAs in Tables 1 and 2). Although consistent with the 5' structures of DCR-1 products, these data do not address whether these trigger-matched products are derived from the initial dsRNA inoculum (Bernstein et al., 2001), or from copies of the original inoculum produced by an RdRP activity.
Table 2.
Small RNAs from various strains captured using the 5'-mono-P-enrichment capture protocol.
| Strain | N2 | rde-4(ne299) | rrf-1(pk1417) glp-4(bn2) | rde-1(ne300) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Total barcoded, linkered reads | 5,370,571 | 643,170 | 2,166,851 | 4,921,175 | |||||
| WS190 cDNAs | sense | 102,558 | 9,431 | 38,351 | 69,174 | ||||
| antisense | 841,472 | 76,879 | 27,914 | 671,455 | |||||
| miRNAs | 922,433 | 358,879 | 1,528,074 | 2,063,689 | |||||
| sel-1 siRNAs/miRNAs: | sel-1 siRNAs | % of N2$ | sel-1 siRNAs | % of N2$ | sel-1 siRNAs | % of N2$ | sel-1 siRNAs | % of N2$ | |
| match target only | 0.008 | [100%] | 0.000 | 1.5% | 0.002 | 24.4% | 0.000 | 0.9% | |
| match trigger only | 0.006 | [100%] | 0.000 | 4.4% | 0.010 | 167.3% | 0.001 | 10.7% | |
| Antisense | |||||||||
| match both | 0.001 | [100%] | 0.000 | 0.6% | 0.002 | 116.9% | 0.000 | 2.8% | |
| total | 0.015 | [100%] | 0.000 | 2.6% | 0.014 | 91.1% | 0.001 | 5.1% | |
| match target only | 0.000 | [100%] | 0.000 | 25.7% | 0.000 | 147.9% | 0.000 | 38.0% | |
| match trigger only | 0.006 | [100%] | 0.000 | 2.2% | 0.009 | 156.4% | 0.002 | 34.0% | |
| Sense | |||||||||
| match both | 0.001 | [100%] | 0.000 | 3.3% | 0.002 | 148.0% | 0.000 | 15.7% | |
| total | 0.007 | [100%] | 0.000 | 2.5% | 0.011 | 155.1% | 0.002 | 31.1% | |
| Total | 0.022 | [100%] | 0.001 | 2.6% | 0.025 | 111.1% | 0.006 | 26.5% | |
In order to control for variability in sRNA capture between samples, sel-1 siRNA counts were normalized to miRNA counts within the same sample.
miRNA-normalized sel-1 siRNA counts are expressed as a fraction of miRNA-normalized counts of the equivalent siRNA class in N2 animals. Particularly significant in these analyses are relative differences between primary and secondary siRNA levels in distinct mutant backgrounds. Differences in primary response can be observed in these experiments but are somewhat sensitive to variation in the character of the initial dsRNA feeding (Timmons et al., 2001); in particular we note that additional experiments with an rde-1(ne300) host showed these animals to be capable of producing a primary response comparable to that seen in wild-type animals. The recovery of target-matched siRNAs in the 5’-monoP-enriched pool is also somewhat variable between experiments, consistent with a degree of technical variability in the 5' enrichment.
ii) Target-matched siRNAs
We found that the majority of the RNAi-induced sel-1 siRNAs were (i) antisense to the target mRNA, (ii) corresponding to sequence within the targeted region of the mRNA (nucleotides 535 to 992 of the mRNA; Table 1, Figure 1D), and (iii) perfectly matched to the target RNA, without the mismatches introduced in the trigger (Figure 1D). Characterization of the target-matched siRNAs revealed distinct length and sequence composition proclivities, with a consensus length of 22nt and frequent appearance of a G at the 5' end (Figure S1). (The 5' G sequence bias was also detected, to a lesser extent, in other classes of siRNAs; a portion of this 5' preference may therefore represent a capture bias; Figure S1.) The secondary siRNAs resemble an abundant class of endogenous siRNAs ('22G') observed from different loci in the absence of an external trigger, also resembling virus-derived siRNAs seen during Orsay virus replication in C. elegans (Ambros et al., 2003; Gu et al., 2009; Han et al., 2009; Gent et al., 2010; Felix et al., 2011). Based on the predominant structure and perfect match to target sequences, we infer that the bulk of the sel-1 siRNA pool is generated by RdRP action on a target RNA template (Sijen et al. 2001, Sijen et al., 2007, and Pak and Fire, 2007). We stress that these overall characteristics do not exclude the presence of siRNAs derived from copying of the trigger population by RdRP, but argue that such trigger copies would be a minority of the total. We note as well that some RNAs without 5’-monoP are, most likely, also captured using our 5’-monoP-enrichment protocol due to modification of 5' termini in vivo or during sample preparation, giving rise to a population of secondary siRNAs in this pool, albeit at reduced levels (Figure 1C, E; Pak and Fire, 2007).
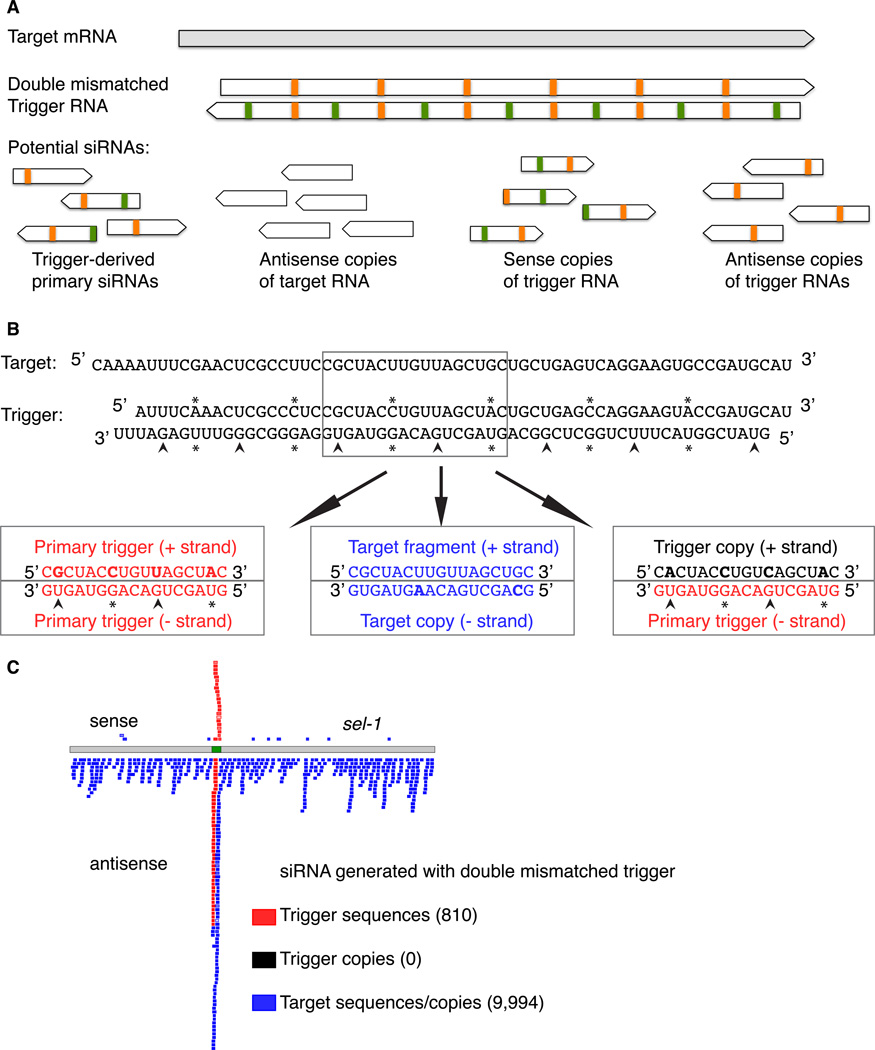
Trigger RNAs guide RdRP copying of target mRNA without themselves being templates for amplification
We used a doubly mismatched (heteroduplex) RNA trigger to address the possibility that the direct activity of the RdRP system on trigger dsRNAs might serve as a component of the amplification (Figure 2). The experiment uses a trigger in which each strand contained mismatches relative to its complementary strand and relative to the target RNA (Figure 2A, B). We used the presence/absence of these mismatches to distinguish between potential siRNA classes; i) primary siRNAs would explicitly retain the mismatches present in the dsRNA trigger, ii) secondary siRNAs generated using the target RNA as template (thereby perfectly complementary to the target RNA), and iii) hypothetical secondary siRNAs that could have been generated by RdRP using the trigger RNA as template. The third class of RNAs were of substantial interest as they address the ability of primary dsRNA or its siRNA products to serve as direct templates for amplification. The manner in which such primary amplification products would be distinguished from trigger, target, and target copies by their mismatch patterns is exemplified in Figure 2A and B.
Figure 2.
A doubly mismatched trigger to test for use of trigger RNA as template for synthesis of secondary siRNAs. (A) Scheme for using a double mismatched trigger to distinguish by sequence between primary siRNAs, target derived molecules and RdRP products, and hypothetical products of RdRP copying of trigger sequences. (B) Sequences of target RNA (wild type), sense and antisense strands of the trigger RNA molecule. Asterisks: Class 1 type mismatches where base pairing is maintained between the two strands of the trigger dsRNA. Arrowheads: Class 2 type mismatches where GU wobble pairs are introduced into the trigger dsRNA. (B) Plot of sel-1 siRNAs captured using 5’-P-independent capture protocol. grey: sel-1 mRNA, green: region of sel-1 mRNA encompassed in double mismatched trigger, blue: siRNA matching only target RNA, red: siRNA matching one strand of trigger RNA. Absent were the hypothetical (black) sequences that would have been derived from copying of the primary trigger by RdRP. Lightened shading indicates multiple incidence.
We sequenced 25,903,899 tags from animals undergoing RNAi with the heteroduplex trigger. Of these tags, 7,203 evidently derived from the original trigger RNA, 19 evidently derived directly from (sense) target RNA, and 136,456 evidently derived as antisense copies of the target sequence. None of the 10,804 sel-1 matched small RNAs had the mismatch pattern expected from RdRP copying of the input dsRNA trigger.
Secondary siRNAs enforce RNAi without re-engaging RdRP activity
Can the primary and secondary siRNAs also be distinguished by the roles that they play during RNAi? Primary siRNAs are required for secondary siRNA generation, presumably by guiding the RdRP to the target RNA template. We asked if this activity could also be performed following recruitment of additional (naive) target messages by secondary siRNAs, resulting in an amplification mechanism that would be sustained as long as additional target is being provided. We addressed this question by analyzing siRNAs that were generated using recipient strains with related target RNAs that allow a distinction between secondary and potential tertiary siRNAs.
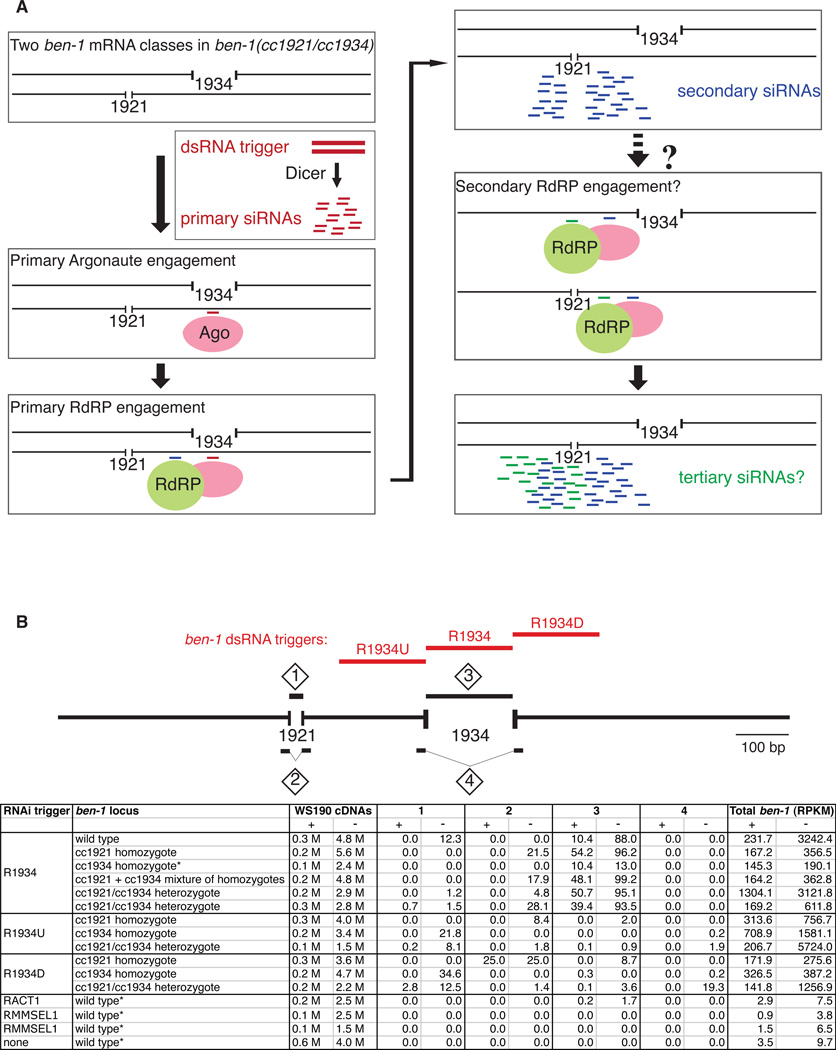
i) siRNAs derived from a heteroallelic configuration
The fact that C. elegans are diploid allows experimental designs in which the two genomic regions for a given locus can carry different and non-overlapping deletions (Figure 3A). This configuration can then be combined with a trigger that engages only one of the two expected transcripts. Secondary siRNAs from such an experiment would correspond only to the targeted transcript, skipping the deletion region present in that transcript. We know from functional experiments (Sijen et al., 2001, Alder et al., 2003) that these secondary siRNAs can interact with additional homologous mRNAs. What is not clear is whether these interactions can spur additional siRNA synthesis by RdRP. An ability of these interactions to trigger tertiary siRNAs through an RdRP engagement would be expected to produce a population of siRNAs corresponding to a region deleted in the initial mRNA target.
Figure 3.
Use of a deletion heterozygote to test for production of tertiary siRNAs. (A) Model for generation of hypothetical tertiary siRNAs in a deletion heterozygote. The ben-1 (cc1934/cc1921) trans-heterozygote contains two populations of ben-1 mRNAs containing either the cc1934 or the cc1921 deletions. dsRNA trigger molecules encompassing the region deleted in the cc1934 allele is introduced when it is subsequently processed into primary siRNAs (red). The primary siRNA guides the Argonaute (presumably RDE-1) to the mRNA derived from the cc1921 allele whence the RdRP is recruited generating a population of secondary siRNAs (blue). Absent from these secondary siRNAs, having been generated on the cc1921 mRNA template, are sequences deleted in the cc1921 allele. If the secondary siRNA, in turn, is capable of recruiting the RdRP to naive mRNAs, the resulting population should include copies made from the cc1934 deletion mRNA, resulting in some siRNAs that would overlap the region deleted in the cc1921 allele. If, however, secondary siRNAs are incapable of recruiting the RdRP, the cc1921 deleted region would be barren. (B) siRNAs from ben-1 (cc1934/cc1921) trans-heterozygote RNAi experiment. siRNAs were analyzed from wild type, ben-1 (cc1921/cc1921), ben-1 (cc1934/cc1934), a mixture of RNA from ben-1 (cc1921/cc1921) and ben-1 (cc1934/cc1934) animals, and from ben-1 (cc1934/cc1921) trans-heterzygotes exposed to dsRNAs corresponding to the R1934, R1934U, and R1934D triggers. In addition, captured RNA datasets from (i) control wild type strains triggered for RNAi in the act-1 gene (RACT1), (ii) the sel-1 gene with the mismatched sel-1 trigger (RMMSEL1), and (iii) no RNAi were included in the analyses (Gent et al., 2010; Maniar and Fire, 2011; Wu et al., 2011). siRNAs were counted as follows: Region 1, all siRNAs that unambiguously overlap the cc1921 deletion; Region 2, all siRNAs that contain sequence spanning the junction of the region deleted in cc1921 with at least 2 nt matches on both sides of the junction; Region 3, all siRNAs that unambiguously overlap the cc1934 deletion; Region 4, all siRNAs that contain sequence spanning the junction of the region deleted in cc1934 with at least 2 nt matches on both sides of the junction. Some variability in overall response is observed in bacterial feeding experiments with C. elegans; the indicated paucity of tertiary siRNAs relative to their neighbors in ben-1 (cc1921/cc1934) heterozygotes has been observed over a variety of different RNAi efficiencies in five independent RNA feeding experiments (bottom panel and data not shown). As the overall efficacy with which RNAi is initiated and amplified can vary between experiments with different triggers (due, in part, to difference in distance between the trigger region and the assayed region), siRNA counts are expressed as a percentage of antisense siRNA reads within a region spanning 50 nt upstream and downstream of the deletion. * Values for cc1934 homozygotes exposed to the R1934 trigger were normalized to equivalent values (as judged by the relative level of sense ben-1 siRNAs) for wild type animals exposed to the R1934 trigger due to a complete lack of an RNAi response in the former. Likewise, counts for control, non-ben-1 RNAi wild type strains were normalized to equivalent values (as judged by the relative level of total antisense siRNAs to all cDNAs) for wild type animals exposed to the R1934 trigger. 'RPKM' is a standard metric of sequence representation calculated as Reads per Kilobase of target per Million sequence instances (in this case antisense cDNA sequences) (Mortazavi et al., 2008). The 5'-P-independent capture method was employed. (See also Figure S2.)
A series of deletions of the ben-1 tubulin locus, isolated in screens for spontaneous resistance to the paralytic drug benomyl (C. Mello, D. Liu, and A. Fire, unpublished), provided an excellent starting point for this analysis. We chose two non-overlapping deletions of 18bp (ben-1(cc1921)) and 108bp (ben-1(cc1934)) which were each in frame (to avoid nonsense-mediated decay of the corresponding mRNAs; Chang et al., 2007) and which each resulted in a null phenotype. For trigger dsRNA, we employed a segment of 109 bp contained completely with the ben-1(cc1934) deletion (R1934; Figure 3). As a positive control, we saw a strong siRNA signal following feeding-based delivery of this dsRNA to either wild type ben-1(+) animals or animals with the ben-1(cc1921) deletion. As expected, only a trigger-limited siRNA signal (presumably from Dicer cleavage of the original trigger but not RdRP amplification) was seen upon delivering this to a homologous ben-1(cc1934) strain.
Our ability to distinguish tertiary siRNAs generated through secondary siRNA interactions with their mRNA targets comes from delivering the R1934 dsRNA trigger to a population of ben-1(cc1921)/ben-1(cc1934) trans-heterozygotes (Figure 3A). In this experiment, the trigger molecules presumably generate primary siRNAs that correspond to the trigger sequence (present in cc1921 but deleted in the cc1934 allele), thus guiding the RdRP to the mRNA derived from the cc1921 allele to generate a population of secondary siRNAs that will skip the cc1921 deletion region. In a hypothetical tertiary round of RNAi following their interaction with new target mRNAs, these siRNAs would presumably generate antisense siRNAs from both alleles. Detection of siRNAs matching the region absent in the initial target cc1921 would be diagnostic of such a population.
From a total of 28,611,376 small RNA sequences from the dsRNA-treated heterozygote, 14,740 matched the ben-1 mRNA on the antisense strand (Figures 3B, S2). Only 7 siRNAs overlapped the cc1921-deleted sequence (average normalized value=1.3; see Figure 3B legend for normalization method). The 1.3 value contrasts to a normalized level of antisense siRNAs of 12.3 from the cc1921 deletion region in a wild type background. 0 siRNAs corresponding to this sequence were observed from both homozygous cc1921 and homozygous cc1934 animals (in which RNAi can not be induced due to the absence of target sequence). These results are consistent with any presence of tertiary siRNAs representing at most a minor fraction of the total siRNA signal in the region surrounding the cc1921 deletion.
In order to control for the possibility that mRNAs generated from these alleles differ in their ability to amplify the RNAi signal, we performed the same experiment with two different ben-1 triggers corresponding to sequence either upstream (R1934U) or downstream (R1934D) of the region deleted in the cc1934 allele (Figure 3B, S2). As mRNAs from either allele in the heterozygotes should, in principle, provide equivalent templates for amplified siRNA products,we would expect higher levels of siRNAs corresponding to the cc1921-deleted region than that observed with the R1934 trigger. Indeed, we found that the ben-(cc1921)/ben-1(cc1934) trans-heterozygotes generated wild type normalized levels of siRNAs overlapping the region deleted in the cc1921 allele (8.1 and 12.5 for the R1934U and R1934D triggers, respectively).
As a summary of the siRNA patterns produced following interference in the deletion heterozygote, we observed primary and secondary siRNA signals, but no evidence of the type of signal expected from a major role of secondary siRNAs in interactions with additional templates to generate a tertiary siRNA response. These results provide support for a model in which signal amplification during an RNAi response is effectively restricted to a single tier of primary triggers.
ii) siRNAs derived from a conserved gene family
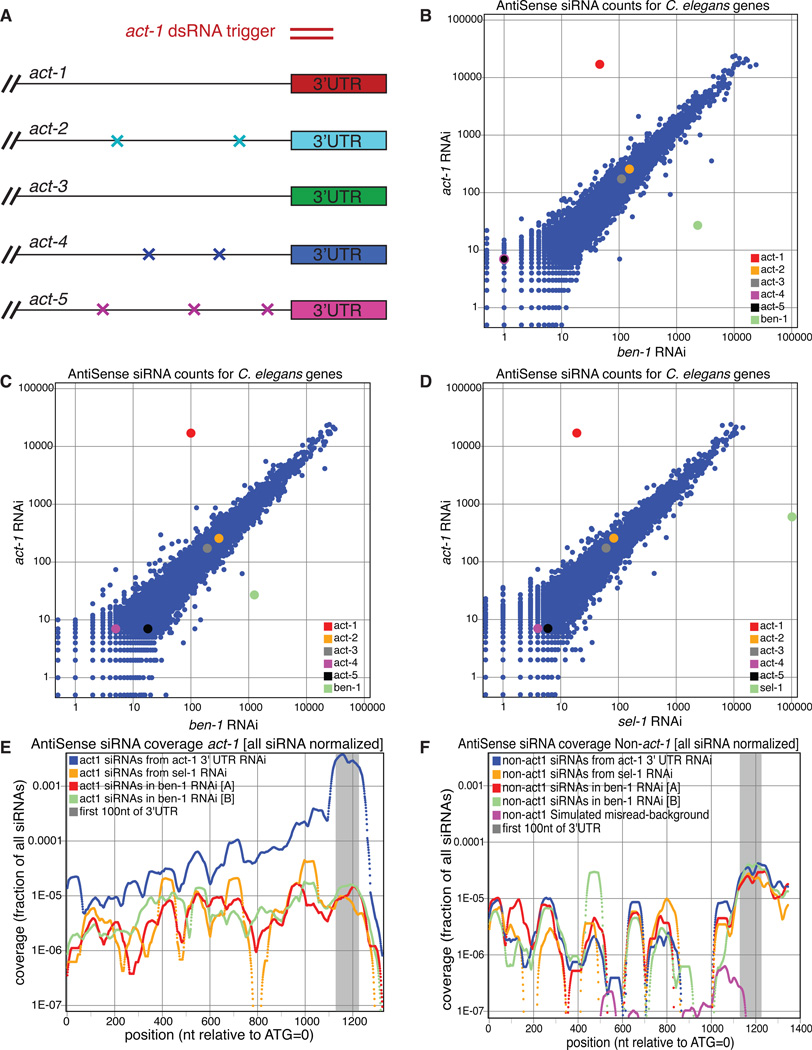
The actin gene family in C. elegans is comprised of 5 genes (act-1 through -5) which display almost perfect peptide sequence identity (Figure S3). Evidence for co-expression of at least four of the genes (act-1, -2, -3, -4), combined with evidence for functional redundancy between act-1, -2, and -3 (Landel et al., 1984; MacQueen et al., 2005; Willis et al., 2006) support the use of this gene family to evaluate the ability of C. elegans to produce tertiary siRNAs.
We reasoned that highly similar secondary siRNAs generated from act-1 mRNAs might elicit tertiary siRNA generation on mRNAs of the other actin genes. In order to exclusively target the primary response to act-1 mRNAs, we induced RNAi with a dsRNA trigger designed against 100 nt of act-1 3'UTR (Figure 4A). This experimental design enabled us to detect putative tertiary siRNAs in any of the other four 3' UTRs (due to the extensive 3' UTR divergence) and in the three actin genes (all except act-3) which have base differences from act-1 interspersed in the coding region (Figure S3; Files et al., 1983).
Figure 4.
Assays for siRNAs emanating from direct and indirect targets of an actin 3' UTR dsRNA trigger. (A) Experimental design. RNAi against a member of a partially-redundant multigene family is induced using a dsRNA trigger uniquely matching the 3' UTR of one of the gene family. For regions of mismatch between the highly similar coding regions, siRNA populations that derive from the direct target gene (act-1 in this case) can be distinguished by sequence from potential tertiary siRNA populations that derive from the remaining genes in the family. (B–D) Genome-wide gene by gene comparison of antisense siRNA matches. Each point represents one C. elegans mRNA transcript from a duplicate-subtracted reference sequence set (Wormbase). Vertical position on each plot represents antisense siRNA counts for that gene in small RNA captured following act-1 3' UTR RNAi as shown in A. Horizontal positions represent antisense siRNA counts from comparable RNAi experiments with non-actin triggers. (B) Horizontal axis: sel-1 mismatched trigger (see Figure 1), (C, D) Horizontal axis: ben-1 triggers R1934U and R1934D (Figure 3). (E) Distribution of antisense siRNAs matching the act-1 transcript following RNAi feeding with the act-1 3' UTR trigger (blue line). Coverage at each base position is normalized to aggregate antisense siRNA counts for all C. elegans genes, and a smoothing window of 61b is used. Enrichment of antisense counts throughout the act-1 gene is evident in comparison to control samples in which RNAi was triggered to a non-actin target (green, red, orange lines). (F) Equivalent composite coverage plot of antisense siRNAs deriving from act-2, act-3, act-4, or act-5 and not from act-1. Given the high signal of act-1 matching siRNAs in E and the known error frequency in high throughput RNA-seq methods, there is some possibility that a fraction of the signal in F represents mis-sequencing of act-1-matched siRNAs. This value is estimated (magenta line, F) by generating a series of 72 "faux-actin" genes (Supplementary Experimental Procedures). (See also Figure S3.)
Using the 5’-P-independent method of siRNA capture, we sequenced 14,759,669 sRNAs of which 462,403 aligned perfectly to at least one of the five actin mRNAs (Figure 4). As expected, we found that many of these mapped to the act-1 3' UTR trigger region (Figure 4E). siRNAs mapping to the 3' UTR regions of the other actin genes were present at low levels comparable to those observed in experiments where animals were grown with non-actin dsRNA triggers (Figures 4B, C, D,). In the coding regions, we saw at most slight increases over background in siRNAs that matched other actin genes but not act-1, while large increases were seen among act-1 matching siRNAs (Figure 4F). These observations provide further support for a dearth of tertiary siRNAs.
Genetic requirements in a two-stage RNAi response
Several conserved protein factors have been shown to be required for RNAi in C. elegans. To place these factors and the resulting siRNA families in a functional pathway for RNAi, we endeavored to assess genetic requirements for primary and secondary siRNA generation (as assessed by sequence and structure) using the fully duplex sel-1 trigger (Figure 1A) carrying mismatches to the target at 25 nt intervals along its length. sRNA pools were captured and sequenced using the Illumina platform and both 5’-P-independent and 5’-monoP -enrichment sRNA capture methods (see above).
i) rde-4 is required for primary siRNA accumulation
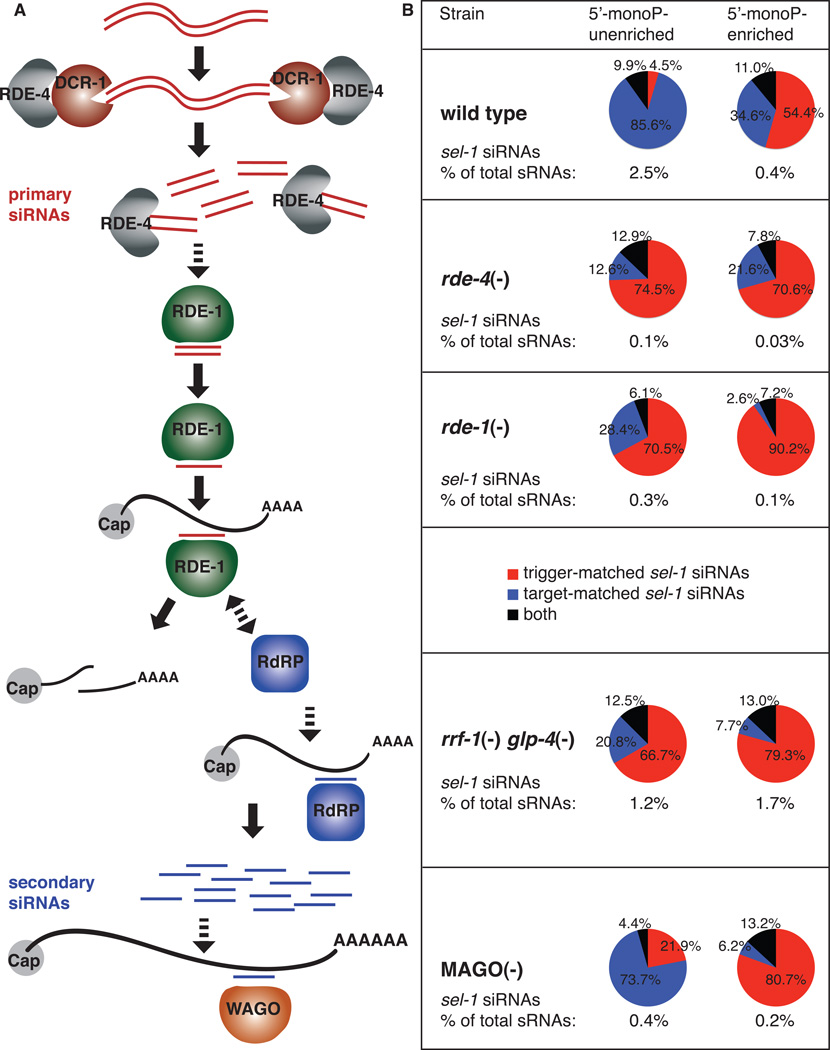
The dsRNA binding protein RDE-4 is predicted to function early in the RNAi pathway with DCR-1 in the generation of primary siRNAs (reviewed in Boisvert and Simard, 2008). Consistent with this role, both trigger-matched (primary) and target-matched (secondary) siRNAs were significantly reduced in rde-4(ne299) mutant animals. rde-4(ne299) is the only mutant tested in which the primary siRNAs were affected (Tables 1 and 2, Figures 5 and S4).
Figure 5.
Genetic requirements for primary and secondary siRNA generation. (A) Model for RNAi in C. elegans. Exogenous dsRNA is processed by DCR-1 and RDE-4 into primary siRNAs. Short dsRNAs are then delivered to RDE-1, potentially through an additional role of the dsRNA binding factor. RDE-1 then cleaves the passenger strand, allowing subsequent maturation into active RISC. The RDE-1 RISC then recruits the RdRP (RRF-1 or EGO-1) to the target RNA where the latter engages in secondary siRNA synthesis using the target as template. Primary siRNA-complexed RDE-1 may endonucleolytically destroy targ et RNAs, while secondary siRNAs complex with the WAGOs and presumably further downregulate the target RNA via an unknown mechanism. (B) Mutations in RNAi factors affect primary and/or secondary siRNA synthesis. Pie charts indicate fraction of antisense sel-1 siRNAs comprised by trigger-matched (red), target-matched (blue), and ambiguous (black) antisense sel-1 siRNAs. Numbers below pie charts indicate percentage of total small RNAs (sRNAs) comprised of sel-1 siRNAs. (See also Figure S4.)
The depletion of rde-4-dependent RdRP products also revealed a population of 28 nt target-matched siRNAs (Figure S1C, right panel). Interestingly, a 28–29 nt peak of antisense matched siRNAs was uncovered in all tested RNAi-defective mutants (Figure S1). Such siRNAs appeared to cluster at the 3’ end of the trigger homology region in the mutants but are distributed throughout this region in wild type animals (Figure S4E).
ii) rde-1 cleavage activity is not required for rde-1-dependent secondary siRNA accumulation
The Argonaute protein RDE-1 sits at a pivotal point in the RNAi mechanism in C. elegans being required for full wild type accumulation of siRNAs but being dispensable for processing of the dsRNA trigger (Sijen et al., 2001; Parrish and Fire, 2001). In addition, Yigit et al. (2006) have shown RDE-1 to complex with primary siRNAs. In our assay with the mismatched sel-1 trigger, we found rde-1 to be required primarily for target-matched siRNA accumulation, consistent with the composition of secondary siRNAs; target-matched siRNAs were reduced 96.5 fold whereas trigger-matched siRNAs were reduced only 1.45 fold in rde-1 (ne300) animals (Table 1, Figure 5). Despite the low level, the siRNA signal in sel-1 dsRNA-triggered rde-1 mutant animals remained trigger dependent, with the residual sel-1 siRNA level >10-fold above that observed in rde-1 animals not subjected to sel-1 RNAi (Table 1 and Gent et al. 2010). Curiously, we found a striking redistribution of the remaining secondary siRNAs in rde-1 animals (Figure S4H). In wild type animals responding to an RNAi trigger, secondary siRNAs corresponding to sequence upstream of the trigger region accumulated to a higher level than those downstream; the ratio of upstream siRNAs per nucleotide to downstream siRNAs per nucleotide is 9.51. In rde-1 animals, this ratio was significantly different at 2.85 (p=0.03). For comparison, a multiple Argonaute deletion strain (MAGO animals; see below) showed a ratio of 10.51 (not significantly different from wild type animals). Furthermore, secondary siRNAs in wild type animals were most abundant nearest the trigger region, tapering off with distance from this region. In rde-1 animals, however, there appeared to be an even distribution of secondary siRNAs throughout the target mRNA (Figure S4H). Thus, the remaining RdRP products do not appear to be biased for the trigger homology region in rde-1 mutants.
We propose, from the above results and prior studies that RDE-1 plays a key role in the recruitment of the amplification machinery rather than an exclusive role in the destruction of target RNAs (Figure 5; Sijen et al., 2001; Parrish and Fire, 2001; Yigit et al., 2006; Pak and Fire, 2007; Sijen et al., 2007). Consistent with this notion, we found that catalytic activity of RDE-1 is not essential for secondary siRNA accumulation; rde-1 (ne300) animals expressing RDE-1 that had been mutated in key putative catalytic residues (RDE-1 AAA; Steiner et al., 2009) resembled wild type animals in the distribution of siRNAs in our mismatched trigger assay (Figure S4J, K), with a retention of 9% of wild type levels in assays for antisense target-matched siRNA accumulation (by contrast RDE-1 null mutants retained less than 1% of wild type antisense target-matched siRNA levels; Table 1). We found that RDE-1 AAA-expressing animals to be compromised, but not completely deficient, in RNAi as assayed by monitoring twitching upon presentation of an extended duplex dsRNA for unc-22 (Figure S4L; Steiner et al., 2009). We conclude that the RDE-1 catalytic residues, although important for robust RNAi, are not essential for the elaboration and pattern of secondary siRNAs in the presence of a homologous target mRNA.
iii) The redundant roles of the RdRPs in C. elegans
Of the four putative RdRPs encoded by the C. elegans genome, rrf-1 and ego-1 were initially shown to be required for RNAi efficacy in the soma and germline, respectively (Smardon et al., 2000; Sijen et al., 2001). By generating rrf-1 mutant animals which lack the bulk of their germline tissue (through raising rrf-1(pk1417Δ);glp-4(bn2ts) animals at a restrictive temperature; Beanan et al., 1992) we could examine dsRNA responses under conditions where combined rrf-1/ego-1 activity was minimized. (The glp-4(bn2ts) mutation results in the genetic ablation of all mature germ cells at the restrictive temperature.) Antisense target-matched siRNAs were reduced 84-fold in this background, while antisense trigger-matched siRNAs were reduced only 1.4-fold (5’-P-independent sequences, Figure 5, Table 1). Apparently RdRP activity is not completely absent in rrf-1 glp-4 animals, giving rise to a subpopulation of secondary siRNAs that are similar to those in wild type animals in distribution along the sel-1 mRNA, in length distribution, and in 5’ nucleotide bias (Figures 5, S4M-P, S1D, N right panel). These findings are consistent with the recent observation of RNAi activity in the intestine in rrf-1 animals (Kumsta and Hansen, 2012).
iv) Nematode-specific Argonautes are required for secondary siRNA accumulation
The C. elegans genome encodes an unusual diversity of putative Argonaute proteins. Eighteen of these 27 homologs fall into a worm-specific clade called the WAGOs. At least four WAGOs (ppw-1, sago-1, sago-2, F58G1.1) were previously shown to function in an additive fashion, downstream of the Argonuate gene rde-1 (see above), bound to secondary siRNAs. If these WAGOs were also required for the accumulation of secondary siRNAs, we would expect to find a decrease in target-matched siRNAs in a strain deleted in these genes. Using the RNAi-insensitive "MAGO" strain fromYigit et al. (2006) (deleting ppw-1, sago-1, sago-2, F58G1.1, C06A1.4, and M03D4.6), we found a 21-fold decrease in accumulation of secondary siRNAs (antisense target-matched siRNAs; Table 1, Figures 5, S4Q-T); in these assays, the MAGO strain also exhibited a modest decrease in primary siRNA accumulation (<4-fold; Figure 5, Table 1).
DISCUSSION
Feed-forward restriction of RNAi amplification
In this work, we demonstrate that signal amplification during dsRNA-triggered gene silencing in C. elegans is attenuated through a scrupulous distinction between RNA triggers, templates, and products of RdRP-driven amplification.
In C. elegans exposed to external dsRNA, Dicer cleavage of the trigger produces a surprisingly small number of siRNAs (Parrish et al., 2000; this work). These appear insufficient for a full gene silencing response, serving instead as guides to recruit RdRPs to target messages. The participation of RdRP enzymes that can physically produce novel populations of RNA effectors provides the process of RNAi both an opportunity for increased efficacy (through greatly increased populations of effectors) and increased danger (through the possibility of large populations of unwanted effector RNAs, potentially replicating through the activity of the RdRP) (Sijen et al., 2001; Alder et al., 2003; Pak and Fire, 2007; Sijen et al., 2007; Aoki et al., 2007; Bergstrom et al., 2003). The worm evidently employs a rather remarkable set of controls to take advantage of the amplification possibilities inherent in RdRP activities without succumbing to them.
As a first protection, despite their engagement with the RNAi machinery, the initial dsRNA trigger and Dicer products were not themselves templates for detectable RdRP activities. Instead, and distinguished perhaps through their 5' structure or route of delivery to the RNAi machinery, the major role of primary siRNAs appears to be the instigation of secondary siRNA generation through RdRP activity on transcripts identified by the primary RISC complexes as targets.
Secondary siRNAs likewise appear to play a highly restricted role in the silencing reaction. Incapable of instigating further siRNA generation, these effectors target homologous transcripts for degradation in a process that remains to be elucidated.
The model shown in Figure 5 proposes that the restraints on the RNAi system described in this work are, at least in part, responsible for the combination of high sensitivity and exquisite specificity of responses to foreign RNA in C. elegans. First, in allowing only primary siRNAs the role of RdRP recruitment, indiscriminate generation of novel siRNAs is minimized and potential exponential signal amplification following RdRP engagement avoided. This goal is enhanced by restricting RdRP activity to the target RNA; in particular, only effective (antisense) siRNAs are produced in the secondary response. Second, the secondary siRNAs that are produced are allowed to serve as guides for destruction of target transcripts, but not to serve as either templates for RdRP or guides for RdRP recruitment. This dual restriction prevents a feed-forward situation that could result in ballooning amplification of secondary siRNAs under circumstances in which a continuous population of target was being synthesized.
While our results argue that secondary siRNAs (those templated on target molecules interacting with a primary siRNA) comprise the bulk of the siRNA response radiating from an initial interaction, the observations do not rule out a population of tertiary siRNAs, either small in number, radiating much less substantially from the initial interactions sites, or derived elsewhere from the transcriptome, e.g. from regions with limited homology.
A model for RNAi in C. elegans
One aspect of this analysis has been to refine and critically test current models for RNAi responses in C. elegans based on the explicit sequence-based detection of primary and secondary siRNAs. Our data support a model for RNAi in C. elegans comprising (i) processing of exogenous dsRNA trigger molecules into a pool of primary siRNAs by a complex including the dsRNA-binding factor RDE-4 and the DCR-1 nuclease (Grishok et al., 2001, Knight and Bass 2001, Tabara et al., 2002, Parrish and Fire, 2002, Duchaine et al., 2006). (ii) transfer of 5'-monoP siRNA to RDE-1 (Yigit et al. 2006), (iii) interactions between the RDE-1::primary siRNA complex with target RNA that guide RdRP machinery to synthesize short antisense transcripts (secondary siRNAs) from target RNA (Sijen et al., 2007, Pak and Fire 2007), (iv) transfer of secondary siRNAs to WAGO Argonautes, which then prosecute a guided destruction of the pool of target RNAs (Yigit et al., 2006). We discuss each step in turn below.
(i) Primary siRNA generation
Primary siRNA generation has been proposed to involve a complex including DCR-1 and RDE-4. Of the RNAi factors we tested, only RDE-4 was required for full accumulation of trigger-matched siRNAs. Available rde-4 mutants retain residual ability to trigger silencing (Parrish et al., 2002; Habig et al., 2008), consistent with a low level of trigger-matched siRNAs that we observed in the rde-4 mutant. Whether this residual level represents an alternative pathway for siRNA generation or simple leakiness of the mutants remains to be determined. Because DCR-1 is required for viability at diverse stages (Grishok et al., 2001; Maniar and Fire, unpublished), explicit confirmation of a DCR-1 role in primary siRNA generation was not possible using the mismatched trigger assay. A lack of dramatic decrease in trigger-matched siRNA levels in our analysis of RDE-1 and WAGO mutant backgrounds suggests that primary siRNAs may be stable without protection by Argonaute complexes.
(ii) The transition from primary to secondary siRNA generation
Data fromYigit et al. (2006) have suggested a model in which primary siRNAs carry out their action in the context of complexes with the RDE-1 Argonaute. Consistent with this model, the pools of primary siRNAs produced in rde-1 mutant backgrounds are not capable of generating a secondary siRNA response (Sijen et al., 2001; this work). It is interesting to note that the residual level of primary siRNAs in the absence of rde-4 is not sufficient to trigger substantial secondary siRNA accumulation. This is consistent with an additional role for rde-4 in this transition. Indeed, in Drosophila, the dsRNA binding protein and Dicer binding partner, R2D2, assists the loading of the siRNA into the Argonaute-containing complex RISC (Liu et al., 2003; Marques et al., 2010; Okamura et al., 2010). In an analogous manner, RDE-4 may steer the newly processed primary siRNA into RDE-1 over the other Argonautes thus engendering the exclusivity RDE-1 exhibits in its choice of primary over secondary siRNA. Alternatively, but not mutually exclusively, it is possible that the selectivity of RDE-1 is based on structural constraints which might only accommodate 5’-monoP siRNAs (primary siRNAs) and not 5’-PPP species (secondary siRNAs). To this end, structure determination of RDE-1 and the WAGOs would be of great interest.
How does RDE-1, once charged with a primary siRNA, recruit the RNA copying machinery? One model that can now be rejected (based on the retention of secondary siRNAs in the absence of the RDE-1 cleavage triad) is one in which the RDE-1 RNAse H activity cleaves the target RNA and the free RNA termini somehow recruit the RdRP. As a working model, a physical interaction between RDE-1 and the RdRP (Blanchard et al., 2006) may tow the RdRP on to the target RNA. As an alternative, it is possible that RDE-1 somehow modifies the template RNA in a manner that does not require its cleavage, creating accessible sites for RdRP entry. Both of these models allow for a non-exclusive direction bias of RdRP recruitment such that secondary siRNAs are generated predominantly upstream of the original trigger but also downstream to a lesser extent.
Interestingly, we also found that a low level of secondary siRNAs was generated in an rde-1-independent fashion. This siRNA pool lacks the characteristic distribution of rde-1-dependent secondary siRNAs as a function of position within the mRNA, suggesting a mechanism that is less efficient than RDE-1 in tethering the RdRP to the vicinity of the trigger region.
(iii) Secondary siRNA generation
Secondary siRNAs are most likely generated by the RdRP as short 5'-PPP transcripts on an RNA template (Aoki et al., 2007; Pak and Fire, 2007; Sijen et al., 2007). In this work, we have demonstrated a strong preference for C. elegans to use the primary target RNA as the template for RdRP.
(iv) Secondary siRNA function
The WAGOs or secondary Argonautes have previously been shown to bind secondary siRNAs and we show here that they are required for secondary siRNA accumulation (Yigit et al., 2006). After the WAGOs bind and stabilize the secondary siRNAs, the resulting complexes can evidently target naive transcripts (Sijen et al., 2001; Alder et al., 2003). The fate of the secondary siRNA-WAGO targeted mRNAs is not clear; curiously none of the WAGOs contain the DDH residues believed to mediate cleavage of the target RNA strand.
EXPERIMENTAL PROCEDURES
Induction of RNAi
Feeding and soaking-based RNAi was carried out using standard procedures (see Supplementary Experimental Procedures).
Small RNA capture
sRNAs were captured using a 5'-monoP-enrichment method (Lau et al., 2001) and a 5'-P-independent method (Gent et al., 2010) as described. Numerical values for siRNA coverage in figures and tables are normalized as noted. Small RNA capture and sequencing protocols have the capability of detecting numerous noncanonical or non-siRNA small RNA populations including tRNA, rRNA, and mRNA fragments and other yet-to-be-characterized species. We have in general not included such RNAs in our counting and analysis, due to some differences in recovery in different samples. Of particular note related to sel-1 was a 29 nt small RNA sequenced 15,261 times in the MAGO mutant background; we have not further characterized the origin of this RNA.
Characterization of secondary siRNAs
Analysis of 5’ nucleotide composition of target-matched siRNAs reveals a distinct preference for G that is not observed at adjacent positions or in the other sRNA classes (Figure S1, right panel; Figure S2). Moreover, the majority of these siRNAs appear to be 22 nt long (Figure S3A, right panel). Interestingly, both secondary siRNAs and endogenous 22G siRNAs also appeared to be 5’-PPP as the former are depleted in sequenced pools generated using the 5’-monoP-enrichment protocol (Figure 1B and C, Figure S3A and F, right panel; Gent et al., 2010); antisense target-matched siRNAs make up 87.9% and 50.3% of total antisense sel-1 siRNAs in the 5’-P-independent and 5’-monoP-enriched data sets, respectively. These results indicate that the majority of the antisense target-matched siRNAs share the salient features of secondary endogenous siRNAs, with a triphosphorylated 5' end and biases of 5'G and 22 nt length (Han et al., 2009; Gent et al., 2010).
Supplementary Material
HIGHLIGHTS.
RNAi amplification triggers are restricted to the initial siRNA population.
Amplification templates are limited to target RNAs.
Restriction is enhanced by limiting the roles of amplification products.
ACKNOWLEDGEMENTS
We thank D. Liu, L. Zhang, M. Stadler, A. Lamm, H. Zhang, D. Wu, L. Gracey, R. Li, I. Gabdank, K. Artiles, S. Gu, J. Gent, C. Kumsta, M. Hansen, P. Parameswaran, C. Smith, Z. Weng, R. Alcazar, W. Lui, P. Lacroute, A. Sidow, A. Jheon, O. Klein, anonymous reviewers and NIGMS (R01GM37706) for help and support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Sequence data are available at SRA (Accession Number SRA057687).
SUPPLEMENTAL INFORMATION
This section contains four supplemental figures (Figures S1 through S4), supplementary experimental procedures and supplementary references.
REFERENCES
- Alder MN, Dames S, Gaudet J, Mango SE. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA. 2003;9:25–32. doi: 10.1261/rna.2650903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bergstrom CT, McKittrick E, Antia R. Mathematical models of RNA silencing: Unidirectional amplification limits accidental self-directed reactions. Proc. Natl. Acad. SciUSA. 2003;100:11511–11516. doi: 10.1073/pnas.1931639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard D, Hutter H, Fleenor J, Fire A. A differential cytolocalization assay for analysis of macromolecular assemblies in the eukaryotic cytoplasm. Mol Cell Proteomics. 2006;5:2175–2184. doi: 10.1074/mcp.T600025-MCP200. [DOI] [PubMed] [Google Scholar]
- Boisvert ME, Simard MJ. RNAi pathway in C. elegans: the argonautes and collaborators. Curr. Top. Microbiol. Immunol. 2008;320:21–36. doi: 10.1007/978-3-540-75157-1_2. [DOI] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, Yates JR, 3rd, Mello CC. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21-and 22-nucleotide RNAs. Genes Dev. 2001b;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001c;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix MA, Ashe A, Piffaretti J, Wu G, Nuez I, Bélicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD, Sanroman M, Miska EA, Wang D. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Files JG, Carr S, Hirsh D. Actin gene family of Caenorhabditis elegans. J.Mol. Biol. 1983;164:355–375. doi: 10.1016/0022-2836(83)90056-6. [DOI] [PubMed] [Google Scholar]
- Fischer SE. Small RNA-mediated gene silencing pathways in C. elegans. IntJBiochem. Cell Biol. 2010;42:1306–1315. doi: 10.1016/j.biocel.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Gent JI, Schvarzstein M, Villeneuve AM, Gu SG, Jantsch V, Fire AZ, Baudrimont A. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNA interference. Genetics. 2009;183:1297–1314. doi: 10.1534/genetics.109.109686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, Chen CC, Chaves DA, Duan S, Kasschau KD, Fahlgren N, Yates JR, 3rd, Mitani S, Carrington JC, Mello CC. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig JW, Aruscavage PJ, Bass BL. In C. elegans high levels of dsRNA allow RNAi in the absence of RDE-4. PLoS One. 2008;3:e4052. doi: 10.1371/journal.pone.0004052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. SciUSA. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Hansen M. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One. 2012;7:e35428. doi: 10.1371/journal.pone.0035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landel CP, Krause M, Waterston RH, Hirsh D. DNA rearrangements of the actin gene cluster in Caenorhabditis elegans accompany reversion of three muscle mutants. J. Mol. Biol. 1984;180:497–513. doi: 10.1016/0022-2836(84)90024-x. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lipardi C, Paterson BM. Identification of an RNA-dependent RNA polymerase in Drosophila involved in RNAi and transposon suppression. Proc. Natl. Acad. SciUSA. 2009;106:15645–15650. doi: 10.1073/pnas.0904984106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Baggett JJ, Perumov N, Bauer RA, Januszewski T, Schriefer L, Waddle JA. ACT-5 is an essential Caenorhabditis elegans actin required for intestinal microvilli formation. Mol. Biol. Cell. 2005;16:3247–3259. doi: 10.1091/mbc.E04-12-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar JM, Fire AZ. EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr. Biol. 2011;21:449–459. doi: 10.1016/j.cub.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Kim K, Wu PH, Alleyne TM, Jafari N, Carthew RW. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol. 2010;17:24–30. doi: 10.1038/nsmb.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacambe AM, Nikic S, Picault N, Rémoué K, Sanial M, Vo TA, Vaucheret H. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Nowotny M, Yang W. Structural and functional modules in RNA interference. Curr. Opin. Struct. Biol. 2009;19:286–293. doi: 10.1016/j.sbi.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics. 2009;183:1283–1295. doi: 10.1534/genetics.109.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen K, Plasterk RH. econdary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. econdary siRNAs result from unprimed RNA synthesis and form a distinct class. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2009;16:207–211. doi: 10.1038/nsmb.1541. [DOI] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte DJr. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci USA. 2010;107:3582–3587. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH, Munro E, Lyczak R, Bowerman B. Conditional dominant mutations in the Caenorhabditis elegans gene act-2 identify cytoplasmic and muscle roles for a redundant actin isoform. Mol. Biol. Cell. 2006;17:1051–1064. doi: 10.1091/mbc.E05-09-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lamm AT, Fire AZ. Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat. Struct. Mol. Biol. 2011;18:1094–1101. doi: 10.1038/nsmb.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.