Abstract
Long noncoding RNAs (lncRNAs) have been detected in nearly every cell type and found to be fundamentally involved in many biological processes. The characterization of lncRNAs has immense potential to advance our comprehensive understanding of cellular processes and gene regulation, along with implications for the treatment of human disease. The recent ENCODE (Encyclopedia of DNA Elements) study reported 9,640 lncRNA loci in the human genome, which corresponds to around half the number of protein-coding genes. Because of this sheer number and their functional diversity, it is crucial to identify a pool of potentially relevant lncRNAs early on in a given study. In this review, we evaluate the methods for isolating lncRNAs by immunoprecipitation and review the advantages, disadvantages, and applications of three widely used approaches – microarray, tiling array, and RNA-seq – for identifying lncRNAs involved in gene regulation. We also look at ways in which data from publicly available databases such as ENCODE can support the study of lncRNAs.
Keywords: Immunoprecipitation, ENCODE, Long noncoding RNA, Microarray, RNA-seq, Tiling array
Long noncoding RNAs
Long noncoding RNA (lncRNA) is operationally defined as RNA longer than 200 bases that does not encode mRNA, rRNA or tRNA [1,2]. Although several lncRNAs have been sporadically identified and characterized in the past 20 years, genome-wide identification of lncRNAs has only recently become possible with the advent of high-throughput sequencing technologies of cDNA (RNA-seq). Evidence that this field is gaining momentum can be seen in the most recent report of the ENCODE (Encyclopedia of DNA Elements) project published in September 2012, which described 9,640 lncRNA loci in comparison to 20,687 protein-coding genes in 15 human cell lines [3-5]. This ratio of lncRNAs and protein-coding genes underscores the potential magnitude and diversity of the biological effects mediated by lncRNAs. Indeed, despite the fact that only about 100 lncRNAs have been functionally characterized to date [4], it has become clear that lncRNAs are involved in almost every aspect of cellular and molecular biology. LncRNAs control cell differentiation, development, cancer progression, and cell metabolism, among other cell functions. At the gene expression level, lncRNAs regulate all processes of RNA metabolism including chromatin modification, transcription, splicing, RNA transport, and translation. LncRNAs themselves are transcribed from intergenic regions, exons, introns, and their overlapping regions (Figure 1A and 1B). At the mechanistic level, lncRNAs serve as “scaffolds” providing platforms to assemble RNA-protein complexes, “guides” to recruit RNAprotein complexes to target genes, and “decoys” by binding to and sequestering regulatory proteins away from their target DNA sequences [1,2]. Given the recent appreciation for the biological importance of lncRNAs, it is now clear that, regardless of the research project or field, one needs to ask whether lncRNAs are essential mechanistic components of the biological process under consideration. The first step to addressing this question is to identify lncRNAs that are potentially relevant to the research field. The current review article provides an overview of four widely used approaches to identify lncRNAs involved in gene regulation – immunoprecipitation of RNA and chromatin, microarray, tiling array, and RNA-seq – and discusses the advantages and disadvantages of each approach. These approaches, which are not mutually exclusive and are often combined in a single study, have been successfully used to identify lncRNAs (Table 1). The focus of this review is gene regulation, which has been the main area of functional studies of lncRNAs; however, lncRNAs are also involved in the organization of cellular structure and subcellular organelles. For further information on these and other aspects of lncRNA biology, readers are referred to the following recent reviews: [1,2,6-11].
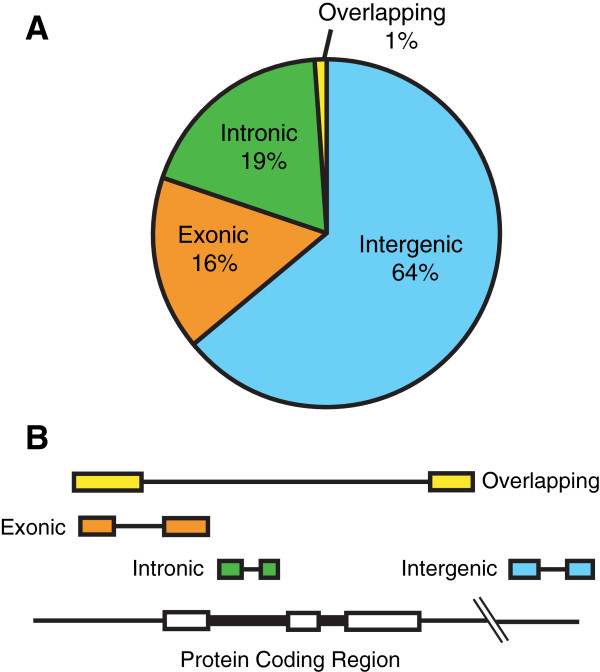
Figure 1.
Overview of lncRNA populations depending on the locations on the genome. LncRNAs can be categorized into subgroups of intergenic, exonic, intronic, and overlapping according to where they are found relative to nearby protein-coding genes. (A) Proportion of lncRNA subgroups [3]. (B) Location of each type of lncRNA.
Table 1.
Examples of lncRNAs discovered with various approaches described in the text
| lncRNA name | Types of RNA | lncRNA identification methods* | Reference |
|---|---|---|---|
| ANRIL |
RNA-ChIP (UV cross-linking) and CLIP |
RT-PCR |
[12] |
| RepA |
RIP (no cross-linking) |
Northern blotting |
[13] |
| TRE-1, -2 and -3 |
RNA-ChIP (cross-linked with formaldehyde) |
RT-PCR |
[14] |
| Xist |
RIP (no cross-linking) |
RT-PCR |
[13] |
| LincRNA-EPS |
Polyadenylated RNA |
Microarrays of 10,802 lncRNAs and RNA-seq |
[15] |
| lincRNA-SFMBT2, lincRNA-RoR, and lincRNA-VLDLR |
Total RNA |
Microarrays of 900 lincRNAs |
[16] |
| lncRNA_ES1, ES2 and ES3 |
Total RNA |
Microarrays of 6,671 lncRNAs |
[17] |
| ncRNA-a1-7 |
Polyadenylated RNA |
Microarrays of 3,019 lncRNAs |
[18] |
| HOTAIR |
Polyadenylated RNA |
Tiling arrays of 39 HOX genes |
[19] |
| RIP (no cross-linking) |
Validation with RT-PCR |
||
| HOTAIRM1 |
Polyadenylated RNA |
Tiling arrays of HOXA genes |
[20] |
| lincRNA-p21 |
Polyadenylated RNA |
Tiling arrays of 400 lincRNAs |
[21] |
| Mistral |
RNA-ChIP (no cross-linking) |
Tiling arrays of chromosomes 6, 8 and 16 |
[22] |
| 300 lincRNAs |
RIP (no cross-linking) |
Tiling arrays of 900 lincRNAs |
[23] |
| 1,600 lincRNAs |
Polyadenylated RNA |
Tiling arrays of 350 K4-K36 domains |
[24] |
| ANCR |
Polyadenylated RNA |
RNA-seq |
[25] |
| BANCR |
Polyadenylated RNA |
RNA-seq |
[26] |
| 5 lncRNAs |
CLIP |
RNA-seq |
[27] |
| 216 lincRNAs |
RIP (no cross-linking) |
RNA-seq |
[28] |
| RIP (UV cross-linking) |
Validation with RT-PCR |
||
| >8,000 lncRNAs | Polyadenylated RNA | Analysis of existing RNA-seq data | [29] |
* LncRNAs are categorized based on the methods of their discovery: gene-specific approaches (RT-PCR and Northern blotting), microarrays, tiling arrays and RNA-seq.
Collection of lncRNAs by immunoprecipitation
The first challenge in studying lncRNAs is how to collect RNA pools that potentially contain lncRNAs of interest. One can prepare RNA pools by simply isolating total RNA from cells or tissues in an unbiased manner; however, immunoprecipitation-based approaches are also commonly used to enrich lncRNAs associated with specific proteins. RNA immunoprecipitation (RIP) can be performed with or without cross-linking whole cellular components before making cell extracts. Without cross-linking, one can isolate lncRNA complexes already existing in soluble form and those that can be readily dissociated from chromatin. Zhao et al. used RIP of polycomb repressive complex 2 (PRC2), a key regulator of epigenetic silencing, without cross-linking and co-immunoprecipitated the lncRNA Xist, which was amplified by RT-PCR [13]. Using the same procedure, they discovered co-immunoprecipitation of the novel lncRNA RepA, which is transcribed within the Xist locus [13]. To identify unknown lncRNAs by RIP, the co-immunoprecipitated RNA pool can be applied to microarray analyses or RNA-seq, as described later [19,23,28]. If one needs to exclude the possibility of indirect interactions between lncRNAs and proteins through their binding to neighboring DNA sequences, the immunoprecipitated materials can be treated with RNase H (digests RNA in RNA-DNA hybrids) and DNase I prior to elution of co-immunoprecipated molecules. As a control, treatment with RNase A, RNase I (both digest single-stranded RNA), and/or RNase V1 (double-stranded RNA) should abolish the co-immunoprecipitation [22,28].
There are several RIP techniques that employ cross-linking. RIP is sometimes performed after ultraviolet (UV) irradiation of cells, which cross-links RNA and protein (pyrimidines and Cys, Lys, Phe, Trp, and Tyr) but not protein and protein [30]. This unique feature allows for the recovery of lncRNAs that directly interact with the immunoprecipitated protein. Taking advantage of this high specificity, UV cross-linking is used to identify the domains within an RNA molecule responsible for the interaction with the protein partner. For instance, Zhao et al. irradiated cells with 254 nm UV prior to making cell extracts and immunoprecipitated PRC2 to identify directly associated lncRNAs [28].
A related variation is called CLIP (cross-linking and immunoprecipitation), which was designed to isolate a protein-interacting domain within a given RNA molecule after using a stringent wash to reduce non-specific binding [30]. In a typical CLIP experiment, extracts are made from cells after UV-irradiation and treated with RNase to retain only the RNA region protected by the interacting protein. The partially digested RNA pool is then tagged with a 3’ linker and also radio-labeled. After purification of the protein with immunoprecipitation, SDS gel electrophoresis, autoradiography, and band excision, the bound protein is removed by proteinase K treatment. The exposed RNA is tagged with a 5’ linker and PCR-amplified to identify the sequence. CLIP was successfully used to immunoprecipitate five intronic lncRNAs directly associated with the PRC2 complex [27].
Cross-linking with UV or formaldehyde followed by fragmentation of chromatin is used to immunoprecipitate RNA-chromatin complexes (RNA-chromatin immunoprecipitation or RNA-ChIP) [12,14,22]. While this approach potentially detects false-positive interactions between RNA and protein through DNA as described above, it can be useful to identify lncRNAs that bind to specifically modified histones which require chromatin fragmentation for extraction.
For any of these immunoprecipitation-based approaches, specificity and affinity of the antibodies are decisive factors for the success or failure of the projects. While the specificity of the antibodies is commonly verified by detecting only one band in western blotting, the antibodies may react with other proteins when detergents are used at a low concentration during immunoprecipitation. One solution to address the specificity issue is to use multiple antibodies against the same protein and select reproducibly co-precipitated lncRNAs for further study. Similarly, immunoprecipitation of several different subunits within a single protein complex is also an option to identify lncRNAs that are likely to be genuinely interacting with the complex.
Identification of lncRNAs with microarrays
Microarray-based approaches and RNA-seq are two of the most commonly used genome-wide screening methods to identify lncRNAs that might be relevant to a specific biological question. Although a tiling array should be included in the microarray section by definition, it will be discussed separately in the next section as it is frequently used for different purposes. Because traditional microarrays can only detect the presence or absence of known lncRNAs in an RNA pool, they are inherently incapable of identifying novel lncRNAs. Inability of distinguishing different splicing variants is another disadvantage of microarrays unless probes encompassing exon-exon junctions are present on the chip. However, given the cost and complexity of the analysis of RNA-seq data, microarray remains the first choice in many applications [15-18]. In particular, since the identification of 9,640 lncRNA loci as part of the ENCODE project, the comprehensiveness of microarrays for human lncRNAs has been drastically improved.
Data generation with microarrays is relatively easy compared to the subsequent step of selecting potentially important lncRNAs from the positive probes on the arrays because the majority of identified lncRNAs remain uncharacterized. Here, the work by Loewer et al. serves as an exemplary case study of how to narrow down lncRNA candidates relevant to one’s interest, in this case, association with pluripotency [16]. Loewer and colleagues designed a microarray containing 900 long intergenic noncoding RNAs (lincRNAs) and hybridized them with total RNA prepared from several different cell lines to identify induced pluripotent stem cell-specific lincRNAs. In their case, the selection criteria included the genomic location (close to the binding sites of pluripotency transcription factors), nearby presence of epigenetic markers for active transcription, behavior of the lincRNA level during differentiation, and consequence of up- and downregulation in terms of the maintenance or acquisition of pluripotency. Similar concepts can be widely applied to selecting lncRNAs in other contexts.
Identification of lncRNAs with tiling arrays
Unlike traditional microarrays, DNA tiling arrays contain oligonucleotide probes encompassing an entire length of a defined DNA region. Resolution of the hybridized genomic DNA sequence can be adjusted by changing the length of the overlapping sequences between two neighboring probes. A major advantage of using tiling arrays is their capacity to identify novel lncRNAs in a selected DNA region without prior knowledge of their precise locations within the region. The DNA region can be defined by the residing genes of interest. For instance, Rinn et al. focused on lncRNAs expressed in the region of the human HOX genes and compared skin fibroblasts isolated from different anatomical regions of the body [19]. They printed 400,000 probes of 50 bases in length with each probe overlapping the next one by 45 bases to cover all four human HOX gene clusters. This configuration allowed for the identification of hybridized DNA sequences at 5-base resolution. Polyadenylated RNAs prepared from fibroblasts were then hybridized to the tiling arrays, resulting in the discovery of the lncRNA HOTAIR transcribed from an intergenic region within the HOXC cluster. A similar HOX tiling array was used to identify lncRNAs specifically expressed in metastatic breast carcinoma [31]. The lncRNA HOTAIRM1 was discovered in the intergenic region between the HOXA1 and HOXA2 genes with commercially available tiling arrays covering the human HOXA gene cluster [20].
The DNA regions of interest can also be determined by the unique epigenetic features of the regions. Actively transcribed genes are enriched with trimethylation of lysine 4 on histone H3 at their promoters and trimethylation of lysine 36 on histone H3 in their coding regions [32], which are collectively called K4-K36 domains. Taking advantage of this knowledge, Guttman et al. prepared DNA tiling arrays with 2.1 million oligonucleotide probes representing 350 K3-K36 domains and hybridized them with polyadenylated RNA to identify 1,600 mouse lincRNAs [24]. A similar tiling array was used to identify 300 lincRNAs in human cells [23]. Thus, the tiling array approach is highly useful to comprehensively detect any transcripts, including lncRNAs, transcribed from a defined DNA region at a high resolution in an unbiased manner. However, unless the target region is reasonably limited, a potential drawback of the tiling array approach is its high cost. Tiling arrays generally need to be custom-made to meet diverse needs, which further raises the cost and slows down manufacturing the arrays.
Identification of lncRNAs with RNA-seq
RNA-seq is a powerful tool based on the principles of next-generation sequencing that can be applied to the detection and quantification of lncRNAs. Some advantages of using RNA-seq over a microarrary-based approach are that RNA-seq works on a genome-wide scale at single nucleotide resolution and is not limited to detecting already known sequences. Thus, it can be used to discover previously unknown lncRNAs in an unbiased manner [33]. However, the time and cost related to the downstream analysis of the data generated by RNA-seq is a considerable disadvantage of this approach.
Before beginning RNA-seq, one must decide whether to use total RNA or polyadenylated RNA. The presence of rRNA (around 80-85% of total RNA) and tRNA (15%) [34,35] can drastically reduce the diversity of a cDNA library during amplification of cDNAs. Polyadenylated RNA is frequently used for RNA-seq to avoid this problem. However, given the prevalence of non-polyadenylated lncRNA in the genome (around 40% of total lncRNAs), the disadvantage of losing this fraction is not negligible [36]. One solution to this problem is to use commercially available kits to remove rRNA from total RNA without losing non-polyadenylated RNA.
After sequencing, the generated reads are typically aligned to the UCSC mouse mm10 or human hg19 reference genomes using software programs such as the short-read mappers Bowtie 2 [37] and Burrows-Wheeler Aligner [38], and the splice-junction identifier TopHat [39]. Next, the reads are used to assemble a transcriptome and discover previously unannotated transcripts with programs such as Cufflinks [40], which relies on a reference annotation database, or Scripture, which builds the transcriptome ab initio[41]. From here, novel lncRNAs can be identified by excluding protein-coding transcripts and annotated lncRNAs based on the databases of RefSeq, ENCODE, and FANTOM (Functional Annotation of the Mammalian Genome) [42], as well as the two databases of experimentally verified lncRNAs generated by the Mattick lab: lncRNAdb [43] and NRED (Noncoding RNA Expression Database) [44].
Novel lncRNAs often undergo further scrutiny to verify that they are not transcriptional noise and that they indeed do not encode proteins. For instance, if the candidate is located within a K4-K36 domain and enriched with RNA polymerase II binding sites and DNase I hypersensitivity sites (a sign of open chromatin) as detected with the ENCODE data, the candidate is likely to be a product of active transcription [25,26,29]. The protein-coding potential of a candidate lncRNA can be evaluated with the Coding Potential Calculator (CPC) algorithm and other programs [45,46]. However, this is not a straightforward task as detailed in a recent review article [47].
Conclusions
The recent identification of the genome-wide human lncRNA loci by the ENCODE project is undoubtedly a milestone toward the long-term goal of understanding the functional significance of lncRNAs in many biological phenomena. Applications of microarrays containing these probes will certainly lower the threshold of launching new studies of lncRNAs. However, the use of tiling arrays and RNA-seq will continue to be required to identify splicing variants and tissue-specific lncRNAs. In addition, because of the low conservation of lncRNA sequences across species, the use of these approaches in new species will remain necessary until their ENCODE equivalents become publicly available. Furthermore, we expect that additional technological innovations geared toward studying lncRNAs will continuously emerge to support the rapid development of this fascinating research field.
Abbreviations
ChIP: Chromatin immunoprecipitation; CLIP: Cross-linking and immunoprecipitation; ENCODE: Encyclopedia of DNA Elements; FANTOM: Functional annotation of the mammalian genome; lincRNA: Large intergenic noncoding RNA; lncRNA: Long noncoding RNA; NRED: Noncoding RNA Expression Database; RefSeq: Reference Sequence; RIP: RNA immunoprecipitation; RNA-seq: RNA sequencing; UV: Ultraviolet.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CL and NK wrote and edited the drafts of the paper. Both authors read and approved the final manuscript.
Contributor Information
Catherine Lee, Email: leex2304@umn.edu.
Nobuaki Kikyo, Email: kikyo001@umn.edu.
Acknowledgements
We thank Michael Franklin for critical reading of the manuscript. This work was supported by Engdahl Funds, the Office of the Vice President for Research of the University of Minnesota, and the National Institutes of Health (R01 GM098294) to N.K.
References
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG. et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L. et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22(9):1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Chang HY. Active chromatin and noncoding RNAs: an intimate relationship. Curr Opin Genet Dev. 2012;22(2):172–178. doi: 10.1016/j.gde.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol. 2010;22(3):357–364. doi: 10.1016/j.ceb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21(3):416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311(5764):1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25(24):2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S. et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31(3):522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113(11):2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43(6):1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE. et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26(4):338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, Kretz M, Khavari PA. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22(6):1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Soler M, Portela A, Carrere J, Fonalleras E, Gomez A, Villanueva A, Esteller M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19(7):664–670. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37(4):376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SR, Marguerat S, Bahler J. Exploring long non-coding RNAs through sequencing. Semin Cell Dev Biol. 2012;23(2):200–205. doi: 10.1016/j.semcdb.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Farrell RJ. Electrophoresis of RNA. 3. Burlington, MA: Elsevier; 2005. pp. 190–237. (RNA methodologies). [Google Scholar]
- Lodish H, Berk A, Kaiser CA, Krieger M, Scott MP, Bretscher A, Ploegh H, Matsudaira P. Molecular Cell Biology. 6. New York: Freeman WH; 2007. Post-transcriptional gene control; pp. 358–367. [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F. et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C. et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Severin J, Lizio M, Forrest AR, van Nimwegen E, Rehli M, Schroder K, Irvine K, Suzuki H, Carninci P. et al. Update of the FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Nucleic Acids Res. 2011;39(Database issue):D856–D860. doi: 10.1093/nar/gkq1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39(Database issue):D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37(Database issue):D122–D126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Fry B, Kamal M, Xie X, Cuff J, Lin MF, Kellis M, Lindblad-Toh K, Lander ES. Distinguishing protein-coding and noncoding genes in the human genome. Proc Natl Acad Sci U S A. 2007;104(49):19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4(11):e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]