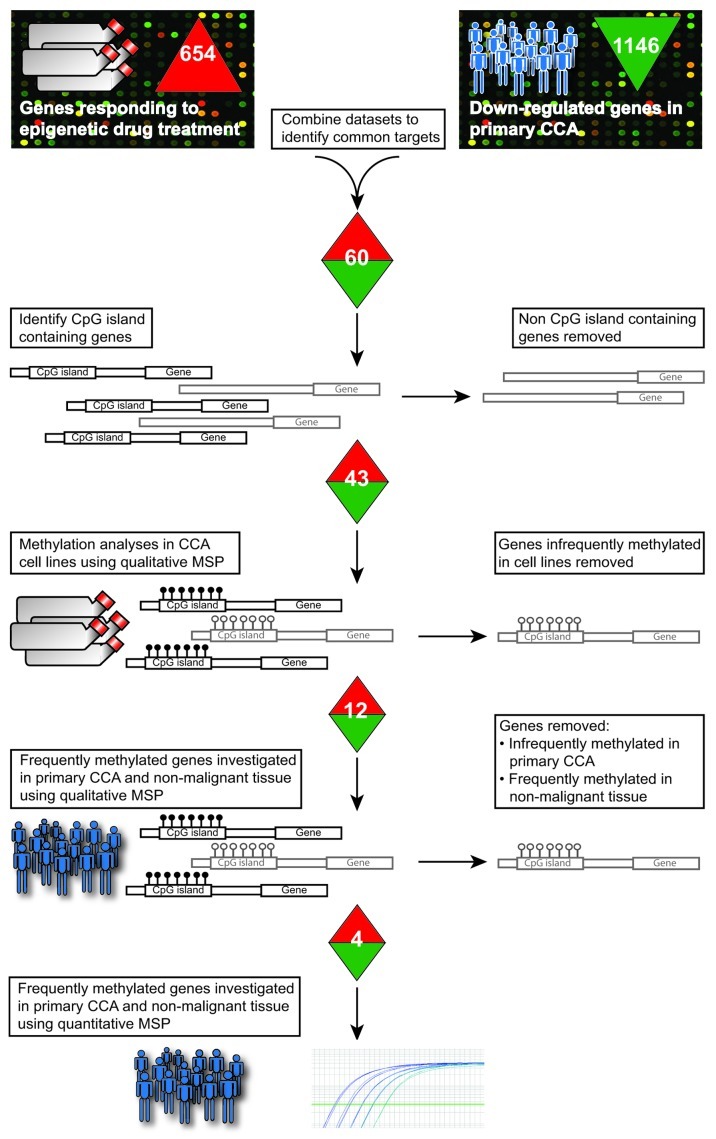
Figure 1. Epigenome-wide experimental approach for identifying methylated genes in cholangiocarcinomas. Six CCA cell lines were cultured with and without a combination of epigenetic drugs (5-aza-2´deoxycytidine and trichostatin A). Array-elements responding to epigenetic drug treatment were compared with previously published downregulated genes in CCAs relative to cancer free tissue. Common genes, harboring a CpG island in the promoter region, were investigated for methylation in cancer cell lines from colon, bile duct, liver, gall bladder and pancreas. Genes frequently methylated in CCA cell lines were subsequently investigated in patient material using MSP. The most promising candidates from this analysis were further evaluated by qMSP. Numbers indicate the number of genes fulfilling the selection criteria in each experimental step and which are subsequently subjected to further analyses.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.