Abstract
Background
Although recent studies have identified genes expressed in human embryonic stem cells (hESCs) that induce pluripotency, the molecular underpinnings of normal stem cell function remain poorly understood. The high mobility group A1 (HMGA1) gene is highly expressed in hESCs and poorly differentiated, stem-like cancers; however, its role in these settings has been unclear.
Methods/Principal Findings
We show that HMGA1 is highly expressed in fully reprogrammed iPSCs and hESCs, with intermediate levels in ECCs and low levels in fibroblasts. When hESCs are induced to differentiate, HMGA1 decreases and parallels that of other pluripotency factors. Conversely, forced expression of HMGA1 blocks differentiation of hESCs. We also discovered that HMGA1 enhances cellular reprogramming of somatic cells to iPSCs together with the Yamanaka factors (OCT4, SOX2, KLF4, cMYC – OSKM). HMGA1 increases the number and size of iPSC colonies compared to OSKM controls. Surprisingly, there was normal differentiation in vitro and benign teratoma formation in vivo of the HMGA1-derived iPSCs. During the reprogramming process, HMGA1 induces the expression of pluripotency genes, including SOX2, LIN28, and cMYC, while knockdown of HMGA1 in hESCs results in the repression of these genes. Chromatin immunoprecipitation shows that HMGA1 binds to the promoters of these pluripotency genes in vivo. In addition, interfering with HMGA1 function using a short hairpin RNA or a dominant-negative construct blocks cellular reprogramming to a pluripotent state.
Conclusions
Our findings demonstrate for the first time that HMGA1 enhances cellular reprogramming from a somatic cell to a fully pluripotent stem cell. These findings identify a novel role for HMGA1 as a key regulator of the stem cell state by inducing transcriptional networks that drive pluripotency. Although further studies are needed, these HMGA1 pathways could be exploited in regenerative medicine or as novel therapeutic targets for poorly differentiated, stem-like cancers.
Introduction
Recent studies have made great strides in discovering a handful of factors important in human embryonic stem cells (hESCs) [1]–[8]. These genes (or pluripotency factors) have been used to “reprogram” normal, adult somatic cells into hESC-like cells, called induced pluripotent stem cells or iPSCs. iPSCs hold enormous promise because they could provide a source of unlimited, patient-specific stem cells for use in regenerative medicine, drug screening, or as disease models. Unfortunately, the derivation of iPSCs is inefficient, and the ability to maintain and differentiate iPSCs remains a technical hurdle in the field. Moreover, iPSCs, and even normal hESCs, can acquire abnormal karyotypes and invasive properties, recapitulating features of cancer cells [9]–[13]. Thus, a better understanding of the molecular mechanisms responsible for normal stem cell properties in hESCs and iPSCs is needed before these cells can be safely used in the clinic. Studies to elucidate the underpinnings of normal hESCs and fully reprogrammed iPSCs should also provide insight relevant to cancer because pluripotent stem cells and cancer cells share a subset of transcriptional networks and properties [9]. It will be critical, however, to identify the molecular mechanisms that distinguish normal stem cells from malignantly transformed, stem-like cells.
The high mobility group A1 (HMGA1) gene is highly expressed during embryogenesis and enriched in hESCs [9], hematopoietic stem cells (HSCs) [13]–[16], and poorly differentiated or refractory cancers [9], [15]–[37], with low or undetectable expression in adult, differentiated tissues. This gene encodes the HMGA1a and HMGA1b protein isoforms [38]–[39], which are members of the HMGA superfamily of chromatin remodeling proteins that include HMGA1a, HMGA1b, and HMGA2 [38]–[43]. HMGA proteins are low molecular weight (thus high mobility group) proteins that bind to AT-rich regions in chromatin and orchestrate the assembly of transcription factor complexes to modulate chromatin structure and regulate gene expression [27], [29], [30], [32], [34]–[36], [42]–[45]. HMGA proteins induce malignant transformation in cultured cells and cause aggressive tumors in transgenic mice [18]–[19], [21]–[37], [40]–[46]. The tumors from HMGA1 mice can be serially transplanted, indicating that they have the stem cell property of long term self-renewal [32]. HMGA1 expression is highest in cultured cells that are derived from poorly differentiated cancers, including breast [21], [45], prostate [23], pancreatic [31], uterine [26], colon [34], and lung [30] cancers as compared to cell lines from more differentiated tumors. Expression of HMGA1 is also associated with poor differentiation status in solid tumors arising from different tissues and embryonic origins [9], [26], [30], [34], [47]–[49]. Moreover, HMGA1 overexpression portends a poor outcome in diverse tumors, including cancers of the pancreas [31], brain [9], [48], bladder [9], lung [49], and breast [9], [47]. HMGA1 is also enriched in refractory hematopoietic cancers [15]–[16], [18]–[19], [29], [33] and in human iPSCs [13]. Together, these studies in cancer and pluripotent stem cells suggest that HMGA1 could function to reprogram cells to a more primitive, undifferentiated, stem-like state.
Previous studies in cancer cells have demonstrated that HMGA1 directly activates specific genes involved in tumor growth and progression, including proliferation, migration, invasion, angiogenesis, genetic instability, resistance to cell death, immune evasion, and an epithelial-mesenchymal transition in cancer cells, although its role in embryonic stem cells is poorly understood [23,26–30,32–36.,45]. Here, we report that HMGA1 promotes the cellular reprogramming of adult somatic cells to undifferentiated, fully pluripotent stem cells (iPSCs). We also identify transcriptional networks induced by HMGA1 to drive the stem cell phenotype in pluripotent stem cells. Our studies provide new insights into the role of HMGA1 in development, stem cells, and cellular reprogramming.
Results
HMGA1 Expression Decreases with Differentiation in hESCs
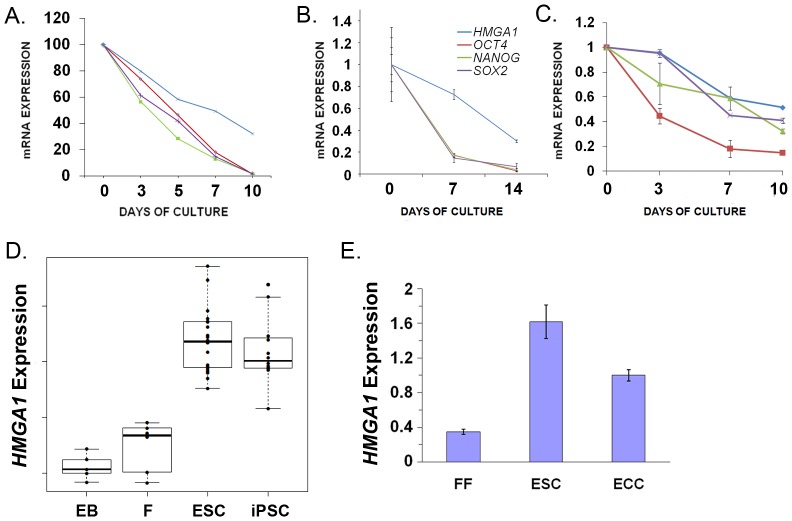
To better define the role of HMGA1 in pluripotent stem cells, we investigated its expression in hESCs during differentiation. First, we assessed HMGA1 expression patterns in H1 hESCs induced to differentiate into blood cells in an established model of hematopoiesis [50]. HMGA1 mRNA was highest at day 0, with levels dropping dramatically as the hematopoietic cells differentiate (day 10; Fig. 1A) by microarray gene expression profile analysis (microarray data found in Gene Expression Omnibus, accession number GSE12531). Notably, the levels of HMGA1 closely parallel those of the embryonic stem cell and pluripotency factors NANOG, OCT4, and SOX2. These results were confirmed by quantitative RT-PCR (qRT-PCR; data not shown). When hESCs are forced to differentiate into neuroectodermal lineages, we also found that HMGA1 expression decreases by qRT-PCR, similar to NANOG, OCT4, and SOX2 (Fig. 1B). Likewise, HMGA1 expression falls and mirrors that of NANOG, OCT4, and SOX2 during mesodermal differentiation, as demonstrated by qRT-PCR (Fig. 1C). To further investigate the role of HMGA1 in pluripotency, we compared HMGA1 expression in embryoid bodies, fibroblasts, hESCs, and iPSCs from a study of global gene expression profile analyses [8]. We found that HMGA mRNA levels were highest in the pluripotent hESCs and iPSCs with lower levels in differentiated cells (embryoid bodies and fibroblasts; Fig. 1D). Using qRT-PCR, we found that cultured cancer cells derived from a germ cell tumor (Tera-2 embryonal cancer cell or ECC line) have ∼50% lower HMGA1 mRNA levels compared to hESCs (Fig. 1E). These findings indicate that HMGA1 expression is similar to that of key pluripotency factors as hESCs differentiate and suggest that HMGA1 could function in maintaining an undifferentiated state in normal hESCs.
Figure 1. HMGA1 expression falls with differentiation in hESCs and parallels that of other pluripotency genes.
A) H1 hESCs were cultured under conditions to promote hematopoietic differentiation. By days 7–10, hESCs differentiate into mesodermal-hematoendothelial (MHE) colonies and fully differentiated progeny of all hematopoietic lineages. (See ref. 50 and GEO accession number GSE12531 for microarray data). B) HMGA1 expression falls in hESCs cultured under conditions to promote neuroectodermal differentiation. HMGA1 decreases with OCT4, NANOG, and SOX2 by day 10, as shown by qRT-PCR. C) Similarly, HMGA1 expression falls when hESCs differentiate into mesoderm as shown by qRT-PCR. D) Expression levels of HMGA1 in embryoid bodies (B), fibroblasts (F), hESCs (H), and iPSCs (I) from a published database [8]. E) HMGA1 expression in fetal fibroblasts, H9 hESCs, and embryonal carcinoma cells (ECC) was assessed by qRT-PCR; HMGA1 expression in the H9 hESCs was arbitrarily assigned a value of 1.0.
HMGA1 Blocks Differentiation in hESCs
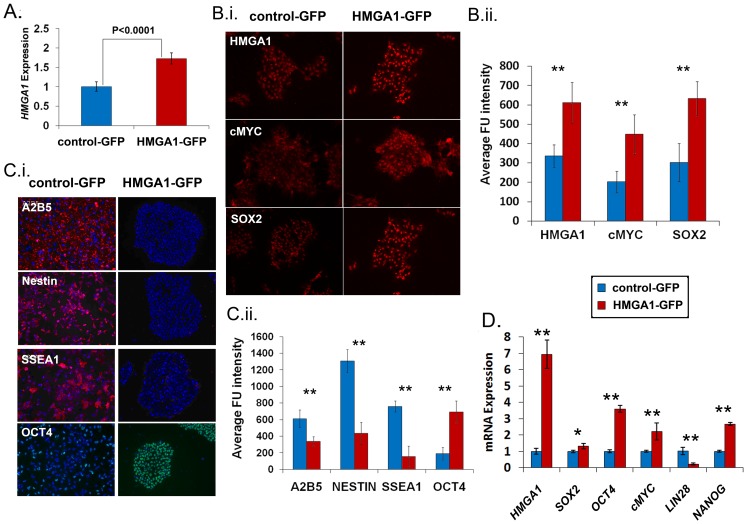
To further investigate the role of HMGA1 in the maintenance of an undifferentiated state, we determined if forced expression of HMGA1 in hESCs will affect differentiation. We therefore engineered H9 hESCs to express HMGA1 by transducing these cells with a lentiviral vector expressing HMGA1 linked to green fluorescent protein (GFP) [26]. Control cells were transduced with a lentiviral vector expressing GFP alone [26]. The hESCs transduced with the HMGA1a lentivector showed a corresponding increase in HMGA1 mRNA levels compared to the control (Fig. 2A). By immunofluorescent cytochemistry, we documented that the HMGA1 protein was increased in the H9 hESCs transduced with the HMGA1 lentivirus compared to the control lentivirus (Fig. 2B, upper panels). In addition, we found that both SOX2 and cMYC proteins were also increased in HMGA1 hESCs compared to controls (Fig. 2B). To better define the role of HMGA1 in stem cells, the transduced HMGA1 and control hESCs were cultured under conditions to promote differentiation into neuroectoderm as described previously [51]–[52]. Strikingly, hESCs expressing exogenous HMGA1 showed no evidence for differentiation into neuroectoderm. The HMGA1-expressing cells maintained normal hESC morphology and expression of pluripotency markers (Fig. 2C–D); there was also no expression of differentiation markers (Fig. 2C). In contrast, the control-GFP cells underwent dramatic morphological changes, growing as a monolayer. Likewise, control cells expressed neuronal markers (A2B5, Nestin, and SSEA1) after culture in differentiation conditions (Fig. 2C). By qRT-PCR, the HMGA1 cells expressed significantly higher levels of pluripotency genes after 7 days in differentiation conditions, including OCT4 (p<0.01), SOX2 (p<0.05), cMYC (p<0.01), and NANOG (p<0.01) compared to the controls (Fig. 2D). In addition, exogenous HMGA1 levels remained high (7-fold) compared to controls. Surprisingly, LIN28 expression was not increased in the HMGA1-GFP cells, indicating that HMGA1 blocks differentiation without maintaining high levels of LIN28. In this setting, the differentiating factors could be the primary regulators of LIN28. To determine if HMGA1 alters growth rates in hESCs, we also performed proliferation (MTT) assays and found no significant difference in growth rates (Fig. S1). These results indicate that constitutive expression of HMGA1 blocks differentiation and maintains hESCs in an undifferentiated, stem-like state.
Figure 2. HMGA1 drives a de-differentiated state.
A) HMGA1 expression is increased in hESCs 3 days after transduction with the HMGA1-GFP lentivirus (HMGA1-GFP) compared to control hESCs (control-GFP). Bars, mean ± standard deviation. B) i. HMGA1, cMYC and SOX2 proteins are up-regulated in HMGA1-GFP (red) compared to control-GFP (blue) hESCs. ii. Fluorescence was assessed quantitatively using MetaMorph (Universal Imaging) version 7.7 (p<0.001 for all genes in HMGA1-GFP compared to control-GFP hESCs). C) i. Control-GFP hESCs (blue) differentiate after treatment with neuroectodermal differentiation factors and express neural markers (left three panels. red: A2B5-top, Nestin–middle, SSEA1-lower panel), while the HMGA1-GFP hESCs (red) remain embryonic stem cell colonies and express hESC markers (OCT4), but no neural markers. DAPI was used to stain nuclei. ii. Quantitative analysis of HMGA1 and the stem cell markers (cMYC and SOX-2) are shown; (p<0.001 for all proteins assessed in HMGA1-GFP compared to control-GFP hESCs). D) The pluripotency genes SOX2, OCT4, cMYC, and NANOG are up-regulated in the HMGA1-GFP hESCs (red) compared to controls (blue) in hESCs cultured in conditions that promote neuroectodermal differentiation. Levels of exogenous HMGA1 are also shown.
HMGA1 Enhances Cellular Reprogramming to Fully Pluripotent iPSCs
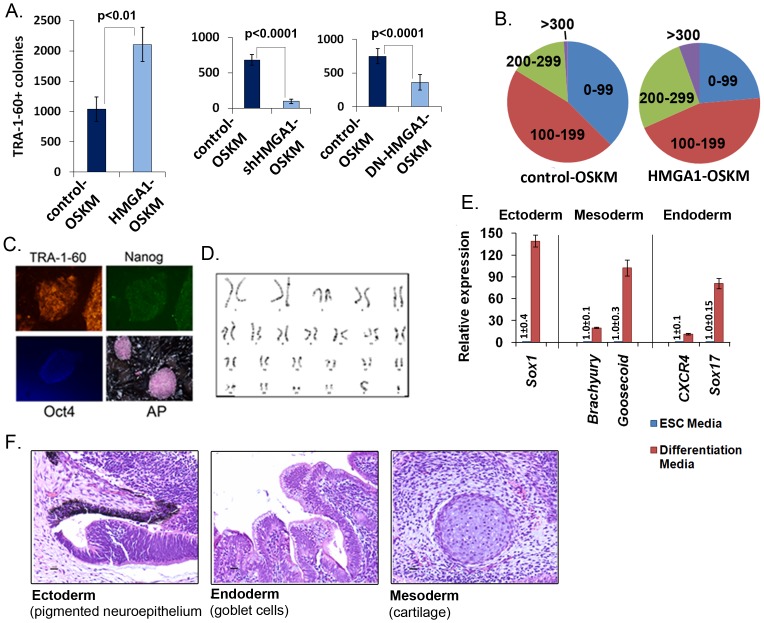
Based on the above findings, we hypothesized that HMGA1 promotes cellular reprogramming and could enhance the derivation of iPSCs. To test this hypothesis, we used standard retroviral reprogramming technology to transduce bone-marrow derived, commercial, adult mesenchymal stem cells (MSCs). MSCs (100,000 cells per reprogramming experiment) were transduced with the four Yamanaka factors (OCT2, SOX4, KLF4, and cMYC or OSKM [1]) plus HMGA1 or control, all expressed by pMX retroviral vectors as previously described [53]–[54]. The addition of HMGA1 to OSKM (denoted HMGA1-OSKM) resulted in a consistent 2-fold increase in TRA-1-60+iPSC colonies compared to the control-OSKM transduction (Fig. 3A). Previous studies showed that TRA-1-60+staining is a reliable early indicator of fully reprogrammed iPSC colonies [10]–[11], [54]–[55]. Moreover, we found that the early HMGA1-OSKM TRA-1-60+colonies were significantly larger than their control-OSKM counterparts (Fig. 3B), indicating that HMGA1 enhances the reprogramming rate, stem cell survival, proliferation, or a combination of these factors during iPSC generation. To determine if HMGA1 is required during cellular reprogramming to iPSCs, we blocked HMGA1 expression or function using a short hairpin RNA (shRNA) to HMGA1 or dominant-negative construct, respectively. MSCs were transduced with OSKM as described above. Forty-eight hours after reprogramming with OSKM, cells were transduced with a lentivirus containing HMGA1 shRNA or control shRNA. Strikingly, we found that there was a marked decrease in TRA-1-60+colonies in the cells treated with HMGA1 shRNA as compared to controls (p<0.0001; Fig. 3A). Similarly, we found that the dominant-negative HMGA1 also blocked cellular reprogramming to iPSCs (p<0.0001; Fig. 3A).
Figure 3. HMGA1 promotes cellular reprogramming of MSCs to fully pluripotent iPSCs.
A) Reprogramming with HMGA1-OSKM results in more TRA-1-60+iPSC colonies compared to controls, while shHMGA1 and dominant-negative HMGA1 (DN-HMGA1) decreases the number of colonies, as assayed on day 16 following retroviral transduction. B) The HMGA1-OSKM TRA-1-60+colonies are significantly larger than the control-OSKM colonies; sizes: µm. C) The HMGA1-OSKM colonies stain positively for standard stem cell markers (TRA-1-60, Nanog, Oct4, and Alkaline Phosphatase or AP). D) Established HMGA1-OSKM colonies have a normal karyotype. E) HMGA1-OSKM colonies express representative markers from 3 germ layers when induced to differentiate in vitro. HMGA1-OSKM colonies were either cultured in standard hESC media or under conditions for differentiation into ectoderm, mesoderm, or endoderm. The appropriate genes for each condition were expressed. F) HMGA1-OSKM colonies form benign teratomas with constituents from all three germ layers.
To determine if the MSC-derived TRA-1-60+colonies are fully reprogrammed and express other standard stem cell markers after transduction with HMGA1-OSKM, we selected and subcultured colonies for further analysis with immunoflourescent intracellular staining after 6 passages. As expected for hESCs and fully reprogrammed iPSCs, the HMGA1-OSKM clones also expressed OCT4, NANOG, and alkaline phosphatase (AP) (Fig. 3C). The HMGA1-OSKM clone (HMGA1-OSKM-4) had a normal karyotype after culturing the cells for>10 passages (Fig. 3D). In addition, the HMGA1-OSKM iPSCs could be fully differentiated into neuronal or meso/endoderm lineages in vitro (Fig. 3E) and generate teratomas with all three germ layers represented (Fig. 3F). There was no detectable expression of exogenous OSKM from the retroviral vectors in the HMGA1-OSKM clone 57 days following transduction, although the vectors were detectable 21 days after transduction (data not shown).
Next, we investigated whether HMGA1 enhances cellular reprogramming of other somatic cells. We therefore transduced fetal lung fibroblasts (IMR90) with the HMGA1-OSKM or control-OSKM retroviruses. Similar to our data in MSCs, we found that HMGA1 significantly enhances the size and number of TRA-1-60+colonies (Fig. S2). We observed an increase in the number of iPSC colonies by about 2-fold with HMGA1 in the reprogramming cocktail. Taken together, our results demonstrate that HMGA1 promotes cellular reprogramming to an undifferentiated, pluripotent stem-like state in somatic cells of different origins (MSCs, IMR90 fetal lung fibroblasts).
HMGA1 Modulates the Expression of Pluripotency Genes
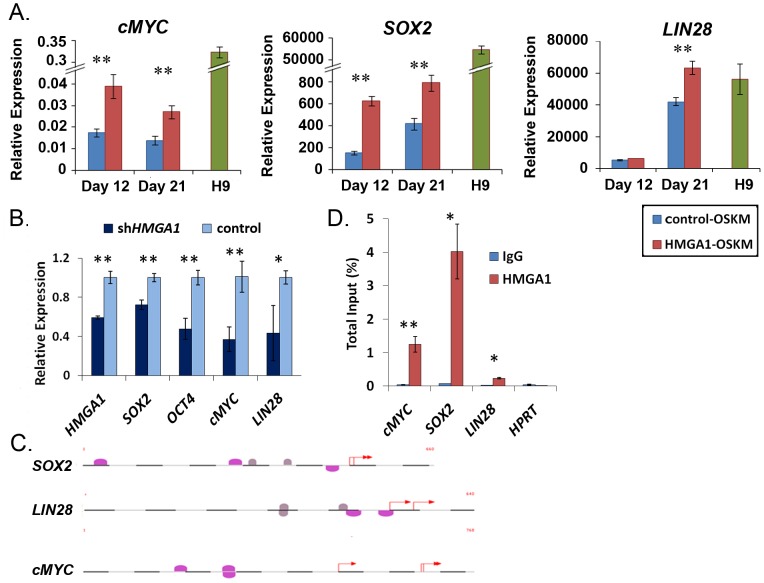
Because HMGA1 functions by modulating gene expression, we hypothesized that it promotes pluripotency by inducing stem cell transcriptional networks. We therefore assessed the expression of a subset of endogenous, human embryonic stem cell/pluripotency genes (OCT4, SOX2, cMYC, NANOG, LIN28, REX1, hTERT) at early stages (day 12 and 21) in the reprogramming pools following transduction of MSCs with HMGA1-OSKM or control-OSKM. At day 12, expression of both SOX2 (p<0.001) and cMYC (p<0.01) were 2–3-fold higher in the HMGA1-OSKM pools compared to the control-OSKM cells (Fig. 4A). At day 21, expression of SOX2 (p<0.01), LIN28 (p<0.01), and cMYC (p<0.01) were all significantly increased in the MSCs reprogrammed with HMGA1-OSKM compared to the control-OSKM pools (Fig. 4A). Surprisingly, the expression of the other stem cell genes was not significantly altered in the HMGA1-OSKM pools compared to controls (data not shown). There were no significant changes in expression of the exogenous, murine OCT4, SOX2, KLF4, or cMYC delivered by retrovirus (see Fig. S3 for qRT-PCR results and Table S1 for primers specific to the murine OSKM delivered by retrovirus). Of note, the established iPSC clones (after 10 passages) generated by transduction of HMGA1-OSKM or control-OSKM had similar expression of all pluripotency genes assessed, and the expression levels were similar to those observed in hESCs (data not shown). These results suggest that HMGA1 enhances the derivation of iPSCs by inducing the expression of a subset of pluripotency-associated genes early in the reprogramming process.
Figure 4. HMGA1 regulates pluripotency genes.
A) Endogenous SOX2, LIN28, and cMYC are induced more in MSCs reprogrammed with HMGA1-OSKM (red) compared to control pools (blue). Gene expression in H9 hESCs is shown as a reference (green). B) Pluripotency genes (SOX2, OCT4, cMYC, LIN28) are repressed following shRNA-mediated knockdown of HMGA1. *p<0.01, **p<0.00001. C) The promoter regions of pluripotency genes contain putative HMGA1 DNA binding sites (pink ovals) located near putative NFκB sites (grey circles). D) Chromatin immunoprecipitation in hESCs shows enrichment in HMGA binding in the promoters of SOX2, cMYC and LIN28. IgG was used as a negative control; cMYC antibody and primers to the B23 promoter were used as a positive control. *p<0.01, **p<0.001.
To determine if HMGA1 also modulates expression of pluripotency genes in hESCs, we knocked-down HMGA1 expression in H9 hESCs using shRNA (shHMGA1) [56] and assessed the expression of seven embryonic stem cell/pluripotency genes (OCT4, SOX2, cMYC, NANOG, LIN28, REX1, hTERT). We found that SOX2, OCT4, cMYC, and LIN28 were all significantly repressed in the hESCs 96 hours following shRNA-mediated knock-down of HMGA1 (Fig. 4B). These results were compared to H9 hESCs treated with a control shRNA vector. To further rule-out off-target effects of the shHMGA1, we also assessed the expression of these genes after knocking down HMGA1 using HMGA1 siRNA (Dharmacon), which targets a different sequence in the HMGA1 mRNA [30]. Using this approach, SOX2, OCT4, cMYC, and LIN28 were also repressed after transduction with the siHMGA1 after 24 hours (Fig. S4). Expression of the other pluripotency genes did not change significantly, although both approaches (shRNA and siRNA) resulted in significant knock-down of HMGA1 mRNA. Of note, there were no gross changes in colony morphology or proliferation at these early time points using siRNA or shRNA. Together, our results indicate that HMGA1 modulates a specific subset of stem cell/pluripotency genes during the generation of iPSCs and in fully pluripotent hESCs.
HMGA1 Binds to Pluripotency Gene Promoters
Because HMGA1 binds chromatin to modulate gene expression [32], we hypothesized that HMGA1 induces an undifferentiated, pluripotent state by binding to DNA and enhancing transcriptional networks downstream of the stem cell gene targets, such as cMYC, SOX2, and LIN28 genes. Using MatInspector [57], we found that the promoter regions of cMYC, SOX2, and LIN28 contain AT-rich regions with putative HMGA1 binding sites (Fig. 4C). To determine if HMGA1 binds to the promoters of these genes in vivo, we performed chromatin immunoprecipitation in H9 hESCs. We found that the promoter regions of cMYC, SOX2, and LIN28 with the putative HMGA1 binding sites were enriched in HMGA1-binding (Fig. 4D). Furthermore, there was no demonstrable binding of HMGA1 to the negative control promoter, HPRT, which was shown in previous studies to lack HMGA1 binding [29]. These results indicate that HMGA1 binds directly to the cMYC, SOX2, and LIN28 promoters and suggest that HMGA1 enhances pluripotency by inducing expression of cMYC, SOX2, and LIN28 early in reprogramming.
HMGA1 iPSCs have Promoter Methylation Patterns similar to hESCs
Prior studies suggest that epigenetic reprogramming is involved in the induction of pluripotent stem cells [12]. We therefore investigated promoter methylation patterns in the HMGA1 iPSCs and reprogramming pools using the Illumina Infinium Methylation27 platform, which includes probes for 27,576 loci (Fig. S5). Promoter methylation was assessed from genomic DNA isolated from a fully characterized iPSC clone (HMGA1-OSKM-A4) and MSC reprogramming pools 12 and 21 days after transduction with HMGA1-OSKM or control-OSKM. For comparison, we also included cancer cells, hESCs, fetal fibroblasts, MSCs, and previously characterized iPSCs for which promoter methylation was assessed in a previous study [12]. The promoter methylation patterns in the fully reprogrammed iPSCs generated by HMGA1–OSKM or control-OSKM were similar to that of hESCs and other iPSCs. There were no significant differences in the relative number of CpG sites with high or low levels of methylation in the HMGA1-OSKM reprogramming pools compared to the OSKM controls. These findings indicate that the iPSCs reprogrammed with HMGA1 have similar methylation patterns to hESCs and other iPSCs and suggest that the enhanced reprogramming by HMGA1 does not occur through large changes in global methylation patterns.
Discussion
Here, we provide compelling evidence that HMGA1 plays a key role in cellular reprogramming to a pluripotent stem cell and the maintenance of the undifferentiated state: 1.) HMGA1 expression is enriched in hESCs and fully reprogrammed iPSCs, with intermediate levels in cancer cells and lower levels in differentiated fibroblasts, 2.) HMGA1 enhances cellular reprogramming of somatic cells to a pluripotent state, while forced expression blocks differentiation in hESCs, and 3.) HMGA1 binds to promoters and induces expression of other pluripotency factors, whereas knock-down of HMGA1 represses pluripotency factors. In multiple settings (bone marrow-derived MSCs, fetal lung fibroblasts), more iPSC colonies formed when HMGA1 was added to the OSKM reprogramming cocktail, and the colony size was greater in most cases. Interestingly, mice that are null for HMGA1 have normal early development [58], while mice deficient in the HMGA family member, HMGA2, have a pygmy phenotype, but otherwise normal early development [59]. Because both HMGA1 and HMGA2 proteins have a high level of homology and similar functions in experimental models [18], [45], it is possible that there is functional redundancy, and knock-out of HMGA1 is partially compensated for by HMGA2. Genetic experiments are underway to address this issue.
We also discovered that germ cell tumor cells express less HMGA1 than fully reprogrammed iPSCs and hESCs, suggesting that a critical level of HMGA1 may be required for a fully reprogrammed, pluripotent, stem-like phenotype in contrast to a malignant phenotype. Perhaps the addition of HMGA1 in the reprogramming cocktail results in a greater proportion of cells that cross a critical HMGA1 gene threshold and produce fully reprogrammed iPSCs. Prior studies have shown that reprogramming cancer cell lines with retroviral delivery of pluripotency genes results in ES-like cells with slower growth rates as well as the ability to form benign teratomas in vivo and respond to differentiating agents in vitro [60]–[63]. iPSCs derived from a colon cancer cell line (DLD1) also became more sensitive to the cytotoxic agent, 5-FU [60], although subsequent studies found that iPSCs generated from another cancer cell line (HuCC-T1 choriocarcinoma cells) became invasive and lost their response to differentiating or cytotoxic agents in vitro as well as their ability to form benign teratomas in vivo when cultured for longer time periods (>120 days following reprogramming) [63]. These cells had activation of endogenous cMYC when they lost their potential for pluripotency, indicating that the reprogramming was reversible. Together, these studies and our findings presented here suggest that strategies to induce expression of HMGA1 and that of other pluripotent genes could reprogram malignant tumors into cells that respond to differentiating agents. Further studies are needed to test this hypothesis and could lead to the development of novel therapeutic strategies to reprogram cancer cells.
The enhanced reprogramming by HMGA1 could also be exploited to generate patient-specific iPSCs for disease modeling, drug testing, or regenerative medicine with patient-derived cells. Surprisingly, we did not find evidence for malignant transformation in the iPSCs reprogrammed by HMGA1-OSKM. Rather, there was an increase in the number of TRA-1-60+staining clones when HMGA1 was included in the reprogramming cocktail, which is a reliable marker for fully pluripotent iPSCs [54]. Moreover, the teratoma assay showed differentiated tissues from all three germ layers, further documenting the pluripotent state of the HMGA1-OSKM iPSCs. The iPSCs could also be differentiated into embryoid bodies, neuroectoderm, or mesoderm, indicating that the addition of HMGA1 did not interfere with the pluripotent/differentiation potential of the iPSCs. The karyotype was normal and promoter methylation patterns were similar to those observed in hESCs. In preliminary studies, we also found that HMGA1 significantly enhances reprogramming of mononuclear blood cells (MBCs) to TRA-1-60+colonies using an episomal vector approach (unpublished data). This latter approach has several advantages for potential clinical uses. Most notably, the reprogramming vectors do not integrate into the genome, thus avoiding the complication of activating (or inactivating) critical loci or otherwise disrupting the genome. Episomal vectors are also ultimately lost as the cells successively divide and the iPSCs generated from this approach may be less immunogenic [64].
To determine how HMGA1 promotes an undifferentiated, pluripotent stem cell state, we investigated the expression of stem cell transcriptional networks and found several key genes that are induced by HMGA1 in the iPSC pools early in reprogramming, including SOX2, LIN28, and cMYC. We also discovered that HMGA1 binds to the promoters of these genes in vivo in hESCs, suggesting that HMGA1 directly induces their expression. Interestingly, most HMGA1 transcriptional targets have a consensus DNA binding site for NF-κB in the promoter regions near the AT-rich site where HMGA1 binds [32], indicating that these factors could function together in activating cellular pathways in stem cells. Based on the MatInspector computational algorithm and published literature, the pluripotency genes induced by HMGA1 also include NF-κB sites near the HMGA1 binding sites, including LIN28, SOX2, cMYC, and OCT4, (Fig. 4C), indicating that both HMGA1 and NF-κB could promote cellular reprogramming by inducing expression of pluripotency genes. A recent study identified HMGA1 as a gene whose translation is induced by Lin28 in hESCs [65]. This suggests that a positive feedback loop could exist whereby Lin28 enhances HMGA1 translation and HMGA1, in turn, feeds back to induce expression of LIN28, along with other pluripotency genes, to activate stem cell networks. Surprisingly, not all pluripotency genes with predicted HMGA1 DNA binding sites in their promoters were induced in the HMGA1 reprogramming pools, indicating that HMGA1 induces a specific stem cell signature in this setting. Further studies are needed to elucidate all of the critical networks related to HMGA1 that drive a fully pluripotent state in iPSCs and stem cells.
In summary, we demonstrate for the first time that HMGA1 enhances cellular reprogramming of somatic cells to a fully pluripotent state. We also discovered that HMGA1 promotes an undifferentiated, pluripotent state and blocks differentiation. These findings provide insight into HMGA1 function in hESCs that could be exploited for patient-derived iPSCs for use in regenerative medicine. Although additional studies are needed, our findings also suggest that HMGA1 transcriptional networks are important in reprogramming normal cells into stem-like, malignant cancer cells and that these pathways could be targeted in therapy.
Materials and Methods
Ethics Statement
All animal experiments were conducted in accordance with a protocol approved by the Johns Hopkins University Animal Care and Use Committee (protocol# MO11M279). All mice were housed in a sterile environment where they had free access to food and water as outlined in our institutional guidelines.
Cell Culture
MSC1640 mesenchymal stem cells (AllCells, LLC) were maintained in DMEM (low glucose) with 10% FBS, 1× NEAA, 1× L-Glutamine, 1× Antibiotic/Antimycotic (Invitrogen) and bFGF (1 ng/ml). IMR90 fetal lung fibroblast cells (ATCC) and cultured similar to MSC1640, but without bFGF. iPSCs and hESCs (H1 and H9 from WiCell) were maintained in ES media: DMEM/F12, 20% Knockout Serum Replacement, 1× NEAA, 1× L-Glutamine, 1× Antibiotic-Antimycotic, 1 mM 2-Mercaptoethanol, bFGF (10 ng/ml for iPSCs or 8 ng/ml for hESCs). The human embryonal carcinoma line, NTERA-2 cl.D1 (ATCC) was cultured on matrigel-coated plates under conditions as previously described [66].
Transduction and Reprogramming Vectors
The pMXs-HMGA1 vector was made by restricting the pMXs retroviral vector [48] at the BstXI site and subsequently made blunt using Klenow. The human HMGA1a cDNA was inserted into pMXs following amplification from pooled human RNA with the following primers: (F) 5′-AGCCAATCCTATGGACCTGCTCCTTAGAGAAGGGAA-3′; (R) 5′-AGCCAATCCTATGGAAAGCTGTCCAGTCCCAGAA-3′. The correct HMGA1 sequence construct was confirmed by sequencing. The pMXs-DN-HMGA1 vector was made by isolating HMGI (mII,III) from pcDNA3.1/zeo.HMGI(mII,mIII) (a generous gift from Raymond Reeves, Washington State University, and described in detail in [67]) by restricting with HINDIII/BamHI; blunt end cloning was used to introduce the cDNA into pMXs (as described above). The empty pMXs vector was used as a negative control.
RNA Interference
The short-hairpin RNA interference vector for HMGA1 targets 5′-CAACTCCAGGAAGGAAACCAA-3′ and has been described elsewhere [56]. The empty vector was used as a negative control in knockdown experiments similar to a previous study [34]. The HMGA1 siRNA targets 5′-AGCGAAGTGCCAACACCTA-3′ and was obtained from Dharmacon [26]. siCONTROL (Dharmacon), which contains 4 siRNAs without matches to human, mouse, or rat genes, was used as the negative control for the siRNA experiments.
Retroviral Infection and Reprogramming Protocols
DNA vectors pMX-OCT4, pMX-SOX2, pMX-cMYC, and pMX-KLF4 (M. musculus genes) were generously provided by Linzhao Cheng (Johns Hopkins University). Retrovirus containing pMX-OCT4, pMX-SOX2, pMX-cMYC, pMX-KLF4, pMX-HMGA1, pMX-DN-HMGA1, pMX-shHMGA1, or pMX empty vector were produced and used for infection as previously described [49]. During days 7–16 following transduction, cells were maintained in ES media+0.5 mM sodium butyrate as previously reported [11].
Immunostaining
Live staining for TRA-1-60 to identify fully reprogrammed colonies was performed as described [11] using anti-TRA-1-60 (Millipore) and a mouse secondary antibody at a dilution of 1∶200 and 1∶400, respectively, premixed in hESC media. The TRA-1-60+putative iPSC colonies were further characterized for stem cell markers after fixation with paraformaldehyde as previously described [11]. To assess stem cell markers, the putative iPSC colonies were stained for immunoflourescence with the following antibodies: TRA-1-60 (1∶200, Millipore), NANOG (1∶1000, Abcam), OCT4 (1∶100, Santa Cruz Biotechnology), SOX2 (1∶100), cMYC and HMGA1 (1∶100) followed by secondary antibodies conjugated to a Alexa Probes (Molecular Devices) as previously described [68]. Immunofluorescence intensity calculations were performed using MetaMorph (Universal Imaging) version 7.7. Alkaline phosphatase staining was performed using the Alkaline Phosphatase detection kit (Millipore).
MTT Cell Proliferation Assays
Cells (5,000) were plated onto 96 well plates coated in matrigel with conditioned media obtained from mouse embryonic fibroblasts as described [29]. The media was replaced daily. MTT assays (Invitrogen) were performed daily for 5 days using 100 µl of MTT solution (5 mg/ml) added to each well and incubated for 3 h at 37°C according to manufacturer’s instructions. The formed MTT formazan crystals were dissolved with 500 µL DMSO, and the spectrophotometric assay was carried out at 590 nm as described. Each condition was done in quadruplicate, and 2 independent experiments were performed.
Gene Expression Analysis with Quantitative, Reverse Transcription PCR
Total RNA was isolated using the miRNeasy kit (Qiagen) and analyzed by quantitative reverse transcription PCR (qRT-PCR) as we previously described [29]. The sequences for the forward and reverse primers are listed in Table S1. For transgene expression analysis, one primer was designed with sequence from the pMX retroviral vector and the other primer was designed with sequence from the gene of interest. The expression level of each gene was normalized to the TATA-binding protein (TBP) gene.
Chromatin Immunoprecipitation (ChIP)
H9 hESCs cells (approximately 5 million) were washed twice with PBS and collected following incubation in trypsin (0.25%). Protein was cross-linked to DNA by treatment with formaldehyde for 8 minutes, after which the reaction was stopped with glycine. Cells were pelleted and resuspended in cell lysis buffer along with a protease inhibitor cocktail (Roche). After 10 minutes on ice, the nuclei were pelleted and resuspended in 200 µl nuclei lysis buffer with protease inhibitors. Chromatin was sheared by sonication using the BioRupter® (Diagenode) for two runs of 10 cycles. ChIP buffer was added to the sonicated samples to a final volume of 1 ml. ChIP was performed either by using the Auto-histone ChIP-seq kit on the SX-8G IP-Star® Compact platform (Diagenode) or the SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology) according to the manufacturers’ instructions.
The sheared DNA-protein complexes were immunoprecipitated using antibodies to HMGA1 as we described [29]. An IgG antibody was used as a negative control. Sequence from an approximately 200 base pair regions of the promoter of the pluripotency genes containing the predicted HMGA1 binding site was amplified using PCR. As a positive control, we amplified the 200 bp region containing the cMYC DNA binding site in the B23 promoter and performed immunoprecipitation with the cMYC antibody as previously described [69].
Genome-wide DNA Methylation Analysis
To assess global promoter methylation, we used the Infinium (Illumina, Inc.) platform to analyze bisulfate-treated DNA (EX DNA Methylation kit, Zymo Research) containing 27,578 informative sites near promoter regions as previously described [10]. Briefly, β values are generated as the signal of methylation-specific probe over the sum of the signals of the methylated and unmethylated-specific probes. The score of 1.0 is assigned for full methylation of a specific CpG site, 0 for the absence of methylation, with 0≤β≤1.0 for all signals. Probes with poor overall signals (p>0.05) were removed from analysis. Only probes positioned from −1,000 to+200 base pairs around transcription start sites (TSS) were analyzed. Heat maps were based on hierarchical clustering of β values using Euclidean distance and Ward’s algorithm, and all probes mapped to the genome (National Center for Biotechnology Information Build 36.3) using the bowtie algorithm and ultrafast and memory-efficient alignment of short DNA sequences (Genome Biology, 10, R25) with genome annotation matching the release of the Ensembl database. X-linked genes were removed from the analysis.
Teratoma Assay
iPSCs were expanded to>80% confluency on 6-well plates. For each teratoma, cells from 6 wells were treated with trypsin, washed in PBS, and resuspended in 100 µl PBS. Mice (NOD/SCID) were injected subcutaneously with a mixture of 100 µl cells+100 µl hESC qualified Matrigel (BD Biosciences). The mice underwent necropsy when teratomas became evident (after 6–8 weeks). Tumors were excised and tissues stained with hematoxylin and eosin (H & E) to identify the various germ layers.
Supporting Information
HMGA1 does not alter proliferation in hESCs. The MTT cell proliferation assay shows that the H9 hESCs transduced to express HMGA1 grow at a similar rate to that observed in the control H9 hESCs, transduced with the GFP vector alone. This assay was done in triplicate; each time point shows the mean+/− the standard deviation.
(DOCX)
HMGA1 promotes cellular reprogramming of IMR90. A) Reprogramming with HMGA1-OSKM results in more TRA-1-60+ iPSC colonies compared to controls. B) The HMGA1-OSKM TRA-1-60+ colonies are significantly larger than the control-OSKM colonies. Numbers represent µm diameters.
(DOCX)
Transgene expression in early stage reprogramming pools. Expression levels of the OCT4, SOX2, cMYC, and KLF4 transgenes were analyzed by qRT-PCR at day 12 and day 21 following the start of reprogramming in MSCs.
(DOCX)
HMGA1 KD targets pluripotency genes. Pluripotency genes (SOX2, OCT4, cMYC, LIN28) are repressed following knockdown of HMGA1, assessed 24 hours following siRNA transfection.
(DOCX)
Global promoter DNA methylation signatures in HMGA1-OSKM or control-OSKM iPSCs. Unsupervised hierarchical clustering of CpG loci shows the greatest variation across cell types. The 2D-hierachial cluster analysis, performed using the Euclidean distance on 38 cell lines, and 414 loci, places the cell lines described in this study into context in the complex network of methylation changes described in Ohm et al. [12]. The HMGA1-OSKM lines are marked in the top margin in blue, while the control-OSKM lines are marked in green. The partially reprogrammed cells collected at days 12 and 21 cluster on the right side with fibroblasts and other partially reprogrammed iPSCs, while the late passage HMGA1-OSKM or control-OSKM lines are found on the left with hESCs and other fully reprogrammed iPSC lines. Methylation patterns for most of the cancer cells (colon, breast, osteosarcoma, fibrosarcoma) located in the middle of the heat map are distinct from both the fibroblasts and pluripotent cells, with more extensive methylation globally and patterns that are negatively correlated with the methylation patterns observed in pluripotent cells. Dark blue – low methylation, light blue – high methylation.
(DOCX)
Primers used in this study.
(DOCX)
Acknowledgments
We thank Drs. Linzhao Cheng and Prashant Mali for advice with the iPSC experiments and Dr. Steven Baylin for sharing methylome data from iPSCs and hESCs.
Funding Statement
This work was funded by the National Institutes of Health (LR, CK, DLH), Alex's Lemonade Stand Foundation (LR, AB, JH), St. Baldrick's Foundation (LR) and the Maryland Stem Cell Research Fund (SNS, LC, EZ, AB, LR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. [DOI] [PubMed] [Google Scholar]
- 2. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. (2007) Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 318: 1917–1920. [DOI] [PubMed] [Google Scholar]
- 3. Zhao R, Daley GQ (2008) From fibroblasts to iPS cells: Induced pluripotency by defined factors. Journal of Cellular Biochemistry 105: 949–955. [DOI] [PubMed] [Google Scholar]
- 4. Wang Y, Mah N, Prigione A, Wolfrum K, Andrade-Navarro M, et al. (2008) A Transcriptional Roadmap to the Induction of Pluripotency in Somatic Cells. Stem Cell Reviews and Reports 6: 282–296. [DOI] [PubMed] [Google Scholar]
- 5. Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, et al. (2008) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 26: 101–106. [DOI] [PubMed] [Google Scholar]
- 6. Park IH, Zhao R, West JA, Yabuuchi A, Huo H, et al. (2008) Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451: 141–146. [DOI] [PubMed] [Google Scholar]
- 7. Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, et al. (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8: 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, et al. (2011) Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144: 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, et al. (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mali P, Ye Z, Hommond HH, Yu X, Lin J, et al. (2008) Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells 26: 1998–2005. [DOI] [PubMed] [Google Scholar]
- 11. Mali P, Chou BK, Yen J, Ye Z, Zou J, et al. (2010) Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 28: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohm JE, Mali P, Van Neste L, Berman DM, Liang L, et al. (2010) Cancer-Related Epigenome Changes Associated with Reprogramming to Induced Pluripotent Stem Cells. Cancer Research 70: 7662–7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chou B-K, Mali P, Huang X, Ye Z, Dowey S, et al. (2011) Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res 21: 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou G, Chen J, Lee S, Clark T, Rowley JD, et al. (2001) The pattern of gene expression in human CD34(+) stem/progenitor cells. Proc Natl Acad Sci U S A 98: 13966–13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karp JE, Smith BD, Resar LS, Greer JM, Blackford A, et al. (2011) Phase 1 and pharmacokinetic study of bolus-infusion flavopiridol followed by cytosine arabinoside and mitoxantrone for acute leukemias. Blood 117: 3302–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nelson DM, Joseph B, Hillion J, Segal J, Karp JE, et al. (2011) Flavopiridol induces BCL-2 expression and represses oncogenic transcription factors in leukemic blasts from adults with refractory acute myeloid leukemia. Leuk Lymphoma 52: 1999–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dang CV, Resar LM, Emison E, Kim S, Li Q, et al. (1999) Function of the c-Myc oncogenic transcription factor. Exp Cell Res 253: 63–77. [DOI] [PubMed] [Google Scholar]
- 18. Wood LJ, Maher JF, Bunton TE, Resar LM (2000) The oncogenic properties of the HMG-I gene family. Cancer Res 60: 4256–4261. [PubMed] [Google Scholar]
- 19. Wood LJ, Mukherjee M, Dolde CE, Xu Y, Maher JF, et al. (2000) HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol Cell Biol 20: 5490–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pedulla ML, Treff NR, Resar LM, Reeves R (2001) Sequence and analysis of the murine Hmgiy (Hmga1) gene locus. Gene 271: 51–58. [DOI] [PubMed] [Google Scholar]
- 21. Dolde CE, Mukherjee M, Cho C, Resar LMS (2002) HMG-I/Y in human breast cancer cell lines. Breast Cancer Research and Treatment 71: 181–191. [DOI] [PubMed] [Google Scholar]
- 22. Dhar A, Hu J, Reeves R, Resar LM, Colburn NH (2004) Dominant-negative c-Jun (TAM67) target genes: HMGA1 is required for tumor promoter-induced transformation. Oncogene 23: 4466–4476. [DOI] [PubMed] [Google Scholar]
- 23. Takaha N, Resar LMS, Vindivich D, Coffey DS (2004) High mobility group protein HMGI(Y) enhances tumor cell growth, invasion, and matrix metalloproteinase-2 expression in prostate cancer cells. The Prostate 60: 160–167. [DOI] [PubMed] [Google Scholar]
- 24. Hommura F, Katabami M, Leaner VD, Donninger H, Sumter TF, et al. (2004) HMG-I/Y is a c-Jun/activator protein-1 target gene and is necessary for c-Jun-induced anchorage-independent growth in Rat1a cells. Mol Cancer Res 2: 305–314. [PubMed] [Google Scholar]
- 25. Xu Y, Sumter TF, Bhattacharya R, Tesfaye A, Fuchs EJ, et al. (2004) The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia. Cancer Res 64: 3371–3375. [DOI] [PubMed] [Google Scholar]
- 26. Tesfaye A, Di Cello F, Hillion J, Ronnett BM, Elbahloul O, et al. (2007) The high-mobility group A1 gene up-regulates cyclooxygenase 2 expression in uterine tumorigenesis. Cancer Res 67: 3998–4004. [DOI] [PubMed] [Google Scholar]
- 27. Fusco A, Fedele M (2007) Roles of HMGA proteins in cancer. Nat Rev Cancer 7: 899–910. [DOI] [PubMed] [Google Scholar]
- 28. Di Cello F, Hillion J, Kowalski J, Ronnett BM, Aderinto A, et al. (2008) Cyclooxygenase inhibitors block uterine tumorigenesis in HMGA1a transgenic mice and human xenografts. Molecular Cancer Therapeutics 7: 2090–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hillion J, Dhara S, Sumter TF, Mukherjee M, Di Cello F, et al. (2008) The high-mobility group A1a/signal transducer and activator of transcription-3 axis: an achilles heel for hematopoietic malignancies? Cancer Res 68: 10121–10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hillion J, Wood LJ, Mukherjee M, Bhattacharya R, Di Cello F, et al. (2009) Upregulation of MMP-2 by HMGA1 promotes transformation in undifferentiated, large-cell lung cancer. Mol Cancer Res 7: 1803–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hristov AC, Cope L, Di Cello F, Reyes MD, Singh M, et al. (2010) HMGA1 correlates with advanced tumor grade and decreased survival in pancreatic ductal adenocarcinoma. Mod Pathol 23: 98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Resar LMS (2010) The High Mobility Group A1 Gene: Transforming Inflammatory Signals into Cancer? Cancer Research 70: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schuldenfrei A, Belton AK, Kowalski J, Talbot CC, Di Cello F, et al. (2011) HMGA1 Drives Stem Cell, Inflammatory Pathway, and Cell Cycle Progression Genes During Lymphoid Tumorigenesis. BMC Genomics 12: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Belton A, Gabrovsky A, Iacobuzio-Donahue C, Huso DL, Resar LMS (2012) HMGA1 induces intestinal polyposis in transgenic mice and drives tumor progression and stem cell properties in colon cancer cells. PLoS One 7: e30034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shah SN, Resar LMS (2012) High Mobility Group A1 and Cancer: Potential Biomarker and Therapeutic Target. Histol Histopathol 27: 567–579. [DOI] [PubMed] [Google Scholar]
- 36. Hillion J, Smail S, Di Cello F, Belton A, Huso T, et al. (2012) The HMGA1a-COX-2 axis: A molecular pathway leading to tumor progression in human pancreatic adenocarcinoma Pancreatology. 12: 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belton A, Paz-Priel I, Friedman AD, Huso D, Resar L (2012) The high mobility group A1 oncogene & NF-κB cooperate in malignant transformation. AACR 2012 [Abstract #3039].
- 38. Johnson KR, Cook SA, Davisson MT (1992) Chromosomal localization of the murine gene and two related sequences encoding high-mobility-group I and Y proteins. Genomics 12: 503–509. [DOI] [PubMed] [Google Scholar]
- 39. Johnson KR, Lehn DA, Elton TS, Barr PJ, Reeves R (1988) Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG-I(Y). J Biol Chem 263: 18338–18342. [PubMed] [Google Scholar]
- 40. Di Cello F, Hillion J, Hristov A, Wood LJ, Mukherjee M, et al. (2008) HMGA2 participates in transformation in human lung cancer. Mol Cancer Res 6: 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hristov AC, Cope L, Reyes MD, Singh M, Iacobuzio-Donahue C, et al. (2009) HMGA2 protein expression correlates with lymph node metastasis and increased tumor grade in pancreatic ductal adenocarcinoma. Mod Pathol 22: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Resar LM, Brodsky RA (2011) “Let”-ing go with clonal expansion? Blood 117: 5788–5790. [DOI] [PubMed] [Google Scholar]
- 43. Reeves R, Beckerbauer L (2001) HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta 1519: 13–29. [DOI] [PubMed] [Google Scholar]
- 44.Moliterno A, Resar L (2011) AKNA: Another AT-hook transcription factor hooking up with inflammation? Cell Res 21L 1528–1530. [DOI] [PMC free article] [PubMed]
- 45. Reeves R, Edberg DD, Li Y (2001) Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol Cell Biol 21: 575–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fedele M, Pentimalli F, Baldassarre G, Battista S, Klein-Szanto AJ, et al. (2005) Transgenic mice overexpressing the wild-type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene 24: 3427–3435. [DOI] [PubMed] [Google Scholar]
- 47. Flohr AM, Rogalla P, Bonk U, Puettmann B, Buerger H, et al. (2003) High mobility group protein HMGA1 expression in breast cancer reveals a positive correlation with tumour grade. Histol Histopathol 18: 999–1004. [DOI] [PubMed] [Google Scholar]
- 48. Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, et al. (2002) Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415: 436–442. [DOI] [PubMed] [Google Scholar]
- 49. Sarhadi VK, Wikman H, Salmenkivi K, Kuosma E, Sioris T, et al. (2006) Increased expression of high mobility group A proteins in lung cancer. J Pathol 209: 206–212. [DOI] [PubMed] [Google Scholar]
- 50. Zambidis ET, Park TS, Yu W, Tam A, Levine M, et al. (2008) Expression of angiotensin-converting enzyme (CD143) identifies and regulates primitive hemangioblasts derived from human pluripotent stem cells. Blood 112: 3601–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chaerkady C, Kerr CL, Marimuthu A, Kelkar DS, Kashyap MK, et al. (2009) Temporal analysis of neural differentiation using quantitative proteomics. J Proteome Res 8: 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Letzen BS, Liu C, Thakor NV, Gearhart JD, All AH, et al. (2010) MicroRNA expression profiling of oligodendrocyte differentiation from human embryonic stem cells. PLoS One 5: e10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, et al. (2003) Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol 31: 1007–1014. [PubMed] [Google Scholar]
- 54. Mali P, Ye Z, Chou BK, Yen J, Cheng L (2010) An improved method for generating and identifying human induced pluripotent stem cells. Methods Mol Biol 636: 191–205. [DOI] [PubMed] [Google Scholar]
- 55. Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, et al. (2009) Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotech 27: 1033–1037. [DOI] [PubMed] [Google Scholar]
- 56. Liau SS, Jazag A, Whang EE (2006) HMGA1 is a determinant of cellular invsiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res 66: 11613–11622. [DOI] [PubMed] [Google Scholar]
- 57. Quandt K, Frech K, Karas H, Wingender E, Werner T (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23: 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Battista S, Pentimalli F, Baldassarre G, Fedele M, Fidanza V, et al. (2003) Loss of HMGA1 gene function affects embryonic stem cell lympho-hematopoietic differentiation. FASEB J 17: 1496–1498. [DOI] [PubMed] [Google Scholar]
- 59. Zhou X, Benson KF, Ashar HR, Chada K (1995) Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376: 771–774. [DOI] [PubMed] [Google Scholar]
- 60. Miyoshi N, Ishii H, Nagai Ki, Hoshino H, Mimori K (2010) Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci U S A 107: 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin SL, Chang DC, Chang-Lin D, Lin CH, We DT (2008) Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 14: 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Utikal J, Maherali N, Kulahert W, Hochedlinger K (2009) Sox2 is dispensible for the reprogramming of melanocytes and melanoma cells into induce pluripotent stem cells. J Cell Sci 122: 3502–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nagai Ki, Ishii H, Miyoshi N, Hoshino H, Saito T, et al. (2011) Long-term culture following ES-like gene-induced reprogramming elicits an aggressive phenotype in mutated cholangiocellular carcinoma cells. Biochem Biophys Res Commun 395: 258–263. [DOI] [PubMed] [Google Scholar]
- 64. Zhao T, Zhang ZN, Rong Z, Xu Y (2011) Immunogenicity of induced pluripotent stem cells. Nature 474: 212–215. [DOI] [PubMed] [Google Scholar]
- 65. Peng S, Chen LL, Lei XX, Yang L, Lin H, et al. (2011) Genome-wide studies reveal that Lin28 enhances the tanslation of genes important for growth and survival of human embryonic stem cells. Stem Cells 29: 496–504. [DOI] [PubMed] [Google Scholar]
- 66. Andrews PW, Damjanov I, Simon D, Banting GS, Carlin C, et al. (1984) Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Invest 50: 147–162. [PubMed] [Google Scholar]
- 67. Himes SR, Reeves R, Attema J, Nissen M, Li Y, et al. (2000) The role of high-mobility group I(Y) proteins in expression of IL-2 and T cell proliferation. J Immunol 164: 3157–3168. [DOI] [PubMed] [Google Scholar]
- 68. Chaerkady R, Kerr CL, Kandasamy K, Marimuthu A, Gearhart JD, et al. (2010) Comparative proteomics of human embryonic stem cells and embryonal carcinoma cells. Proteomics 10: 1359–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zeller KI, Haggerty TJ, Barrett JF, Guo Q, Wonsey DR, et al. (2001) Characterization of nucleophosmin (B23) as a Myc target by scanning chromatin immunoprecipitation. J Biol Chem 276: 48285–48291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HMGA1 does not alter proliferation in hESCs. The MTT cell proliferation assay shows that the H9 hESCs transduced to express HMGA1 grow at a similar rate to that observed in the control H9 hESCs, transduced with the GFP vector alone. This assay was done in triplicate; each time point shows the mean+/− the standard deviation.
(DOCX)
HMGA1 promotes cellular reprogramming of IMR90. A) Reprogramming with HMGA1-OSKM results in more TRA-1-60+ iPSC colonies compared to controls. B) The HMGA1-OSKM TRA-1-60+ colonies are significantly larger than the control-OSKM colonies. Numbers represent µm diameters.
(DOCX)
Transgene expression in early stage reprogramming pools. Expression levels of the OCT4, SOX2, cMYC, and KLF4 transgenes were analyzed by qRT-PCR at day 12 and day 21 following the start of reprogramming in MSCs.
(DOCX)
HMGA1 KD targets pluripotency genes. Pluripotency genes (SOX2, OCT4, cMYC, LIN28) are repressed following knockdown of HMGA1, assessed 24 hours following siRNA transfection.
(DOCX)
Global promoter DNA methylation signatures in HMGA1-OSKM or control-OSKM iPSCs. Unsupervised hierarchical clustering of CpG loci shows the greatest variation across cell types. The 2D-hierachial cluster analysis, performed using the Euclidean distance on 38 cell lines, and 414 loci, places the cell lines described in this study into context in the complex network of methylation changes described in Ohm et al. [12]. The HMGA1-OSKM lines are marked in the top margin in blue, while the control-OSKM lines are marked in green. The partially reprogrammed cells collected at days 12 and 21 cluster on the right side with fibroblasts and other partially reprogrammed iPSCs, while the late passage HMGA1-OSKM or control-OSKM lines are found on the left with hESCs and other fully reprogrammed iPSC lines. Methylation patterns for most of the cancer cells (colon, breast, osteosarcoma, fibrosarcoma) located in the middle of the heat map are distinct from both the fibroblasts and pluripotent cells, with more extensive methylation globally and patterns that are negatively correlated with the methylation patterns observed in pluripotent cells. Dark blue – low methylation, light blue – high methylation.
(DOCX)
Primers used in this study.
(DOCX)