Abstract
The aim of this study was to better understand the altered functional modules in breast cancer at pathway and network levels. An integrated bioinformatics analysis of differentially expressed proteins in human breast cancer was performed. Breast cancer protein profiles were constructed by data mining proteins in literature and public databases, including 1031 proteins with 153 secretory and 69 cell surface proteins. An experimental investigation was performed by two-dimensional electrophoresis, and 4 proteins were further validated by western blotting. Enriched bioinformatics functions were clustered. This study may be used as a reference in further studies to help identify the underlying biological interactions associated with breast cancer and discover potential cancer targets.
Keywords: bioinformatics, biomarker, breast cancer, cancer/testis antigen, proteomes
Introduction
Breast cancer is the most common neoplasia in women and its pathogenesis is related to an acquired or inherited genetic disorder influenced by environmental, behavioral or reproductive factors (1). Cancer biomarker discovery is important for both cancer biology and clinical applications. These markers may come from DNA, RNA, miRNA or proteins (2), with proteins being the most significant (3).
The development and improvement of biotechnologies has allowed researchers to perform high-throughput analyses of genomes, transcriptomes and proteomes in health and disease, and identify hundreds of potential biomarkers (4), offering the potential to discover diagnostic, prognostic or therapeutic targets. However, less than two dozen cancer biomarkers are currently approved by the Food and Drug Administration (FDA) (5), including only 9 protein biomarkers identified in the blood (6). Due to the lack of sensitivity and specificity of these known biomarkers (7), researchers continue to search for more significant targets. Proteomics is a promising approach for the discovery of cancer targets and biomarkers (8). The mapping of proteome profiles and differential proteomics has been widely performed in breast cancer to identify potential biomarkers (9). The identified proteins were reported to have potential clinical significance, and certain proteins may be used as potential diagnostic, prognostic or predictive biomarkers (10,11,12,13,14). However, due to the heterogeneity in the different studies, including experimental design, sample collection and classification and analytical method (15), these results lack good reproducibility and require further validation before they can be used in clinical detection and to explain the underlying mechanisms of breast cancer. In addition, few protein candidates were warranted to be specific to breast cancer, and were often differentially expressed in other cancer types (16). Hence, research encountered the challenge of how to decipher and use these individual results and bring them into clinical applications. In addition, an understanding of the underlying biological mechanisms of carcinogenesis and the altered molecular events in breast cancer at integrated pathway levels is necessary. Proteins do not act alone to perform biological functions, but through complex biological pathways. The discovery of these intricate pathways is essential to understanding biological mechanisms. A wide variety of cancers may also link to the same pathways that affect tumorigenesis and progression through altering protein expressions. Therefore, once these pathways are known, it may be easier to monitor different aspects of cancer progression and develop a therapeutic strategy by focusing on pathways instead of individual proteins. The enriched pathways or functions may be the most probable cause of cancer (17), and the enriched proteins involved in these processes could in turn serve as target agents in diagnosis or treatment. Several monoclonal antibodies and small molecular inhibitors have been developed to target certain molecular pathways involved in cell growth, survival and metastasis in breast cancer (18,19,20). Therefore, integrated bioinformatics should be applied when discovering cancer-associated pathways and networks to improve our understanding of cancer biology, as well as cancer diagnosis and therapeutics.
In the present study, we integrated protein profiles in breast cancers from literature and public databases to perform bioinformatical analyses. Differential serum proteomics between human breast cancer patients and healthy volunteers were performed, and western blotting was used to validate the differential expression of secretory proteins in breast cancer. Our aim was not to discover all the breast cancer-associated proteins but rather to focus on screening enriched signaling pathways in breast cancer which may provide new insights into breast cancer research. This bioinformatical insight into breast cancer-associated protein profiles may potentially provide clues for identifying new functional modules in breast cancer and may be used to understand the underlying tumorigenesis process.
Materials and methods
Patient characteristics and serum collection
Blood samples were collected from 25 breast cancer patients and 20 healthy volunteers at the Yantai Yu-Huang-Ding Hospital, China, with written informed consent and approval of the Yu-Huang-Ding Hospital research and ethics committee. Venous blood was drawn from each subject into 10-ml fasting blood tubes and allowed to clot at room temperature for 1 h. Serum was separated by centrifugation at 2000 g for 15 min at 4°C. Proteins in the supernatant were precipitated by mixing with 4 volumes of ice-cold acetone and allowing it to stand at −20°C for 1 h. After being centrifuged at 12000 g for 1 h, washed with 90% acetone and dried, the proteins were taken up in 2 ml lysis solution (7 M urea, 2 M thiourea and 65 mM dithiothreitol) and stored at −80°C.
Collection of breast cancer-associated proteins
Breast cancer protein profiles were constructed by retrieving public databases (Uniprot, Release 2011_11, http://www.uniprot.org) and literature on differential proteomes in breast cancer (dbDEPC 2.0, http://lifecenter.sgst.cn/dbdepc/index.do). All human proteins in the Uniprot database were downloaded with all annotations. Breast cancer-associated proteins were further selected manually from the downloaded data using the keywords ‘cancer’, ‘tumor’ or ‘carcinoma’. The regulation levels were recorded as ‘over-regulation’, ‘downregulation’ and ‘no-annotation’. All proteins extracted from the literature were further grouped into over- and downregulation clusters, and certain proteins with controversial expression in different reports were clustered into the no-annotation group.
Bioinformatics analysis
Ontological analysis
All breast cancer-associated proteins were classified into different protein classes according to Interpro (www.ebi.ac.uk/interpro) and Gene Ontology (GO) annotation (www.geneontology.org), and significantly enriched functions were further selected according to Panther (http://www.pantherdb.org/).
Pathway analysis
Ingenuity Pathway Analysis v8.0-2803 (Ingenuity Systems, Redwood City, CA, USA) was used to analyze pathways and networks involving the breast cancer-associated proteins. The following settings were used: reference set, Ingenuity Knowledge Base (genes only); network analysis, direct and indirect relationships; molecules per network, 35; networks per analysis, 25. All species, tissues and cell lines were used for the analysis. IPA uses Fisher’s exact test to determine which pathways (canonical pathways, toxicity pathways or biological functions) are significantly linked to the input protein set compared with the whole Ingenuity Knowledge Base.
Selection of secretory and cell surface proteins
Secretory proteins and cell surface proteins are promising biomarkers. All cancer-associated proteins were compared with the serum/plasma proteome (21) to select secretory proteins, and compared with the cell surfaceome (22) to select cell surface proteins. GO was used to further filter the results.
Proteomic analysis of human serum from patients with breast cancer and healthy volunteers
Two-dimensional gel electrophoresis and mass spectrometric analyses were performed as described in our previous study (23). Gels were made in triplicate to confirm the spot patterns and were scanned with a Z320 scanner (Founder, Beijing, China). The gel images were processed with ImageMaster software (GE Healthcare, Piscataway, NJ, USA). Images were briefly checked manually to eliminate artefacts. Following spot detection, a match set was built including all the experimental and control gels. The significance of the differential expression of protein spots between experimental and control groups was estimated by mean ratio >2.0 and the independent samples t-test; P<0.05 was considered to indicate a statistically significant result.
Western blot analysis
A total of 50 μg of pooled proteins were separated at 12% SDS-PAGE by electrophoresis and then transferred onto a nitrocellulose membrane (Millipore, Bedford, MA, USA). The membranes were blocked with 5% milk TBST (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Tween-20) overnight at 4°C and then hybridized with the following primary antibodies: anti-ORM2 (sc-51020), anti-APOA1 (sc-69755), anti-GC (sc-18706) and anti-CLU (sc-166907), all from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). The immune complexes were visualized with a DAB staining kit (Zhongshan Jinqiao Technology, China).
Results
Overview of breast cancer-associated proteins
The breast cancer-associated protein profile was mainly comprised of proteins screened from the Uniprot database and literature reports. A total of 803 breast cancer-associated proteins were screened from literature reports and 243 were selected in the Uniprot database. Finally, 1031 breast cancer-associated proteins were obtained including 514 (49.6%) upregulated and 318 (30.8%) downregulated proteins. One hundred and seventy-two proteins required further confirmation of expression.
Bioinformatics analysis
From protein lists to biological functions
According to GO analysis, Interpro and literature annotations, the enriched biological functions are listed in Table I. Metabolic process, chaperone transcription and catalytic activity-associated proteins were more significantly enriched in breast cancer-associated proteins. The comparison of up- and downregulated proteins demonstrated that more chaperone, cell adhesion, transporter and antioxidant-related proteins were present in the upregulated group, while defense/immunity and extracellular proteins were present in the downregulated group.
Table I.
Broad biological functions of breast cancer protein profiles.
| Protein profiles a
|
Upregulated proteins
|
Downregulated proteins
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Biological functions | Observed nos. | Expected nos. | P-value | Observed nos. | Expected nos. | P-value | Observed nos. | Expected nos. | P-value |
| Antioxidant activity | 4 | 0.75 | 7.10e−3 | 10 | 1.41 | 2.31e−6 | 3 | 0.43 | 9.28e−3 |
| Apoptosis | 36 | 24.84 | 1.81e−2 | 73 | 46.96 | 1.78e−4 | 24 | 14.17 | 8.92e−3 |
| Binding | 158 | 173.60 | 7.86e−2 | 358 | 328.21 | 2.39e−2 | 126 | 99.01 | 6.36e−4 |
| Calmodulin | 7 | 1.60 | 1.34e−3 | 1 | 0.85 | 5.72e−1 | 1 | 0.48 | 3.84e−1 |
| Catalytic activity | 213 | 137.21 | 3.29e−13 | 375 | 259.42 | 4.35e−16 | 104 | 78.25 | 5.80e−4 |
| Cell adhesion | 13 | 4.52 | 8.11e−4 | 12 | 2.39 | 7.51e−6 | 0 | 1.36 | 2.55e−1 |
| Cell-cell signaling | 19 | 34.23 | 2.61e−3 | 43 | 64.71 | 2.04e−3 | 15 | 19.52 | 1.74e−1 |
| Chaperone | 28 | 6.32 | 1.56e−10 | 22 | 3.34 | 8.96e−12 | 5 | 1.91 | 4.40e−2 |
| Cytoskeletal protein | 55 | 21.44 | 5.64e−10 | 27 | 11.34 | 4.24e−5 | 16 | 6.47 | 9.47e−4 |
| Defense/immunity protein | 10 | 5.20 | 3.93e−2 | 2 | 2.75 | 4.81e−1 | 8 | 1.57 | 2.15e−4 |
| Extracellular matrix protein | 7 | 3.50 | 6.50e−2 | 1 | 1.85 | 4.47e−1 | 5 | 1.06 | 4.50e−3 |
| G-protein coupled receptor | 1 | 21.73 | 6.62e−9 | 0 | 11.49 | 8.94e−6 | 0 | 6.56 | 1.32e−3 |
| Hsp70 family chaperone | 5 | 0.63 | 4.94e−4 | 4 | 0.33 | 3.95e−4 | 1 | 0.19 | 1.74e−1 |
| Isomerase activity | 12 | 4.86 | 4.19e−3 | 26 | 9.19 | 3.65e−6 | 6 | 2.77 | 6.18e−2 |
| Lyase activity | 17 | 5.30 | 3.54e−5 | 24 | 10.01 | 1.11e−4 | 6 | 3.02 | 8.50e−2 |
| Metabolic process | 283 | 212.58 | 2.58e−10 | 529 | 401.91 | 1.39e−16 | 157 | 121.24 | 1.64e−5 |
| Metalloprotease | 16 | 7.05 | 2.47e−3 | 7 | 3.73 | 8.36e−2 | 4 | 2.13 | 1.66e−1 |
| Oxidoreductase activity | 59 | 18.08 | 3.86e−15 | 105 | 34.18 | 1.40e−23 | 31 | 10.31 | 7.17e−8 |
| Peptidase activity | 28 | 18.44 | 2.06e−2 | 53 | 34.86 | 2.10e−3 | 14 | 10.51 | 1.72e−1 |
| Peptidase inhibitor activity | 11 | 4.55 | 6.96e−3 | 22 | 8.61 | 8.61e−5 | 6 | 2.60 | 4.79e−2 |
| Peroxidase activity | 4 | 0.69 | 5.55e−3 | 8 | 1.31 | 6.73e−5 | 2 | 0.40 | 6.04e−2 |
| Protease | 42 | 23.14 | 2.23e−4 | 22 | 12.24 | 6.84e−3 | 11 | 6.98 | 9.46e−2 |
| Protease inhibitor | 15 | 5.30 | 3.93e−4 | 6 | 2.80 | 6.48e−2 | 5 | 1.60 | 2.32e−2 |
| Protein transport | 67 | 42.33 | 1.43e−4 | 111 | 80.02 | 3.43e−4 | 23 | 24.14 | 4.58e−1 |
| Receptor activity | 30 | 46.49 | 4.84e−3 | 56 | 87.90 | 9.95e−5 | 19 | 26.51 | 7.17e−2 |
| Structural molecule activity | 62 | 38.26 | 1.39e−4 | 133 | 72.34 | 1.34e−11 | 48 | 21.82 | 2.42e−7 |
| Transcription factor | 44 | 100.49 | 3.02e−11 | 20 | 53.15 | 5.28e−8 | 17 | 30.31 | 4.39e−3 |
| Transfer/carrier protein | 29 | 12.06 | 2.12e−5 | 10 | 6.38 | 1.11e−1 | 7 | 3.64 | 7.51e−2 |
| Transferase activity | 59 | 40.96 | 3.27e−3 | 96 | 77.45 | 1.85e−2 | 25 | 23.36 | 3.92e−1 |
| Translation factor | 10 | 2.72 | 5.21e−4 | 6 | 1.44 | 3.61e−3 | 3 | 0.82 | 5.03e−2 |
| Transport | 99 | 73.47 | 1.18e−3 | 183 | 138.90 | 5.80e−5 | 50 | 41.90 | 1.04e−1 |
| Vesicle coat protein | 6 | 1.46 | 3.86e−3 | 6 | 0.77 | 1.48e−4 | 0 | 0.44 | 6.44e−1 |
| Unclassified | 93 | 171.80 | 6.09e−15 | 95 | 173.91 | 7.31e−15 | 45 | 97.98 | 2.04e−12 |
Protein profiles included all the breast cancer proteins.
From biological functions to pathways
To explore the enrichment pathways of these proteins with different biological functions, a pathway analysis was performed using Ingenuity Pathway Analysis tools and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Thirty-three pathways were identified in breast cancers, including 25 pathways enriched in upregulated proteins and 8 pathways with downregulated proteins (Table II).
Table II.
Canonical pathways in up- and downregulated breast cancer proteins.
| Pathway | Ratio | Number | P-value |
|---|---|---|---|
| Upregulated | |||
| Glycolysis | 0.18 | 15 | 4.88e−8 |
| Pyruvate metabolism | 0.18 | 11 | 3.52e−6 |
| Protein ubiquitination pathway | 0.09 | 23 | 1.30e−5 |
| RhoA signaling | 0.12 | 13 | 5.00e−5 |
| NRF2-mediated oxidative stress response | 0.09 | 17 | 6.71e−5 |
| PI3K/AKT signaling | 0.10 | 13 | 8.75e−5 |
| Fatty acid metabolism | 0.13 | 13 | 2.62e−4 |
| Integrin signaling | 0.08 | 16 | 2.80e−4 |
| ILK signaling | 0.08 | 15 | 2.82e−4 |
| 14-3-3-mediated signaling | 0.10 | 12 | 3.10e−4 |
| RAN signaling | 0.28 | 5 | 3.16e−4 |
| Aryl hydrocarbon receptor signaling | 0.12 | 17 | 3.48e−4 |
| Clathrin-mediated endocytosis signaling | 0.08 | 14 | 6.40e−4 |
| Valine, leucine and isoleucine degradation | 0.13 | 8 | 7.71e−4 |
| IGF-1 signaling | 0.10 | 10 | 1.12e−3 |
| VEGF signaling | 0.10 | 9 | 1.75e−3 |
| Arginine and proline metabolism | 0.11 | 8 | 2.10e−3 |
| EIF2 signaling | 0.07 | 14 | 2.19e−3 |
| Actin cytoskeleton signaling | 0.07 | 15 | 2.96e−3 |
| ERK5 signaling | 0.11 | 7 | 3.55e−3 |
| GABA receptor signaling | 0.13 | 6 | 4.00e−3 |
| Regulation of actin-based motility by Rho | 0.09 | 8 | 5.16e−3 |
| LPS/IL-1 mediated inhibition of RXR function | 0.06 | 13 | 9.42e−3 |
| HER-2 signaling in breast cancer | 0.09 | 7 | 1.10e−2 |
| Downregulated | |||
| Citrate cycle | 0.09 | 5 | 1.25e−4 |
| Acute phase response signaling | 0.06 | 10 | 6.53e−4 |
| P53 signaling | 0.07 | 7 | 1.13e−3 |
| Primary immunodeficiency signaling | 0.09 | 5 | 1.80e−3 |
| Urea cycle and metabolism of amino groups | 0.13 | 4 | 2.76e−3 |
| Cdc42 signaling | 0.06 | 8 | 3.40e−3 |
| Glyoxylate and dicarboxylate metabolism | 0.18 | 4 | 1.12e−3 |
| Autoimmune thyroid disease signaling | 0.08 | 3 | 1.02e−3 |
From biological pathways to networks
Network generation was performed using Ingenuity Pathway Analysis tools. This generated 25 networks using all the breast cancer-associated proteins and 13 networks using the upregulated proteins. Several functions were linked to more than 3 of the networks in the upregulated group and the main ones included cancer, genetic disorder, cell death and cellular movement.
Cancer/testis proteins, secretory proteins and cell surface proteins
Cancer/testis antigens are potential cancer biomarkers with restricted expression in the testis and certain cancer cells. The earlier comparison of breast cancer proteins showed that 119 testis proteins including 11 testis-specific proteins were included in this study. Twenty-one proteins were common to both the breast cancer proteins and cancer/testis antigens. The comparison of breast cancer proteins with human serum/plasma and cell surface proteins (surfaceome) revealed that the cancer proteins included 153 secretory proteins and 69 cell surface proteins
Experimental analysis by 2D-electrophoresis and western blotting
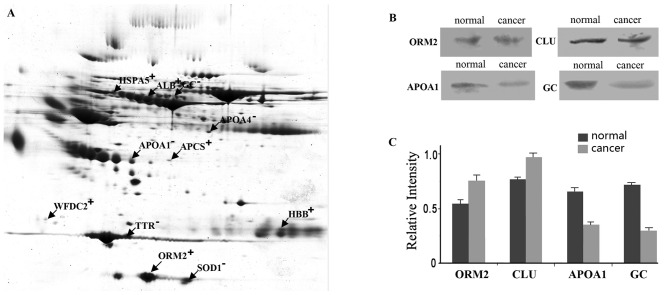
All gels were statistically analyzed through ImageMaster software, and 11 proteins were shown to be differentially expressed between breast cancer patients and healthy donors (Fig. 1A). Of the 4 proteins tested by western blotting, ORM2 and CLU were verified to be highly expressed in the serum of breast cancer patients, and APOA1 and GC were expressed at low levels (Fig. 1B).
Figure 1.
Western blot analysis of ORM2, CLU, APOA1 and GC. (A) Separation of breast cancer serum proteins by 2D-PAGE; (B) Western blot analysis was performed for the serum of breast cancer patients and healthy volunteers; (C) Western blot images were quantified by densitometric scanning and quantified using ImageQuant software.
Discussion
With the development and improvement of proteomic technology, cancer proteomes are evolving quickly and aid in the search for biomarkers and therapeutic targets. Breast cancer proteomes were widely analyzed and certain important proteins were identified (9,10). However, these data were heterogeneous and many of them lacked experimental verification and validation. In our preliminary study, 11 serum proteins were found to be differentially expressed between breast cancer patients and healthy individuals. The results provided insight into certain aspects of breast cancer, but had little correlation with all the breast cancer proteomes available. Thus, the integration of protein lists containing breast cancer proteomes to understand the correlations involved in regulation functions and networks may aid in the screening of diagnostic markers or therapeutic targets.
In the present study, integrated bioinformatical tools were used to analyze the enriched pathways and networks that are associated with breast cancer. The breast cancer protein profile was constructed, including proteins in the published literature of differential proteomics and the Uniprot database. Protein expressions in breast cancer were evaluated, and broadly categorized into three groups: upregulated, downregulated and those that had contradictory expressions in breast cancer. An ontological analysis indicated that most of these breast cancer-associated proteins were classified into different enrichment functional groups that may be involved in different aspects of cancer biology. The pathway networks analysis showed that several signaling pathways and networks were significantly associated with human breast cancer. These pathways and functions were relevant to the cancer microenvironment, invasion and metastasis processes as briefly described below.
Previous studies have shown that the cancer microenvironment is deficient in oxygen, low in glucose (24), and is is usually followed by highl glycolytic activity (25,26). Studies of breast cancers showed that hypoxic conditions may alter the expression of certain proteins that serve as hypoxic markers (27). Fifteen glycolytic enzymes were investigated in the present study which may be involved in tumor proliferation in hypoxic conditions. The protein ubiquitination pathway and the PI3K/Akt signaling pathway have been reported to be associated with the development of human breast cancers (28,29). Certain drugs have been applied to affect these pathways in the treatment of breast cancer (30,31,32,33). The RhoA signaling pathway was strongly correlated with tumor cell invasion and metastasis (34), and the Nrf2-mediated oxidative stress response activated genes encoding detoxification enzymes and antioxidant proteins to protect cells from oxidative stress. In breast cancer cells, NRP/B was able to enhance oxidative stress responses via the Nrf2 pathway (35). More than 13 cell adhesion proteins in the present study may play roles in the intercellular and cell-extracellular matrix interactions of cancer, leading to cancer invasion or metastasis (36), or participating in signal transduction, cell growth and differentiation (37). E-cadherin is one prominent adhesion molecule that forms the E-cadherin-catenin complex which plays a role in epithelial cell-cell adhesion and differentiation (38), in particular serving as a potent invasion/tumor suppressor of breast cancer (39). Previous studies indicated that the downregulation of E-cadherin was relevant to several pathways including the integrin-linked kinase (ILK) signaling pathway (40,41). The ILK signaling pathway was one prominent enrichment pathway in breast cancer, and may play an important role in hormonal cancer progression (42).
Some of these canonical pathways are known to participate in cancer processes, but the mechanism and the altered individual members are not well studied. We hypothesized that the altered expressions of different members or activators in these enriched pathways may be associated with the tumor process and its microenvironment alteration, and may serve as important targets or agents in the diagnosis and treatment of breast cancer.
More than 200 cancer/testis antigens have been listed in the cancer/testis database (http://www.cta.lncc.br/). The significant feature of these proteins is their restricted expression in the testis and low or no expression in normal tissues (43); therefore, they may be used as potential cancer vaccine targets. Twenty-one cancer/testis antigens (8%) and 119 testis proteins (11 testis-specific proteins) were included in the breast cancer protein profiles and these proteins warrant further study. Among these proteins, a recent study showed that MAGE-A3 and A4 in the peripheral blood of breast cancer patients may have potential prognostic and predictive implications (44). All of these proteins are promising specific tumor markers of breast cancer.
Due to the limitations of protein identification using current proteomic techniques, some molecules in certain pathways may be missing. However, the present study gives a new bioinformatical insight into breast cancer at systems biology levels by integrating the individual studies to identify enriched biological and molecular pathways, providing vital evidence for future research. Further studies are warranted to substantiate the enriched functions and pathways. The studies may advance our understanding of cancer biomarker discovery, and also facilitate the biological interpretation of cancer biology in a network context.
References
- 1.Warner E. Clinical practice. Breast-cancer screening. N Engl J Med. 2011;365:1025–1032. doi: 10.1056/NEJMcp1101540. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal C, Somaiah N, Simon GR. Biomarkers with predictive and prognostic function in non-small cell lung cancer: ready for prime time? J Natl Compr Canc Netw. 2010;8:822–832. doi: 10.6004/jnccn.2010.0059. [DOI] [PubMed] [Google Scholar]
- 3.Tan Y, Ma SY, Wang FQ, Meng HP, Mei C, Liu A, Wu HR. Proteomic-based analysis for identification of potential serum biomarkers in gallbladder cancer. Oncol Rep. 2011;26:853–859. doi: 10.3892/or.2011.1353. [DOI] [PubMed] [Google Scholar]
- 4.Abu-Asab MS, Chaouchi M, Alesci S, Galli S, Laassri M, Cheema AK, Atouf F, VanMeter J, Amri H. Biomarkers in the age of omics: time for a systems biology approach. OMICS. 2011;15:105–112. doi: 10.1089/omi.2010.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn BK, Wagner PD, Anderson D, Greenwald P. Molecular markers for early detection. Semin Oncol. 2010;37:224–242. doi: 10.1053/j.seminoncol.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 7.Wistuba II, Gelovani JG, Jacoby JJ, Davis SE, Herbst RS. Methodological and practical challenges for personalized cancer therapies. Nat Rev Clin Oncol. 2011;8:135–141. doi: 10.1038/nrclinonc.2011.2. [DOI] [PubMed] [Google Scholar]
- 8.Schmitz-Spanke S, Rettenmeier AW. Protein expression profiling in chemical carcinogenesis: a proteomic-based approach. Proteomics. 2011;11:644–656. doi: 10.1002/pmic.201000403. [DOI] [PubMed] [Google Scholar]
- 9.Lau TY, O’Connor DP, Brennan DJ, Duffy MJ, Pennington SR, Gallagher WM. Breast cancer proteomics: clinical perspectives. Expert Opin Biol Ther. 2007;7:209–219. doi: 10.1517/14712598.7.2.209. [DOI] [PubMed] [Google Scholar]
- 10.Hondermarck H, Tastet C, El Yazidi-Belkoura I, Toillon RA, Le Bourhis X. Proteomics of breast cancer: the quest for markers and therapeutic targets. J Proteome Res. 2008;7:1403–1411. doi: 10.1021/pr700870c. [DOI] [PubMed] [Google Scholar]
- 11.Böhm D, Keller K, Wehrwein N, Lebrecht A, Schmidt M, Kölbl H, Grus FH. Serum proteome profiling of primary breast cancer indicates a specific biomarker profile. Oncol Rep. 2011;26:1051–1056. doi: 10.3892/or.2011.1420. [DOI] [PubMed] [Google Scholar]
- 12.Kadowaki M, Sangai T, Nagashima T, Sakakibara M, Yoshitomi H, Takano S, Sogawa K, Umemura H, Fushimi K, Nakatani Y, Nomura F, Miyazaki M. Identification of vitronectin as a novel serum marker for early breast cancer detection using a new proteomic approach. J Cancer Res Clin Oncol. 2011;137:1105–1115. doi: 10.1007/s00432-010-0974-9. [DOI] [PubMed] [Google Scholar]
- 13.Balluff B, Elsner M, Kowarsch A, Rauser S, Meding S, Schuhmacher C, Feith M, Herrmann K, Röcken C, Schmid RM, Höfler H, Walch A, Ebert MP. Classification of HER2/neu status in gastric cancer using a breast-cancer derived proteome classifier. J Proteome Res. 2010;9:6317–6322. doi: 10.1021/pr100573s. [DOI] [PubMed] [Google Scholar]
- 14.Lai TC, Chou HC, Chen YW, Lee TR, Chan HT, Shen HH, Lee WT, Lin ST, Lu YC, Wu CL, Chan HL. Secretomic and proteomic analysis of potential breast cancer markers by two-dimensional differential gel electrophoresis. J Proteome Res. 2010;9:1302–1322. doi: 10.1021/pr900825t. [DOI] [PubMed] [Google Scholar]
- 15.Issaq HJ, Waybright TJ, Veenstra TD. Cancer biomarker discovery: Opportunities and pitfalls in analytical methods. Electrophoresis. 2011;32:967–975. doi: 10.1002/elps.201000588. [DOI] [PubMed] [Google Scholar]
- 16.Weigelt B, Pusztai L, Ashworth A, Reis-Filho JS. Challenges translating breast cancer gene signatures into the clinic. Nat Rev Clin Oncol. 2011;9:58–64. doi: 10.1038/nrclinonc.2011.125. [DOI] [PubMed] [Google Scholar]
- 17.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 18.Schlotter CM, Vogt U, Allgayer H, Brandt B. Molecular targeted therapies for breast cancer treatment. Breast Cancer Res. 2008;10:211. doi: 10.1186/bcr2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukai H. Targeted therapy in breast cancer: current status and future directions. Jpn J Clin Oncol. 2010;40:711–716. doi: 10.1093/jjco/hyq037. [DOI] [PubMed] [Google Scholar]
- 20.Bernard-Marty C, Lebrun F, Awada A, Piccart MJ. Monoclonal antibody-based targeted therapy in breast cancer: current status and future directions. Drugs. 2006;66:1577–1591. doi: 10.2165/00003495-200666120-00004. [DOI] [PubMed] [Google Scholar]
- 21.Li SJ, Peng M, Li H, Liu BS, Wang C, Wu JR, Li YX, Zeng R. Sys-BodyFluid, a systematical database for human body fluid proteome research. Nucleic Acids Res. 2009;37:D907–D912. doi: 10.1093/nar/gkn849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Cunha JP, Galante PA, de Souza JE, de Souza RF, Carvalho PM, Ohara DT, Moura RP, Oba-Shinja SM, Marie SK, Silva WA, Jr, et al. Bioinformatics construction of the human cell surfaceome. Proc Natl Acad Sci. 2009;106:16752–16757. doi: 10.1073/pnas.0907939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Liu F, Wang H, Liu X, Liu J, Li N, Wan F, Wang W, Zhang C, Jin S, Liu J, Zhu P, Liu Y. Systematic mapping and functional analysis of a family of human epididymal secretory sperm-located proteins. Mol Cell Proteomics. 2010;9:2517–2528. doi: 10.1074/mcp.M110.001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dvorak HF, Weaver VM, Tlsty TD, Bergers G. Tumor microenvironment and progression. J Surg Oncol. 2011;103:468–474. doi: 10.1002/jso.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 26.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 27.Bando H, Toi M, Kitada K, Koike M. Genes commonly upregulated by hypoxia in human breast cancer cells MCF-7 and MDA-MB-231. Biomed Pharmacother. 2003;57:333–340. doi: 10.1016/s0753-3322(03)00098-2. [DOI] [PubMed] [Google Scholar]
- 28.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 29.Ghayad SE, Cohen PA. Inhibitors of the PI3K/Akt/mTOR pathway: new hope for breast cancer patients. Recent Pat Anticancer Drug Discov. 2010;5:29–57. doi: 10.2174/157489210789702208. [DOI] [PubMed] [Google Scholar]
- 30.Orlowski RZ, Dees EC. The role of the ubiquitination-proteasome pathway in breast cancer: applying drugs that affect the ubiquitin-proteasome pathway to the therapy of breast cancer. Breast Cancer Res. 2003;5:1–7. doi: 10.1186/bcr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi S, Loda M. The role of the ubiquitination-proteasome pathway in breast cancer: use of mouse models for analyzing ubiquitination processes. Breast Cancer Res. 2003;5:16–22. doi: 10.1186/bcr542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipkowitz S. The role of the ubiquitination-proteasome pathway in breast cancer: ubiquitin mediated degradation of growth factor receptors in the pathogenesis and treatment of cancer. Breast Cancer Res. 2003;5:8–15. doi: 10.1186/bcr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McAuliffe PF, Meric-Bernstam F, Mills GB, Gonzalez-Angulo AM. Deciphering the role of PI3K/Akt/mTOR pathway in breast cancer biology and pathogenesis. Clin Breast Cancer. 2010;10:S59–S65. doi: 10.3816/CBC.2010.s.013. [DOI] [PubMed] [Google Scholar]
- 34.Struckhoff AP, Rana MK, Worthylake RA. RhoA can lead the way in tumor cell invasion and metastasis. Front Biosci. 2011;16:1915–1926. doi: 10.2741/3830. [DOI] [PubMed] [Google Scholar]
- 35.Seng S, Avraham HK, Jiang S, Yang S, Sekine M, Kimelman N, Li H, Avraham S. The nuclear matrix protein, NRP/B, enhances Nrf2-mediated oxidative stress responses in breast cancer cells. Cancer Res. 2007;67:8596–8604. doi: 10.1158/0008-5472.CAN-06-3785. [DOI] [PubMed] [Google Scholar]
- 36.Martin TA, Mason MD, Jiang WG. Tight junctions in cancer metastasis. Front Biosci. 2001;16:898–936. doi: 10.2741/3726. [DOI] [PubMed] [Google Scholar]
- 37.Kim SH, Turnbull J, Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J Endocrinol. 2011;209:139–151. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- 38.Wijnhoven BP, Dinjens WN, Pignatelli M. E-cadherincatenin cell-cell adhesion complex and human cancer. Br J Surg. 2000;87:992–1005. doi: 10.1046/j.1365-2168.2000.01513.x. [DOI] [PubMed] [Google Scholar]
- 39.Berx G, Van Roy F. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 2001;3:289–293. doi: 10.1186/bcr309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan C, Costello P, Sanghera J, Dominguez D, Baulida J, de Herreros AG, Dedhar S. Inhibition of integrin linked kinase (ILK) suppresses beta-catenin-Lef/Tcf-dependent transcription and expression of the E-cadherin repressor, snail, in APC−/− human colon carcinoma cells. Oncogene. 2001;20:133–140. doi: 10.1038/sj.onc.1204052. [DOI] [PubMed] [Google Scholar]
- 41.Bravou V, Klironomos G, Papadaki E, Taraviras S, Varakis J. ILK over-expression in human colon cancer progression correlates with activation of beta-catenin, down-regulation of E-cadherin and activation of the Akt-FKHR pathway. J Pathol. 2006;208:91–99. doi: 10.1002/path.1860. [DOI] [PubMed] [Google Scholar]
- 42.Cortez V, Nair BC, Chakravarty D, Vadlamudi RK. Integrin-linked kinase 1: role in hormonal cancer progression. Front Biosci (Schol Ed) 2011;3:788–796. doi: 10.2741/s187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grigoriadis A, Caballero OL, Hoek KS, da Silva L, Chen YT, Shin SJ, Jungbluth AA, Miller LD, Clouston D, Cebon J, Old LJ, Lakhani SR, Simpson AJ, Neville AM. CT-X antigen expression in human breast cancer. Proc Natl Acad Sci. 2009;106:13493–13498. doi: 10.1073/pnas.0906840106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussein YM, Gharib AF, Etewa RL, El-Shal AS, Abdel-Ghany ME, Elsawy WH. The melanoma-associated antigen-A3, -A4 genes: relation to the risk and clinicopathological parameters in breast cancer patients. Mol Cell Biochem. 2011;351:261–268. doi: 10.1007/s11010-011-0734-4. [DOI] [PubMed] [Google Scholar]