Summary
In contrast to RNA viruses, double-stranded DNA viruses have low mutation rates, yet must still adapt rapidly in response to changing host defenses. To determine mechanisms of adaptation we subjected the model poxvirus vaccinia to serial propagation in human cells, where its anti-host factor K3L is maladapted against the anti-viral Protein Kinase R (PKR). Viruses rapidly acquired higher fitness via recurrent K3L gene amplifications, incurring up to 7-10% increases in genome size. These transient gene expansions were necessary and sufficient to counteract human PKR and facilitated the gain of an adaptive amino acid substitution in K3L that also defeats PKR. Subsequent reductions in gene amplifications offset the costs associated with larger genome size while retaining adaptive substitutions. Our discovery of viral ‘gene-accordions’ explains how poxviruses can rapidly adapt to defeat different host defenses despite low mutation rates and reveals how classical Red Queen conflicts can progress through unrecognized intermediates.
Introduction
Rapidly diversifying host immune repertoires pose significant barriers to successful transmission of viruses within and between species. Many RNA viruses counter host immunity with high mutation rates coupled with short generation times and large effective population sizes, allowing rapid exploration of mutational space for adaptation. Previous studies have elucidated how the high mutation rates of RNA viruses are required for tropism in animal species (Pfeiffer and Kirkegaard, 2005; Vignuzzi et al., 2006), despite driving them perilously close to an ‘error catastrophe’ threshold (Crotty et al., 2001; Vignuzzi et al., 2005). Much less is known about the adaptive strategies of large double-stranded DNA viruses that despite short generation times, seem to evolve more slowly than RNA viruses (Drake et al., 1998; Drake and Holland, 1999; Li et al., 2007; Lynch, 2010) yet comprise highly successful lineages.
The poxviruses are large double-stranded DNA viruses that are notable because they can infect most vertebrates (chordopoxvirinae) and insects (entomopoxvirinae) (Harrison et al., 2004; Moss, 2007). Poxviruses encode a large array of genes that hijack or antagonize components of the host cell to promote viral fitness (Bahar et al., 2011; Werden et al., 2008). Strong natural selection at these host-virus interfaces can drive rapid adaptations, which are reflected by the fixation of non-synonymous substitutions in coding regions. These substitutions can either enhance or weaken interactions between host and viral factors (Emerman and Malik, 2010). The Red Queen hypothesis postulates that such interactions evolve as an ongoing series of counter-adaptations (Dawkins and Krebs, 1979; Meyerson and Sawyer, 2011; Van Valen, 1973). Several retrospective evolutionary studies comparing the sequences of open reading frames among poxvirus genomes provide ample evidence of adaptive changes to overcome rapidly evolving host defenses (Bratke and McLysaght, 2008; McLysaght et al., 2003).
One of the most potent innate defenses against viruses is encoded by Protein Kinase R (PKR), which is activated upon sensing double-stranded RNA that accumulates in host cytoplasm during many viral infections (Weber et al., 2006). Active PKR phosphorylates the translation initiation factor eIF2α to inhibit protein production, which strongly impairs viral replication (Kaufman, 2000). Antagonism by a variety of viruses over the course of primate evolution has driven the rapid evolution of PKR in primates (Elde et al., 2009; Rothenburg et al., 2009). As a result of this diversification, primate variants of PKR are differentially susceptible to inhibitors encoded by different viruses. Vaccinia virus encodes two such antagonists, K3L and E3L, which each specifically inhibit PKR by distinct mechanisms in different species (Davies et al., 1993; Langland and Jacobs, 2002). The rapid evolution of PKR has led to species-specific resistance to K3L among primates.
A partial explanation for flexibility in poxvirus host range is provided by several ‘host-range’ factors like K3L and E3L, which can overcome species-specific blocks to infection. Left unanswered, however, is the fundamental question of how poxviruses, despite their low mutation rate, efficiently explore mutational space and overcome rapidly evolving immune factors such as PKR as they move between host species. For example, some poxviruses such as cowpox, vaccinia, and monkeypox infect both animals and humans in zoonotic infections, with the latter cases being of considerable biomedical importance. The fact that these viruses can infect divergent host species despite diverse mechanisms of immunity suggests that they employ different means of evolutionary adaptation from those known for rapidly evolving RNA viruses to overcome low mutation rates. Moreover, even within the same host populations, poxviruses must adapt rapidly to persist despite low mutation rates. Gaining insight into the currently unknown basis of adaptation in this biomedically and evolutionarily important class of viruses is essential for understanding how these viruses infect a wide range of hosts.
Fundamental insights into evolutionary mechanisms have been derived from laboratory and experimental evolution of various microbial populations (Beaumont et al., 2009; Meyer et al., 2012; Montville et al., 2005; Vignuzzi et al., 2006; Wichman et al., 1999). To date, however, nearly all protocols involving experimental evolution of viruses have used strains with high mutation rates (e.g. small RNA viruses) and/or ones that quickly generate large effective population sizes under laboratory conditions (e.g. bacteriophages). Here we devised an experimental evolution protocol to determine how poxviruses adapt using serial vaccinia infections at relatively small population sizes. By passaging a vaccinia strain deleted for the host range gene E3L, we mimicked a host-switching event, which placed strong selective pressure on K3L to inhibit the antiviral PKR. Our studies reveal mechanisms of poxvirus adaptation to defeat host defenses via rapid episodes of transient gene amplification. This simple and recurrent mechanism of adaptation on a genomic scale also reveals a new general model for mitigating fitness costs associated with large changes in genome structure. Our experiments not only reveal an important means by which poxviruses adapt, but also highlight a mechanism by which radical adaptations might be explored in many organisms that have large genomes and low mutation rates.
Results
Experimental evolution of vaccinia virus reveals adaptation by gene expansion
We used the fact that K3L from vaccinia virus is a poor inhibitor of human PKR to determine how poxviruses can adapt to overcome an initially insurmountable host defense. By starting with the Copenhagen strain of vaccinia virus deleted for E3L (ΔE3L)(Beattie et al., 1995), we placed strong selective pressure on K3L (Figure 1A). Three replicate virus populations were repeatedly propagated in HeLa cells at a low multiplicity of infection of 0.1 plaque-forming units/cell. After 48 hours of infection, we collected virus, measured titers, and repeated the protocol for 10 passages (Figure 1B and Experimental Procedures).
Figure 1. Experimental evolution of vaccinia virus.
(A) Vaccinia encodes two host-range factors, E3L and K3L that inhibit antiviral Protein Kinase R (PKR). (B) Vaccinia was repeatedly passaged through HeLa cells in triplicate (see methods). After each infection, virus titer was measured, an aliquot was reserved for the “fossil record”, and fresh cells were infected. Red dots represent viruses in which a theoretical adaptation arises in the population and increases in frequency. (C) With passaging of ΔE3L virus, replicate populations A-C gained the ability to replicate in HeLa cells as judged by viral titers assayed over the course of the experiment. Dotted horizontal lines show the average titer of parental virus (ΔE3L) and the wildtype Copenhagen strain of vaccinia (WT). Because E3L was replaced by lacZ we could also use β-gal activity assays as a proxy for measuring virus replication (inset). Gains in replication were corroborated by measurements of β-gal activity, shown in arbitrary units, from parental virus and replicates A-C at passage 10 (p10; inset). Data are represented as mean +/− SEM.
ΔE3L virus initially replicated much less efficiently than wildtype virus in HeLa cells, consistent with previous reports (Langland and Jacobs, 2002). By six rounds of passaging in HeLa cells, each of three replicate virus populations consistently produced 10-fold more infectious viruses than the parental virus strain as measured by plaque assays (Figure 1C) and β-gal activity assays (Figure 1C, inset). Further gains in virus production were not evident during the remaining four passages. Thus, despite a low mutation rate, and relatively small effective population sizes starting at 5×105 viruses/passage, all three replicates of vaccinia rapidly gained increased fitness over the course of passaging.
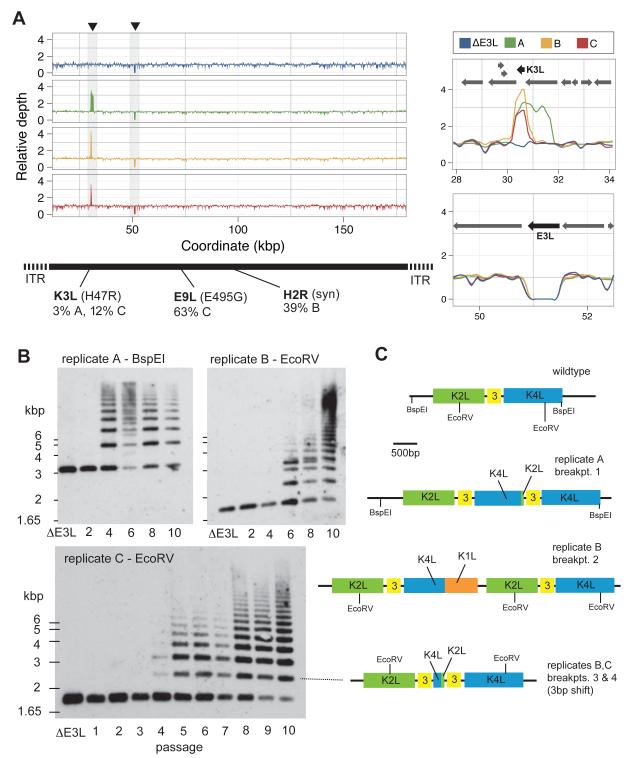
To determine the genetic basis of rapid adaptation, we performed deep sequencing of all virus populations to approximately 1,000-fold genome coverage. We found only two non-synonymous and one synonymous polymorphism in the genomes of passage 10 virus replicates at a frequency higher than 1% (Figure 2A). This paucity of mutations highlights the low rate of nucleotide substitution in vaccinia genomes. In addition, none of the substitutions were found at high frequency or in all replicates. In contrast, by scanning sequencing coverage among mapped reads we found a substantial spike in relative depth that mapped exclusively over the K3L gene which is consistent with a striking increase in copy number of the K3L locus in all three replicate strains (Figure 2A). The increased read-depth corresponds to an average of three (replicates A, C) to four (replicate B) copies of K3L per viral genome (Figure 2A).
Figure 2. Rapid evolution of copy number amplification.
(A) Genomic DNA traces of parent and virus replicates A-C at passage 10, showing relative depth of sequence coverage across the entire ~180kb vaccinia genome, except for the inverted terminal repeat regions (ITR; ITRs are shown in Figure S4). Increased K3L sequence coverage was observed in replicates A-C (top right panel; also see Figure S1, S2) while coverage of E3L was missing in all populations (lower right panel). Polymorphisms were detected in the K3L, E9L, and H2R genes as indicated (also see Table S2). We also detected 15 nucleotide differences between the parent and reference Copenhagen strain, which were unrelated to our protocol (Goebel et al., 1990)(Table S6). (B) Southern blots of the K3L locus of viral replicates A, B, and C during the course of experimental evolution from passages 1-10 reveals rapid gene copy number amplification of K3L. Amplification may be a consequence of recombination-driven tandem duplications (Figure S3). (C) Diagrammed is the parental K3L locus and the major duplication breakpoints near K3L that led to copy number amplification (also see Table S3). We also found duplications at the highly repetitive termini of the vaccinia genomes, consistent with previous observations (Moss et al., 1981)(see Figure S4). However, these terminal expansions were also present in the parental strain and therefore not a consequence of our experiments.
To further determine the character of these K3L expansions, we performed Southern blot analysis of viral genomes sampled from points throughout the passaging protocol, using the K3L gene as a probe. These analyses revealed a highly heterogeneous pool of viral genomes in each replicate, with some harboring 15 or more copies of K3L by passage 10 (Figure 2B). Intriguingly, the appearance of multiple copies of K3L paralleled the pronounced increases in virus replication, which occurred by passage 4 for both replicates A and C, and passage 6 for replicate B (compare Figures 1C and 2B). Therefore, increased copy number of K3L directly correlated with increased viral fitness. Additional analysis of discordantly mapping paired-end reads revealed a complex pattern of duplications at the K3L locus exclusive to the adapted strains (Figures S1, 2C, and Table S1). Notably, analysis of the duplication breakpoints confirmed the independent origins of the K3L duplications in each of the three replicate populations of adapted vaccinia viruses.
Re-sequencing of parental genomes to nearly 1,000-fold coverage failed to reveal any pre-existing duplications of K3L in this homogenous virus population (Table S1). PCR confirmed the independent emergence of tandem duplications of K3L at several distinct breakpoints in the genes adjacent to K3L in the evolved populations. In addition to our initial experiment, three additional ΔE3L virus replicates passaged under the same conditions all yielded expansions of K3L as judged by PCR to detect K3L associated breakpoints (Figure S2), demonstrating the reproducibility of gene amplification as a means of adaptation.
To determine if genomic amplification of K3L accounted for enhanced viral replication, we first compared K3L protein expression between parental and passaged viruses (Figure 3A). Each passage 10 replicate expressed considerably more K3L protein than even wildtype virus, which still replicates more efficiently than any of the adapted strains due to the presence of E3L (Figure 3A). To determine whether overexpression of K3L alone is sufficient to explain the robust increase in viral fitness, we specifically knocked down K3L expression using siRNA transfections. Successful knockdown of K3L reduced virus replication to levels indistinguishable from pre-passaged, parental ΔE3L virus (Figure 3B). Therefore, K3L copy number amplification was necessary and sufficient for increased expression of K3L, which caused the substantial gain in virus fitness we observed. Because K3L mimics PKR’s substrate eIF2α and inhibits PKR by direct binding (Dar and Sicheri, 2002), it appears that an increased abundance of even weakly active K3L antagonizes PKR by mass action, providing the basis for an immediate evolutionary advantage due to genomic expansion.
Figure 3. Overexpression of K3L protein in adapted strains is necessary and sufficient for increased viral fitness.
(A) Western blot analysis of vaccinia and mock infected HeLa cells with wildtype (WT), parental (ΔE3L), and virus replicates A-C at passage 10 for levels of K3L, vaccinia proteins, and actin. (B) siRNA knock-down of K3L in infected HeLa cells. Viral replication was measured by β-gal activity assay of virus and mock infected cells transfected with K3L-specific siRNAs (1-3) or a non-targeting control. Western blot shows levels of K3L, vaccinia proteins, and actin. (C) β-gal activity from HeLa cells infected with various vaccinia viruses. Wildtype vaccinia virus, ΔE3L parental virus, and passage 10 virus from replicate C were compared to plaque-purified clones with either a single copy H47R-K3L (1), a single copy WT-K3L (3), or a multi-copy K3L virus with both WT and the H47R substitutions (2). Numbers correspond to plaque-purified viruses described in Figure 6. Data are represented as mean +/− SEM.
Acquisition of an adaptive substitution following gene expansion
While K3L copy number amplification provided a rapid, recurrent basis for adaptation in all three replicate populations, most studies of virus evolution identify adaptive point mutations as the basis of fitness gains. We found that two of the replicates from our study possessed a single amino acid substitution, H47R, in K3L at frequencies of 3 and 12 percent (Table S2). Remarkably, this exact substitution was previously identified in a genetic screen for K3L mutants with enhanced activity against human PKR using a yeast assay interrogating the K3L-PKR interaction (Kawagishi-Kobayashi et al., 1997). We isolated this variant from the adapted passage 10 replicate C vaccinia population containing a single copy of K3L with the H47R mutation and tested its fitness relative to viruses with gene expansions of wildtype K3L sequence. Both the single copy H47R-bearing virus and the multi-copy K3L virus had marked improvements in fitness over the parental single-copy wildtype K3L virus (Figure 3C). These findings also strongly support the idea that fitness gains realized by K3L amplification and acquisition of H47R in serially propagated virus populations reflect specific antagonism of PKR. In addition, although only 12% of the K3L genes at passage 10 of replicate C contain the H47R mutation (541 reads out of 4146 total sequencing reads), 47% of single-copy K3L genes from the same virus population harbor this substitution (24 cases with H47R out of 51 K3Ls sampled via PCR and sequencing; p<0.0001, Fisher’s exact test). This result implies that low copy number viruses are strongly selected to possess the adaptive H47R allele of K3L compared to K3L expanded viruses in the same pool. Thus, our experimental evolution strategy uncovered two responses to the same selective challenge: adaptive copy number expansions seen in all replicate populations and an adaptive non-synonymous mutation seen at low frequency in two of three replicates.
The emergence of the H47R substitution raised the possibility that K3L expansions facilitated the appearance of the variant by providing additional targets for mutation to generate increased variation, including beneficial substitutions like H47R. Examination of parental ΔE3L strain sequences mapping to codon 47 of the K3L coding region failed to reveal H47R in the starting population (Table S2). Also consistent with K3L expansions preceding the acquisition of H47R was our observation that replicate B, the slowest population to undergo K3L amplification (Figure 1C), lacked any detectable H47R K3L alleles by passage 10. Together these data suggest that H47R appeared after K3L gene expansion.
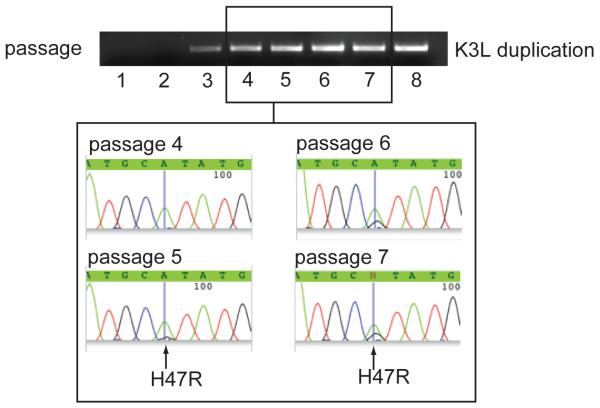
To determine when H47R emerged during the course of passaging, we amplified K3L from genomic samples of replicate C at each passage point and sequenced amplicon populations. K3L expansions, as judged by PCR amplification across K3L duplications, preceded the appearance of the H47R substitution during viral passaging (Figure 4). Sequencing of PCR products spanning K3L duplications at late passage points revealed the presence of H47R in both single and multi-copy K3L arrays. We further sampled the composition of multi-copy arrays by sequencing 34 clones bearing two K3L gene copies. Of these, ten had both wildtype and H47R-containing copies of K3L. Together these data strongly suggest that K3L expansions facilitated the emergence and increased frequency of the H47R substitution. Thus, not only are the K3L gene expansions immediately advantageous due to their ability to provide enhanced antagonism of human PKR, but they also facilitate the acquisition of single Darwinian ‘gain-of-function’ changes by increasing the number of mutational targets despite an otherwise modest mutation rate.
Figure 4. K3L expansions precede the appearance of the H47R substitution.
PCR with genomic samples from the “fossil record” of replicate C with primers amplifying duplications of K3L is consistent with Southern blot analysis. Sequencing of duplicated K3L regions reveals that the appearance of the mutation leading to the H47R substitution is only appreciable after duplications of K3L appeared.
A general mechanism of gene expansion in vaccinia genomes
Two previously proposed mechanisms could account for facile gene expansion in vaccinia genomes. One study proposed unequal crossover as a mechanism to account for variation in the distal inverted terminal repeat regions of the vaccinia genome (Moss et al., 1981). A second study pointed to the possibility that gene expansions could occur episomally, because vaccinia can promote rapid amplification of non-viral plasmids. To determine whether the K3L locus was being expanded by a mechanism involving episomes containing K3L, we used RecBCD enzyme, a complex helicase and nuclease, which rapidly digests all linear genomic DNA, but spares any potential circular episomes (Eichler and Lehman, 1977; Smith et al., 1995). Southern blot analysis of undigested genomes as well as those treated with RecBCD to eliminate linear DNA forms, that might have otherwise obscured circular forms, failed to reveal the presence of circular DNA intermediates containing K3L in genomes of viruses passaged in HeLa cells (Figure S3). Thus, we can exclude an episomal mode of amplification, which suggests that recombination-driven K3L gene expansions within linear genomes of each replicate virus account for the rapid adaptation of vaccinia virus.
Recombination-mediated gene expansions, which can facilitate the acquisition of adaptive substitutions could represent a general mechanism for rapid adaptation of poxviruses. To determine if gene duplications similar to K3L were present at low frequency in virus populations, we assayed for the presence of additional duplications in virus populations. Twelve sets of PCR primers were designed to amplify products across potential duplication breakpoints at locations spaced roughly evenly throughout the genome (Table S3). These probes for duplications were used to query three vaccinia strains (Western Reserve, Copenhagen-VC2, and the parental strain used in this study, Copenhagen-VC2-ΔE3L). Despite our limited sampling, we found that each strain harbored at least one duplication breakpoint in virus populations (Figure 5). We did not detect any shared sequence motifs or consistent patterns of homology at sites where duplications arose that might promote genomic expansions. However, each of the duplications we uncovered maps toward the distal regions of the vaccinia genome, which have a higher frequency of recombination and are comprised of many genes that are dedicated to the modulation of host factors (Moss, 2007). In contrast, genes towards the center of the vaccinia genome are essential for virus-specific functions in the cytoplasm of the host cell. Four of the twelve primer sets tested amplified duplications in the Western Reserve strain, which included several breakpoints from duplications of K3L (Table S4). Further examination of deep sequencing reads also revealed rare duplication breakpoints that were confirmed for two cases by PCR in populations of adapted strains at genomic locations outside the K3L locus (Table S5). These data reveal that virus populations contain a variety of low-frequency breakpoints that could rapidly seed genomic expansions under suitable selective conditions, such as changing hosts. Much like random nucleotide substitutions that can start adaptive sweeps driven by selection, we propose that these low-frequency duplication events across the vaccinia genome can drive rapid gene expansions to overcome host defenses, followed by expanded sampling of point mutations. Given the low rate of nucleotide substitutions uncovered by our sequencing analyses, the frequency of these duplications may be on par with or exceed the frequency of nucleotide substitutions, as has been previously observed in some eukaryotic genomes (Lipinski et al., 2011; Lynch et al., 2008).
Figure 5. Low frequency gene duplications in vaccinia genomes.
Of the primers tested (Table S3), duplications at several loci near the termini of the genome were amplified and sequenced (Table S4). Breakpoint regions are drawn to scale highlighting the location of each duplication point. One breakpoint observed in K3L leads to a fusion product in place of the wildtype copy. The C20L and B26R regions are mirrored at each end of the chromosome, such that the reported breakpoints could be at either end of the genome. Additional low frequency duplications were detected in deep sequencing reads of parental and adapted virus genomes (Table S5).
Genomic accordions reveal tradeoffs associated with genomic expansion
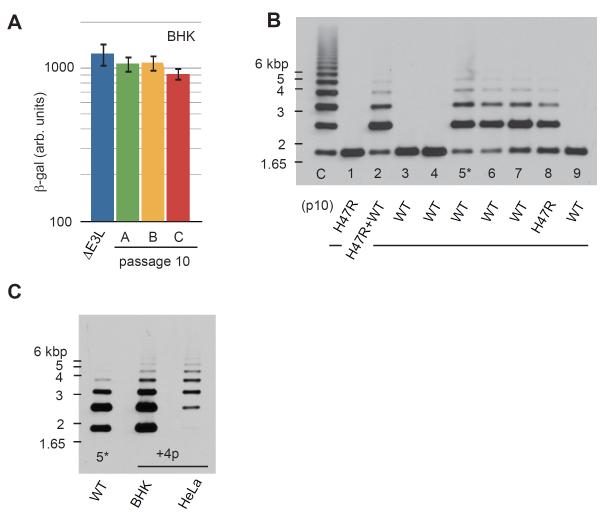
K3L copy number expansions represent a significant increase in vaccinia genome size. Although comprising a minor fraction of the population, viruses harboring 15-fold amplifications of K3L have genomes 7-10% larger than the parental strain (Figure 2B). Vaccinia can package similarly sized genomic additions with a seemingly negligible impact on replication efficiency (Smith et al., 1983), but poxvirus genomes also undergo adaptive size reductions under a variety of conditions (Hendrickson et al., 2010; Meisinger-Henschel et al., 2007). Therefore, to determine if there was an evolutionary trade-off associated with large genomic expansions of K3L, we removed selective pressure from amplifications of K3L. Unlike in human cells, single copy K3L from vaccinia virus defeats PKR in BHK (hamster) cells (Figure 6A)(Beattie et al., 1995; Langland and Jacobs, 2002), therefore passaging virus in these permissive cells directly tests whether the expansions incurred a fitness tradeoff.
Figure 6. Evolution of transient copy number amplification.
(A) β-gal activity from BHK cells infected with parent virus (ΔE3L) and replicates A-C at passage 10. Data are represented as mean +/− SEM. (B) Southern blot of the K3L locus from plaque-purified vaccinia viruses obtained from serial plaque purification of passage 10 replicate C virus in BHK cells. The presence or absence of the H47R substitution in K3L is indicated for each strain. (C), Southern blot of multi-copy plaque-purified virus (clone 5* from panel B) after four additional passages (+4p) in BHK or HeLa cells. Viruses passaged in HeLa cells showed re-expanded K3L copy number relative to those passaged in BHK cells.
We plaque purified nine vaccinia clones from the population of passage 10 viruses from replicate C by passaging four times in BHK cells. Southern analyses of these viral genomes revealed that these viruses contained either only a single copy of K3L or nearly identical multi-copy arrays with considerably fewer copies of K3L compared to passage 10 viruses from HeLa cells (Figure 6B). To rule out the possibility that secondary genetic changes had caused contraction of the K3L expansions, we reintroduced selective pressure on one of the plaque-purified multi-copy K3L viruses (5*) with four additional passages in HeLa cells. Re-application of selective pressure led to a clear increase in K3L copy number compared with passaging the same viruses in BHK cells (Figure 6C). Thus, removing the selective pressure that led to gene amplification resulted in a reproducible and reversible reduction in K3L locus expansion, consistent with there being a fitness cost associated with the genomic expansions. Alternatively, it is possible that simply relaxing selective pressure for K3L expansion leads to a relative loss of K3L due to a bias in which recombination-driven losses exceed gains. It is notable, however, that replicate B virus populations lacking the H47R substitution continue to show ongoing increases in K3L copy number at passage 10 as judged by Southern analysis (Figure 2B). This suggests that copy number of K3L may continue to expand until a beneficial mutation provides an adaptive alternative to genomic expansion.
Discussion
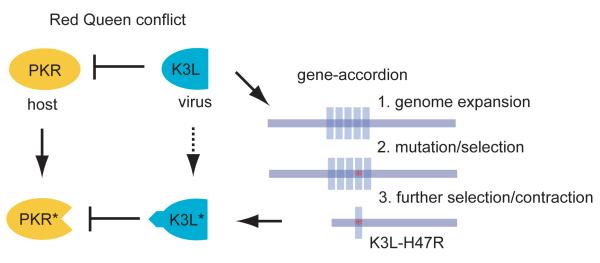
Our experimental evolution studies of vaccinia evoke a “gene accordion” model for genome evolution. Localized genomic regions transiently expand to enhance levels of gene expression, while simultaneously providing additional gene copies for sampling mutational space (Figure 7). Subsequent contractions mitigate the costs of replicating larger genomes while retaining adaptive alleles like H47R that arose during the expansion. Thus, a simple mechanism of recombination-driven genomic expansions and contractions facilitates the rapid evolution of virus populations with otherwise low mutation rates.
Figure 7. Genomic accordion model of poxvirus evolution.
The model depicts Red Queen conflicts proceeding through ‘gene-accordion’ intermediates. Gene copy number amplification provides an immediate fitness advantage and additional gene copies for sampling potentially beneficial mutations. After acquisition of a beneficial mutation, virus genomes are selected that retain advantageous point mutations without the gene expansion, leading to contractions down to a single gene copy.
Gene accordions do not appear unique to the K3L locus. In addition to the low frequency duplications we detected in vaccinia genomes (Figure 5 and Table S3), this model of adaptation through intermediates of transient gene expansion also helps explain a variety of previous observations in the genomes of large double-stranded DNA viruses, including ancient gene family expansions among several poxviruses (McLysaght et al., 2003) and herpesviruses (Searles et al., 1999), chemically induced gene amplification in vaccinia (Slabaugh and Mathews, 1986; Slabaugh et al., 1989), and a potentially adaptive duplication observed in the myxoma poxvirus (Kerr et al., 2010). A particularly clear example of such adaptive gene expansions is evident in the genomes of avipoxviruses, which devote as much as 30% of the genome to a small set of large gene families that includes many ankyrin repeat containing genes (Afonso et al., 2000). Avipoxviruses illustrate an exceptional example of gene duplications facilitating neofunctionalization of individual copies of the amplified gene family, leading to the retention of multiple variants rather than contraction back to a single copy. We propose that sampling a reservoir of low frequency duplications at various genomic regions in the virus population reflects an underappreciated but common mechanism seeding dynamic and adaptive gene expansions in these viruses. Our observations reveal the underlying mechanisms by which such viruses adapt to new, hostile host environments first by rapid gene expansion rather than gene mutation.
These results also reveal that a strength of the experimental evolution strategy is its power to reveal important evolutionary intermediates that could be too fleeting to capture by less frequent sampling or missed all together by studying viruses long after host switches. For example, we could clearly demonstrate that vaccinia genomes underwent a specific K3L gene expansion that exactly correlated with all fitness gains observed during our experiment. However, by 10 passages the K3L gene had already been replaced by the H47R variant in 12% of all K3L genes in one of the replicates. Sampling at a later point could conceivably obscure the accordion-like origins of an H47R-like adaptation as the variant sweeps through the population and replaces large and costlier gene expansions. This duality of adaptation might also be reflected in the different stages of viral transmission between species, including zoonosis. Gene expansions might be favored early in infection when most viral adaptation is centered on overcoming host defenses, as was modeled by our experiment. At later stages of infection, gene expansions are likely to be strongly selected against as viruses compete not only against the initial challenge imposed by host defenses, but also against larger populations of fit viruses carrying the equivalent of an H47R mutation. Future studies will be important to assess not only the generality of this adaptive mechanism, but also to compare the dynamics observed in our experiments to those that occur during natural poxviral infections in future studies.
Despite the rapid gene expansions observed in our study, very few poxviruses sequenced from the wild have been found to harbor recent gene duplications or expansions, with the ankyrin repeat genes in avipoxviruses being a clear exception. While some expansions may be obscured by subsequent contractions, it is also possible that many expansion events have been missed because of an ascertainment bias. Until recently, sequencing methodologies rarely accounted for copy number variation as a source of genetic heterogeneity. Indeed, sequencing of different strains of variola (smallpox) virus with the aim of determining how variola major had a higher fatality rate than variola minor focused primarily on nucleotide substitutions, which revealed less than 2% divergence (Esposito et al., 2006). While this genetic divergence might be sufficient to explain differential pathogenicity, important instances of copy number variation might have gone undetected in these comparisons. New sequencing technologies greatly increase the prospects of capturing such intermediate states from poxviruses that have undergone minimal passaging in the lab.
Our study also highlights the important role that gene copy number expansion plays in adaptation of genomes with low mutation rates. This mode of adaptation may be especially prevalent when an immediate advantage can be gained via mass action. Examples include selection for resistance to drug therapies against cancer (Drummond et al., 1997; Schimke, 1986), for enhanced enzymatic degradation of toxins (Sandegren and Andersson, 2009), or for overcoming a host antiviral protein like PKR. In these cases, the duplication event is itself non-neutral and can be further acted upon by recombination to drastically alter genome architecture. While rapid evolution of copy number variation is known for some non-virus microbial populations, including Plasmodium, yeast, and a variety of bacteria (Bergthorsson et al., 2007; Demuth and Hahn, 2009; Dunham et al., 2002; Kugelberg et al., 2010; Nair et al., 2008; Pränting and Andersson, 2011; Stambuk et al., 2009; Sun et al., 2009), adaptation via gene expansion in poxviruses represents an unappreciated mechanism of evolution in this large, medically relevant class of DNA viruses (Harrison et al., 2004). In the cases cited above, copy number expansions may also be poised for adaptive contractions, but that unfold on longer time scales, thus obscuring a ‘genomic-accordion’ mechanism revealed here by vaccinia virus (but also see Pränting and Andersson, 2011).
Some of the passage 10 viruses we assayed have a 7-10% expansion in genome size purely by virtue of a single gene expansion. In a 200 kb genome, this is a noteworthy adaptation for its major alteration of genomic structure. Our findings also indicate that a diversity of adaptive strategies - copy number variation in viruses with low mutation rates and large genomes versus nucleotide substitution - falls well within the viral kingdom. It seems likely that this mode of adaptation is applicable to many other viruses that have similarly low mutation rates and relatively limited restrictions on genome size. Exceptions are likely to be viruses that have low mutation rates but strict limits on genome size due to a highly constrained capsid structure (e.g., adenoviruses and certain bacteriophages). It will be informative to determine whether other large DNA viruses like herpesviruses, which replicate in the nucleus and also show evidence of gene expansions (Searles et al., 1999), follow similar gene-accordion dynamics as the poxviruses, which replicate in the cytoplasm of host cells.
There are striking parallels between the challenges imposed on viruses by exposure to new batteries of innate defense genes and the challenges imposed on organisms adapting to strong selective pressure, like bacteria grown under stress-inducing conditions (Andersson et al., 1998; Cairns et al., 1988; Foster, 1994; Hastings et al., 2000; Hendrickson et al., 2002). In each of these cases, a significant step in the adaptive process involves rapid and short-lived gene expansions that permit the selection of ‘escape’ variants. Our study thus reveals a previously unappreciated insight into the dynamics of Red Queen conflicts between pathogens and hosts (Elde and Malik, 2009; Emerman and Malik, 2010; Meyerson and Sawyer, 2011). Traditionally viewed primarily as substrates for rapid selection of non-synonymous codon substitutions, these interactions may also depend on adaptive gene amplifications that are overlooked due to their transience or difficulty of detection (Figure 7). Indeed, ‘gene-accordions’ may be a critical feature of many genetic conflicts involving strong selection.
Experimental Procedures
Viruses and reagents
The VC2 (wildtype) Copenhagen strain of vaccinia virus and the E3L deletion mutant (ΔE3L), with the lacZ gene replacing E3L, were a gift from B. Jacobs (Beattie et al., 1995). HeLa and BHK cells were cultivated in DMEM+10% Nu serum (BD Biosciences).
Experimental evolution of vaccinia virus
For each passage of vaccinia, 100mm dishes were seeded with a fresh aliquot of HeLa cells in triplicate (5×106 cells/dish). Cells were infected with ΔE3L virus at a multiplicity of infection of 0.1 pfu/cell. After 48 hours, infected cells were pelleted, washed, and virus was harvested by 3 freeze/thaw cycles in a mixture of dry ice/ethanol and resuspended in 1mL of DMEM+10% Nu serum. Viral titers were calculated by performing plaque assays in BHK cells between each passage. After 10 passages, aliquots of virus reserved from each passage were expanded with a single 48-hour infection of BHK cells and viral titers were determined. To evaluate the replication properties of these viruses, triplicate wells of HeLa cells were infected at 0.1 pfu/cell using viruses from every other passage and viral yield was measured by titering cell lysates made at 48 h post infection. β-gal activity was determined with 4-methylumbelliferyl-β-D-galactopyranoside (MUG) fluorescence assays (Hakki and Geballe, 2005). Genomic DNA was prepared as previously described (Esposito et al., 1981) from viruses obtained at every passage of the protocol.
Deep genome sequencing of vaccinia strains
Illumina sequencing libraries were prepared from genomic DNA from parental ΔE3L virus and each replicate (A-C) at passage 10 using the Nextera sequencing library construction kit (Epicentre). Multiplexing barcodes were added by PCR as previously described (Adey et al., 2010). Barcoded libraries were combined and sequenced across three lanes on Illumina GA-IIx and HiSeq 2000 instruments. Reads were mapped to the vaccinia reference genome with bwa version 0.6.1 (Li and Durbin, 2009); custom python scripts were used to calculate read depth and find breakpoint junctions.
Plaque purification of vaccinia
BHK cells were infected with dilutions of passage 10, replicate C virus. At ~2 days post infection, cells were collected from isolated individual plaques, diluted into medium, and lysed by three cycles of freezing and thawing. Dilutions of these first round plaque-purified viruses were used to infect fresh BHK cells and the process was repeated three more times. After the fourth round of plaque-picking, a stock of each plaque-purified virus was prepared in BHK cells and was used for infections to assess replication and to make viral DNA.
RecBCD digests
Vaccinia replicate C genomes harvested after 10 passages were first digested with restriction enzymes recognizing sites near K3L breakpoints (EcoRV), within K3L (BstZ17I), at sites distant from K3L (SacI), or left intact. Samples were incubated with 5 units of RecBCD +/− 125 μM ATP based on a previously reported method (Eichler and Lehman, 1977), but with a modified reaction buffer containing 1mM DTT, 20mM potassium phosphate, and 10% glycerol. Samples were incubated twice for 2 hours at 37°C with 5 additional units of RecBCD added between incubations. Genomes were resolved by agarose gel electrophoresis at 80V for 10 hours and Southern blots were performed as described in the methods.
Southern and Western blot analysis
For Southern blots, 2-5 micrograms of vaccinia genomic DNA was digested with EcoRV or BspEI (New England Biolabs). Genomes were resolved by agarose gel electrophoresis, DNA was transferred by capillary action and UV-crosslinked to nylon membranes (Nytran SuPerCharge; Whatman). Probes of PCR-amplified K3L, or an adjacent genomic region, were generated by biotin random prime labeling, hybridized, and bands detected with a North2South Chemiluminescent detection kit (Pierce).
For western blots, cells were infected with vaccinia for 48 hours and whole cell lysates were collected in 2% SDS. Proteins were resolved by SDS-PAGE (12% Tris-glycine gel; Invitrogen) and transferred to nitrocellulose membranes (Pierce). Proteins were detected with anti-K3L antibody (1:2,000; gift from J. Tartaglia), serum from rabbits infected with vaccinia virus (1:1,000; gift from B. Moss), and anti-actin antibody (1:1,000; Abcam).
siRNA knockdown of K3L
Three double stranded siRNAs were designed to target expression of K3L (1: 5′-aagauaaauguauagagcauaaucc(uu)-3′; 2: 5′- uucaacauaucuauccauaugcauc(uu)-3′; 3: 5′-uauaaucaacucuaaucacuuuaac(uu)-3′; IDTDNA). HeLa cells were transfected with one of the siRNA molecules (10nM) or a non-targeting control siRNA that has at least 4 mismatches to all ORFs from several genomes (Dharmacon, Thermo Scientific) using Lipofectamine 2000 (Invitrogen). 24 hours after transfection, cells were infected with vaccinia for 48 hours. Expression of K3L was measured by western blot and β-gal activity was measured as a proxy for virus replication.
Sequence data
Genomic sequence files are available from the NCBI Sequence Read Archive under accession SRP013416.
Supplementary Material
Highlights.
Poxviruses rapidly adapt against host defenses via highly specific gene amplifications
Gains in vaccinia fitness occur within only a few serial passages in human cells
Gene expansions precede and can facilitate adaptation via point mutation
Gene ‘accordions’ reveal a new mode of virus adaptation in double-stranded DNA viruses
Acknowledgements
We thank B. Jacobs for virus strains, J. Tartaglia for K3L antibody, B. Moss for vaccinia reactive rabbit serum, and G. Smith for RecBCD and advice on recombination. We thank M. Daugherty, M. Dunham, M. Emerman, A. Frost, J. Kerns, M. Patel, and S. Sawyer for critical reading of the manuscript and B. Stackhouse for technical assistance with sequencing library preparation. This work is supported by NIH K99/R00 award GM090042 (NCE), NIH grant 1R21CA160080-01 (JS), NIH grant AI26672 (APG), a NSF Graduate Research Fellowship (JOK), and an NSF CAREER award (HSM). HSM is an Early Career Scientist of HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, Shendure J. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso CL, Tulman ER, Lu Z, Zsak L, Kutish GF, Rock DL. The genome of fowlpox virus. J Virol. 2000;74:3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson DI, Slechta ES, Roth JR. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science. 1998;282:1133–1135. doi: 10.1126/science.282.5391.1133. [DOI] [PubMed] [Google Scholar]
- Bahar MW, Graham SC, Chen RA, Cooray S, Smith GL, Stuart DI, Grimes JM. How vaccinia virus has evolved to subvert the host immune response. J Struct Biol. 2011;175:127–134. doi: 10.1016/j.jsb.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie E, Denzler KL, Tartaglia J, Perkus ME, Paoletti E, Jacobs BL. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J Virol. 1995;69:499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont HJ, Gallie J, Kost C, Ferguson GC, Rainey PB. Experimental evolution of bet hedging. Nature. 2009;462:90–93. doi: 10.1038/nature08504. [DOI] [PubMed] [Google Scholar]
- Bergthorsson U, Andersson DI, Roth JR. Ohno’s dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci U S A. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratke KA, McLysaght A. Identification of multiple independent horizontal gene transfers into poxviruses using a comparative genomics approach. BMC Evol Biol. 2008;8:67. doi: 10.1186/1471-2148-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J, Overbaugh J, Miller S. The origin of mutants. Nature. 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 2001;98:6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar AC, Sicheri F. X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol Cell. 2002;10:295–305. doi: 10.1016/s1097-2765(02)00590-7. [DOI] [PubMed] [Google Scholar]
- Davies MV, Chang HW, Jacobs BL, Kaufman RJ. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol. 1993;67:1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc Lond B. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW. The life and death of gene families. Bioessays. 2009;31:29–39. doi: 10.1002/bies.080085. [DOI] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci U S A. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond JT, Genschel J, Wolf E, Modrich P. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proc Natl Acad Sci U S A. 1997;94:10144–10149. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig R, Botstein D. Characteristic genome rearrangements in experimenatl evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler DC, Lehman IR. On the role of ATP in phosphodiester bond hydrolysis catalyzed by the recBC deoxyribonulease of Escherichia coli. J Biol Chem. 1977;252:499–503. [PubMed] [Google Scholar]
- Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nat Rev Microbiol. 2009;7:787–797. doi: 10.1038/nrmicro2222. [DOI] [PubMed] [Google Scholar]
- Emerman M, Malik HS. Paleovirology--modern consequences of ancient viruses. PLoS Biol. 2010;8:e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito J, Condit R, Obijeski J. The preparation of orthopoxvirus DNA. J Virol Methods. 1981;2:175–179. doi: 10.1016/0166-0934(81)90036-7. [DOI] [PubMed] [Google Scholar]
- Esposito JJ, Sammons SA, Frace AM, Osborne JD, Olsen-Rasmussen M, Zhang M, Govil D, Damon IK, Kline R, Laker M, et al. Genome sequence diversity and clues to the evolution of variola (smallpox) virus. Science. 2006;313:807–812. doi: 10.1126/science.1125134. [DOI] [PubMed] [Google Scholar]
- Foster PL. Population dynamics of a Lac-strain of Escherichia coli during selection for lactose utilization. Genetics. 1994;138:253–261. doi: 10.1093/genetics/138.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel SJ, Johnson GP, Perkus ME, Davis SW, Winslow JP, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- Hakki M, Geballe AP. Double-stranded RNA binding by human cytomegalovirus pTRS1. J Virol. 2005;79:7311–7318. doi: 10.1128/JVI.79.12.7311-7318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC, Alberts B, Ehrenfeld E, Enquist L, Fineberg H, McKnight SL, Moss B, O’Donnell M, Ploegh H, Schmid SL, et al. Discovery of antivirals against smallpox. Proc Natl Acad Sci U S A. 2004;101:11178–11192. doi: 10.1073/pnas.0403600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Bull HJ, Klump JR, Rosenberg SM. Adaptive amplification: an inducible chromosomal instability mechanism. Cell. 2000;103:723–731. doi: 10.1016/s0092-8674(00)00176-8. [DOI] [PubMed] [Google Scholar]
- Hendrickson H, Slechta ES, Bergthorsson U, Andersson DI, Roth JR. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc Natl Acad Sci U S A. 2002;99:2164–2169. doi: 10.1073/pnas.032680899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson RC, Wang C, Hatcher EL, Lefkowitz EJ. Orthopoxvirus genome evolution: the role of gene loss. Viruses. 2010;2:1933–1967. doi: 10.3390/v2091933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman RJ. In: Double-stranded RNA-activated protein kinase PKR, In Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Mathews MB, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. pp. 503–527. [Google Scholar]
- Kawagishi-Kobayashi M, Silverman JB, Ung TL, Dever TE. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2alpha. Mol Cell Biol. 1997;17:4146–4158. doi: 10.1128/mcb.17.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr PJ, Hone J, Perrin L, French N, Williams CK. Molecular and serological analysis of the epidemiology of myxoma virus in rabbits. Vet Microbiol. 2010;143:167–178. doi: 10.1016/j.vetmic.2009.11.025. [DOI] [PubMed] [Google Scholar]
- Kugelberg E, Kofoid E, Andersson DI, Lu Y, Mellor J, Roth FP, Roth JR. The tandem inversion duplication in Salmonella enterica: selection drives unstable precursors to final mutation types. Genetics. 2010;185:65–80. doi: 10.1534/genetics.110.114074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland JO, Jacobs BL. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology. 2002;299:133–141. doi: 10.1006/viro.2002.1479. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Carroll DS, Gardner SN, Walsh MC, Vitalis EA, Damon IK. On the origins of smallpox: correlating variola phylogenics with historical smallpox records. Proc Natl Acad Sci U S A. 2007;104:15787–15792. doi: 10.1073/pnas.0609268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski KJ, Farslow JC, Fitzpatrick KA, Lynch M, Katju V, Bergthorsson U. High spontaneous rate of gene duplication in Caenorhabditis elegans. Curr Biol. 2011;21:306–310. doi: 10.1016/j.cub.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, Dickinson WJ, Okamoto K, Kulkarni S, Hartl DL, Thomas WK. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci U S A. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Baldi PF, Gaut BS. Extensive gene gain associated with adaptive evolution of poxviruses. Proc Natl Acad Sci U S A. 2003;100:15655–15660. doi: 10.1073/pnas.2136653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger-Henschel C, Schmidt M, Lukassen S, Linke B, Krause L, Konietzny S, Goesmann A, Howley P, Chaplin P, Suter M, Hausmann J. Genomic sequence of chorioallantois vaccinia virus Ankara, the ancestor of modified vaccinia virus Ankara. J Gen Virol. 2007;88:3249–3259. doi: 10.1099/vir.0.83156-0. [DOI] [PubMed] [Google Scholar]
- Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE. Repeatability and contingency in the evolution of a key innovation in page lambda. Science. 2012;335:428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson NR, Sawyer SL. Two-stepping through time: mammals and viruses. Trends Microbiol. 2011;19:286–294. doi: 10.1016/j.tim.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montville R, Froissart R, Remold SK, Tenaillon O, Turner PE. Evolution of mutational robustness in an RNA virus. PLoS Biol. 2005;3:e381. doi: 10.1371/journal.pbio.0030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Poxviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2905–2946. [Google Scholar]
- Moss B, Winters E, Cooper N. Instability and reiteration of DNA sequences within the vaccinia virus genome. Proc Natl Acad Sci U S A. 1981;78:1614–1618. doi: 10.1073/pnas.78.3.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Miller B, Barends M, Jaidee A, Patel J, Mayxay M, Newton P, Nosten F, Ferdig MT, Anderson TJ. Adaptive copy number evolution in malaria parasites. PLoS Genet. 2008;4:e1000243. doi: 10.1371/journal.pgen.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pränting M, Andersson DI. Escape from growth restriction in small colony variants of Salmonella typhimurium by gene amplification and mutation. Mol Microbiol. 2011;79:305–315. doi: 10.1111/j.1365-2958.2010.07458.x. [DOI] [PubMed] [Google Scholar]
- Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat Struct Mol Biol. 2009;16:63–70. doi: 10.1038/nsmb.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandegren L, Andersson DI. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7:578–588. doi: 10.1038/nrmicro2174. [DOI] [PubMed] [Google Scholar]
- Schimke RT. Methotrexate resistance and gene amplification. Mechanisms and implications. Cancer. 1986;57:1912–1917. doi: 10.1002/1097-0142(19860515)57:10<1912::aid-cncr2820571004>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Searles RP, Bergquam EP, Axthelm MK, Wong SW. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabaugh MB, Mathews CK. Hydroxyurea-resistant vaccinia virus: overproduction of ribonucleotide reductase. J Virol. 1986;60:506–514. doi: 10.1128/jvi.60.2.506-514.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabaugh MB, Roseman NA, Mathews CK. Amplification of the ribonucleotide reductase small subunit gene: analysis of novel joints and the mechanism of gene duplication in vaccinia virus. Nucleic Acids Res. 1989;17:7073–7088. doi: 10.1093/nar/17.17.7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GL, Murphy BR, Moss B. Infectious poxvirus vectors have capacity for at least 25,000 base pairs of foreign DNA. Gene. 1983;25:21–28. doi: 10.1016/0378-1119(83)90163-4. [DOI] [PubMed] [Google Scholar]
- Smith GR, Amundsen SK, Dabert P, Taylor AF. The initiation and control of homologous recombination in Escherichia coli. Philos Trans R Soc Lond B Biol Sci. 1995;347:13–20. doi: 10.1098/rstb.1995.0003. [DOI] [PubMed] [Google Scholar]
- Stambuk BU, Dunn B, Alves SL, Jr., Duval EH, Sherlock G. Industrial fuel ethanol yeasts contain adaptive copy number changes in genes involved in vitamin B1 and B6 synthesis. Genome Research. 2009;19:2271–2278. doi: 10.1101/gr.094276.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Berg OG, Roth JR, Andersson DI. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics. 2009;182:1183–1195. doi: 10.1534/genetics.109.103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L. A new evoluitonary law. Evolutionary Theory. 1973;1:1–30. [Google Scholar]
- Vignuzzi M, Stone JK, Andino R. Ribavirin and lethal mutagenesis of poliovius: molecular mechanisms, resistance and biological implications. Virus Res. 2005;107:173–181. doi: 10.1016/j.virusres.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werden SJ, Rahman MM, McFadden G. Poxvirus host range genes. Adv Virus Res. 2008;71:135–171. doi: 10.1016/S0065-3527(08)00003-1. [DOI] [PubMed] [Google Scholar]
- Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ. Different trajectories of parallel evolution during viral adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.