Abstract
Multidrug resistant tuberculosis (MDR TB), defined by resistance to the two most effective first-line drugs, isoniazid and rifampin, is on the rise globally and is associated with significant morbidity and mortality. Despite the increasing availability of novel, rapid diagnostic tools for Mycobacterium tuberculosis (Mtb) drug susceptibility testing (DST), the clinical applicability of these methods is unsettled. Here, we review the mechanisms of action and resistance of Mtb to isoniazid and rifampin as well as the utility, advantages, and limitations of the available Mtb DST tools. We place particular emphasis on molecular methods with rapid turn-around including line probe assays, molecular beacon-based real time-polymerase chain reaction, and pyrosequencing. We conclude that neither rapid molecular drug testing nor phenotypic methods are perfect in predicting MTB drug susceptibility, and therefore must be interpreted within the clinical context of each patient.
Keywords: tuberculosis, multidrug-resistance, drug susceptibility testing, molecular diagnostic tools
INTRODUCTION
Multidrug-resistant tuberculosis (MDR TB), defined by resistance to the two most effective antituberculous first-line drugs, isoniazid and rifampin, is on the rise globally. In 2008 the World Health Organization (WHO) reported an estimated 390,000–510,000 new cases of MDR TB constituting 3.6% (95% CI: 3.0–4.4) of all incident TB cases worldwide that year. Mortality from MDR TB in 2008 was also high, with an estimated 150,000 attributable deaths1. The prognosis for drug-resistant TB is especially poor in HIV-infected patients, with a recent South African study reporting one-year mortality of 71% for MDR and 83% for extensively drug–resistant TB (XDR TB, defined as MDR plus resistance to a fluoroquinolone and an injectable second-line therapy); 40% of the MDR TB and 51% of XDR TB HIV-coinfected cases died within 30 days of sputum collection2. The high global MDR TB prevalence and mortality calls for timely DST and improved therapies.
To provide a framework for our review of the currently available diagnostic modalities for the detection of MDR TB, we first review the mechanisms of resistance to isoniazid and rifampin. In 1998 the circular genome of the best-characterized strain of Mtb, H37Rv, was elucidated and noted to consist of almost 4,000 genes and over 4 million base pairs3,4. A large body of literature has since emerged, describing the association between specific gene mutations and DST. The database with identified TB drug resistance mutations can be found at http://www.tbdreamdb.com.
Isoniazid: Mechanism of action and resistance in Mycobacterium tuberculosis
Isoniazid is a prodrug that requires activation by the mycobacterial catalase peroxidase KatG after it enters the cell by passive diffusion. The activated isoniazid targets two principal enzymes that are involved in the elongation cycle of the fatty acid molecules, an enoyl-acyl carrier protein reductase (inhA) and a beta ketoacyl-acyl carrier protein synthase, resulting in the inhibition of synthesis of the mycolic acids necessary for the mycobacterial cell wall5,6.
Spontaneous mutations responsible for isoniazid resistance, in contrast to rifampin resistance, are not concentrated within one gene. Drug resistance mutations in the katG gene result in loss of the ability of the catalase to activate the prodrug of isoniazid7. Mutations in the inhA gene or its promoters may alter the activated isoniazid binding site or increase InhA production resulting in H resistance8. While katG mutations may confer high-level isoniazid resistance, inhA mutation may cause low-level isoniazid resistance and cross resistance to ethionamide. Although isoniazid mutations most frequently occur in the katG and inhA genes, they also occur in other enzymes coding genes such as ndh, ahpC, and furA 5,9. Between 31–97% of INH resistance has been attributed to katG mutations (at codon 315), with higher frequencies occurring in TB-endemic countries10. In a recent study by Dalla Costa et al of 224 INH-resistant Mtb isolates from Argentina, Brazil, and Peru the frequency of inhA mutations was 11%. Eighty-six percent had either a katG or inhA mutation associated with INH resistance.11
Rifampin: Mechanism of action and resistance in Mycobacterium tuberculosis
Rifampin inhibits transcription and thus protein synthesis by targeting one of the four subunits, the β subunit, of the mycobacterial DNA-dependent RNA polymerase which is coded by the rpoB gene3,12. Certain mutations in the rpoB gene reduce the binding affinity of rifampin for the RNA polymerase, resulting in drug resistance (see Figure 1)3,13. Rifampin resistance is considered a major surrogate marker for MDR TB, since greater than 90% of isolates resistant to rifampin are also resistant to isoniazid14–16. Over 95% of mutations responsible for resistance to rifampin occur within an 81-base pair core region (codon 507 to 533) of the rpoB gene, termed the Rifampin Resistance Determining Region (RRDR)12,17. Furthermore, greater than 92% of the mutations occur at either codon 516 (which codes low-level resistance), 526, or 531 (which code for high-level resistance)18.
Figure 1.
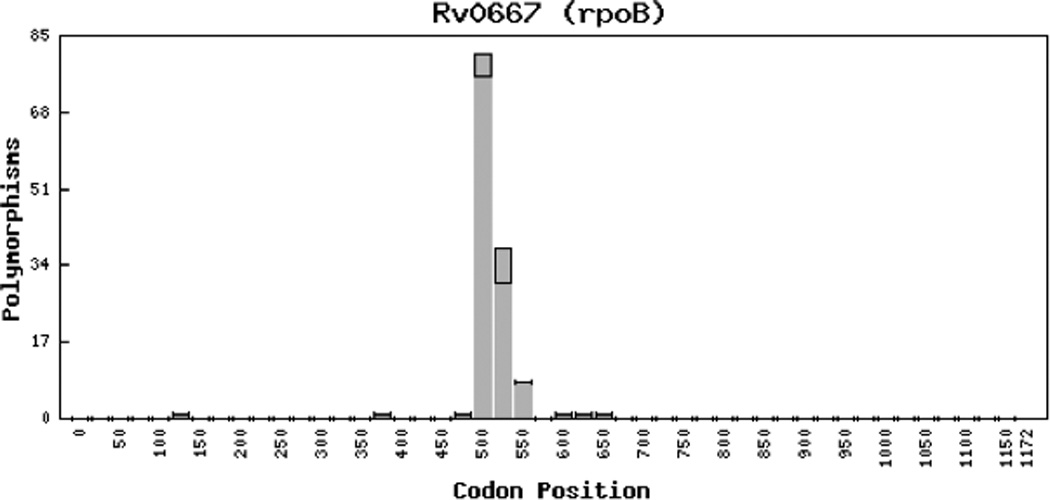
the distribution of known mutations in the Mtb rpoB gene associated with rifampin resistance (Figure adapted from: Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. (2009) Tuberculosis Drug Resistance Mutation Database. PLoS Med 6(2): e1000002. doi:10.1371/ journal.pmed.1000002. Written permission was obtained).
PHENOTYPIC ASSAYS FOR DETECTION OF MDR TB
Despite its recognized limitations, conventional phenotypic DST remains the gold standard for MTB DST. Liquid media is used more commonly for DST than solid media in resource-rich countries (Table 1). Traditional solid media DST uses the agar proportion, absolute concentration, or resistance ratio method on Lowenstein-Jensen (LJ) or Middlebrook 7H10/11 (MB7H10/MB7H11) media. The agar proportion method compares the number of colonies growing on a drug-containing medium at a defined critical concentration to that growing on a drug-free medium 19. For MB7H10 media the critical concentration for the first-line drugs are: H 0.2 µg/mL (low-level resistance) and 1 µg/mL (high-level resistance), R 1µg/mL, and E 10 µg/mL19. The prolonged turn-around-time (TAT) of solid media for culture from specimen collection was significantly shortened when liquid media replaced solid media for DST in the USA (from 8–12 weeks to 3–7 weeks).
Table 1.
Most commonly used phenotypic assays for Mycobacterium tuberculosis drug susceptibility testing
| Methods | DST turn around time | Advantages | Disadvantages |
|---|---|---|---|
| Conventional phenotypic methods | |||
| Solid media (Lowenstein-Jensen or Middlebrook 7H10/11 media) | |||
| Proportion method | 8–12 weeks from sample collection |
Time-consuming Labor-intensive |
|
| Resistance ratio | |||
| Absolute concentration | |||
| Broth-based methods | |||
| BACTEC 460 TB system (radiometric) | 3–7 weeks from sample collection |
Commercially available | Requires radiation safety protocol Cumbersome inoculation |
| BACTEC MGIT 960 TB (non-radiometric) | Automated | ||
| VersaTREK | Automated | ||
| Rapid phenotypic methods | |||
| Microscopic Observation Drug Susceptibility (MODS) | 2 – 4 weeks from sample collection |
Low cost SN 92–96%; SP 96% |
Labor-intensive (frequent exams) |
| Can perform directly on sputum specimens | Requires inverted microscope Bio-safety concerns Difficulty distinguishing Mtb from NTMs |
||
Colorimetric redox indicator methods
|
1–2 weeks after Mtb isolated |
SN=91–100%, SP=98– 100% |
Performed on culture plates Bio-safety concern Not standardized |
|
Nitrate reductase assay (based on GRIESS method) |
1–4 weeks from sample collection |
SN>94%; SP=100% Low cost Can perform directly on smear-positive specimens |
False positive reaction by other nitrate-producing organisms Accuracy limited by mycobacterial metabolic activity Cannot detect nitrate reductase- negative Mtb strains |
DST=drug susceptibility testing; SN=sensitivity; SP=specificity; Mtb=Mycobacterium tuberculosis; NTMs=non-tuberculous mycobacteria
The original system using liquid media for DST was the BACTEC 460 TB system. Limited by issues with handling and disposing of radioactive material, it was supplanted by the BACTEC MGIT 960 system which used fluorescent light emission for detection of TB growth. The sensitivity and specificity of the BACTEC MGIT 960 for detection of isoniazid and rifampin resistance are over 95%20,21. For the MGIT 960, the critical drug concentrations for the first-line drugs are: H 0.1 µg/mL (low-level resistance) and 0.4 µg/mL (high-level resistance), R 2µg/mL, and E 5 µg/mL19. Another automated broth-based method is the Versa TREK system which has the capacity to simultaneously detect mycobacterial growth and conduct phenotypic DST to first-line drugs using measurement of oxygen consumption.
Other novel rapid phenotypic methods include the colorimetric methods that use the color change of a chemical dye (i.e., tetrazolium bromide and resazurin) for culture and DST, the microscopic observation of drug susceptibility, and the nitrate reduction assays. Limitations of rapid phenotypic methods include the uncertain reliability of conventional breakpoints, decreased accuracy in cultures mixed with other mycobacteria, and the possibility of reduced fitness and growth of mutant organisms, which may require a higher inoculum to increase test sensitivity.
GENOTYPIC METHODS FOR DETECTING ISONIAZID AND RIFAMPIN RESISTANCE
DNA Sequencing
While conventional DNA sequencing for detection of mutations associated with Mtb resistance is not routinely available in the commercial setting due to expense, necessary expertise, and time-consuming nature, it is available in some research and public health laboratories and the CDC Molecular Detection of Drug Resistance (MDDR) service. Conventional DNA sequencing utilizes a “chain-termination method” to sequence DNA fragments. Specifically, it first binds a primer to a denatured single strand of DNA. DNA extension then begins at the primer site using a DNA polymerase. It is eventually terminated because a dye-labeled dideoxynucleotide interrupts the phosphodiesterase bond between two subsequent nucleotides. This cycle results in DNA fragments of various lengths, which can be separated by electrophoresis and subsequently sequenced. Conventional DNA sequencing remains the gold-standard of DNA sequencing, is highly accurate, and offers the advantage of being able to read larger amounts of DNA. It is the foundation on which many of the rapid molecular assays, (i.e., line probe assays, molecular beacon-based real time-polymerase chain reactions (RT-PCR), and pyrosequencing) were developed (Table 2). These assays have the capacity to both identify Mtb isolates and evaluate for drug resistance to isoniazid and rifampin.
Table 2.
Summary of clinically available rapid molecular resistance testing assays for Mycobacterium tuberculosis
| Molecular Testing Method |
Turn-around time |
Sensitivity | Specificity | Advantages | Limitations |
|---|---|---|---|---|---|
Line probe assay
|
6 – 48 hours | Rifampin resistance: -culture isolates: 95–100% - clinical specimens: 80–100% Isoniazid resistance: 72–92% |
Rifampin resistance: - culture isolates: 92–100% - clinical specimens: 100% Isoniazid resistance: 95–100% |
- rapid -sensitivity in detecting rifampin resistance mutations |
- inability to differentiate silent mutations from those responsible for rifampin resistance - limited accuracy in detecting isoniazid resistance mutations |
| RT-PCR coupled with molecular beacons |
2 hours | Rifampin resistance: 86–100% Isoniazid resistance: 76–94% |
Rifampin resistance: 95–100% Isoniazid resistance: 100% |
- rapid - accuracy in detecting rifampin resistance mutations within the target region - high throughput - elimination of cross- contamination - hands-free processing - capacity to detect novel mutations |
- expensive - inability to detect mutations outside of pre-specified target region - limited accuracy in detecting isoniazid resistance mutations |
| Pyrosequencing | 2 – 48 hours | Rifampin resistance: 92–100% Isoniazid resistance: 64–81% |
Rifampin resistance: 92–100% Isoniazid resistance: 100% |
- provides exact DNA sequences - high throughput |
- accuracy limited in lengthy DNA sequences - limited accuracy in detecting isoniazid resistance mutations |
Line probe assays (PCR-based)
Line probe assays involve DNA extraction, amplification of a predefined gene region associated with resistance, and reverse hybridization of the PCR products with standard, immobilized probes for gene mutations associated with resistance22. For example, the INNO-LipA® Rif.TB assay contains wild-type “S” probes as well as “R” probes that detect resistance mutations in the RRDR of the rpoB region. The non-detection of one of the “S” probes implies rifampin resistance23. At present the only three commercially available line probe assays for the detection of first-line drug resistance of Mtb are the INNO-LipA® Rif.TB (Innogenetics, Belgium), Genotype® MTBDR, and second-generation Genotype® MTBDRplus (Hain LifeScience GmbH, Germany). While rpoB gene mutations responsible for rifampin resistance are detected by all three assays, Genotype® MTBDR additionally detects katG mutations and Genotype® MTBDRplus detects both katG and inhA mutations24.
Two recent meta-analyses evaluating the accuracy of the line probe assays have demonstrated sensitivity in detecting rifampin resistance mutations to be 94–100% and specificity to be 99–100% in clinical specimens and laboratory isolates22,25. The first meta-analysis, which evaluated the accuracy of the INNO-LipA® Rif.TB assays compared to susceptibility results obtained from either BACTEC 460 or agar proportion method, was conducted using 15 studies and 1,738 specimens from several countries and body sites. Although the sensitivity on the cultured isolates was greater than 95%, it demonstrated higher variability (range 80%–100%) of the assay on direct clinical specimens. The second meta-analysis was similar, but evaluated the accuracy of the Genotype® MTBDR assay (as determined by comparison to the agar proportion method, BACTEC 460, and/or BACTEC MGIT 960), and was comprised of 10 articles and 3,349 laboratory isolates and clinical specimens from various geographic areas. The pooled sensitivity and specificity of the Genotype® MTBDR assay was only 84.3% (76.6%–89.8%) and 99.5% (97.5%–99.9%), respectively, for detecting of INH resistance. The TAT for the line probe assays ranged from 1 to 2 days.
A few studies have conducted a head-to-head comparison of INNO-LipA® Rif.TB to the Genotype® MTBDR in their ability to accurately detect MDR TB. One such study compared the two line probe assays with DST and conventional sequencing on 52 Mtb clinical isolates from Finland and Russia. The two assays had a 100% concordance rate in detecting rifampin resistance, each detecting 51/52 (98.1%) of rifampin resistance detected by DST. The Genotype®MTBDR and INNO-LipA® Rif.TB detected 92.3% and 96.2%, respectively, of the rpoB mutations found by DNA sequencing26.
Thus, although not FDA-approved, line probe assays are rapid and accurate tools for the detection of rifampin resistance provided that the mutations responsible for resistance are within the RRDR of the rpoB gene, as occurs in greater than 95% of rifampin resistant strains. They have less clinical utility in detecting isoniazid resistance, because of the limited number of INH resistance-incurring mutations represented in the assay. Other major limitations of the line probe assays include their inability to differentiate between resistance-inducing and silent mutations and their insensitivity in detecting novel mutations, because they do not rely on DNA sequencing technology. Variability in assay sensitivity can be in part explained by regional differences in rifampin and INH resistance mutation frequencies16.
Molecular beacon-based real time-polymerase chain reaction (RT-PCR)
Another commercially available hybridization method for the detection of MDR TB is the GeneXpert® MTB/Rif TB assay (Cepheid, CA). This real-time PCR assay uses the molecular beacons, probes for hybridization to different target segments within a region of the gene of interest. When there is exact nucleotide concordance between the probe and target sequence, the beacons emit fluorescent signals. The absence of signaling suggests a mutation in the corresponding surveyed segment of the region.
Molecular beacon-based RT-PCR methods have been tested in countries with high and low MDR TB prevalence27–33. Sensitivity and specificity in detection of rifampin resistance in clinical specimens range from 86–100% and 95–100%, respectively, with higher sensitivity in smear-positive cases33. Sensitivity and specificity in detection of isoniazid resistance in clinical isolates range from 76%–94% and 100%, respectively. Reduced sensitivity is often due to presence of drug-resistance incurring mutations outside of the surveyed region, poor-quality sputum specimens, and smear-negative and mixed mycobacterial populations.
Major advantages of molecular beacon-based RT-PCR assays include their high sensitivity and specificity in detection of MDR TB, rapid TAT (of less than 2 hours), hands-free processing, near-patient technology, and high throughput29,33. Cross-contamination is virtually eliminated because amplification, hybridization, and analysis occur within one closed well31. Additionally, these assays are not limited to detection of pre-determined mutations; they have the capacity to detect previously unrecognized mutations within a given region. The major limitations to their use include the cost of equipment, inability to detect resistance-incurring mutations outside of specified target region, and the detection of silent mutations which are falsely interpreted as conferring resistance.
Pyrosequencing
Pyrosequencing is a rapid, automated DNA sequencing technique that has recently been used to detect mutations associated with drug resistance in M. tuberculosis18,34–38. Its “sequencing by synthesis” methodology involves synthesizing a strand of DNA complementary to the DNA segment of interest via a DNA polymerase. When the DNA polymerase integrates a nucleotide complementary to a base pair on the template of the study strand, ATP is generated and provides energy for the light-generating luciferase reaction36.
Like conventional sequencing, pyrosequencing can provide exact DNA sequences, thus detecting both previously known and novel mutations. Advantages of pyrosequencing over conventional DNA sequencing include reduced cost, speed and simplicity of processing, ease of interpretability, and relative high throughput. A major drawback of pyrosequencing is its inaccuracy in reading contiguous, long sequences (i.e., greater than 50 nucleotides)35. It has great utility in detecting rifampin resistance-associated mutations (sensitivity 92–100%, specificity 92–100% among clinical and laboratory specimens). Its utility in detecting isoniazid resistance is poorer (sensitivity 64%–81%, specificity 100% among clinical and laboratory isolates) because many INH-resistance mutations remain unknown and lie outside of the normally studied regions of katG and inhA genes18,34–39.
CONCLUSIONS
The high global prevalence of MDR TB and its associated worldwide morbidity and mortality necessitate rapid DST. Diagnostic methods must not only have high accuracy in detecting rifampin and isoniazid resistance, rapid TATs, and high through-put, but also be available at low cost in low-income, MDR TB-endemic countries.
Rapid molecular diagnostics are well-suited for confirmation of suspected MDR TB and provide a valuable adjunct to conventional phenotypic testing. At present, however, neither rapid molecular drug testing nor phenotypic methods are perfect in predicting MTB drug susceptibility40,41. Clinicians must consider treating patients at high risk for MDR until phenotypic susceptibility results are known even if rapid molecular tests do not predict resistance. DNA sequencing should be reserved for suspected drug-resistant MTB isolates in which phenotypic susceptibility and rapid molecular testing yield discrepant results. Lastly, future guidelines should address a diagnostic algorithm to aid clinicians and clinical laboratories in the management and detection of MDR TB.
ACKNOWLEDGEMENTS
We would like to acknowledge Michael Leonard (Emory University, Atlanta, GA), Jyothi Rengarajan (Emory University, Atlanta, GA), Andrew Vernon (Centers for Disease Control and Prevention, Atlanta, GA), and Aliya Yamin (Fulton County Health Department, Atlanta, GA) for their assistance and input in writing this manuscript.
Source of Funding: This work was supported by the National Institute of Health/National Institute of Allergy and Infectious Diseases [grant number T32AI074492] to ASK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest We have no conflicts of interest to report.
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the supporting agency.
Contributor Information
Ameeta S. Kalokhe, Emory University School of Medicine, Division of Infectious Diseases, 206 Woodruff Research Extension Bldg., 49 Jesse Hill Jr. Drive, Atlanta, GA 30303, Ph: 313-598-8584, Fax: 404-880-9305, akalokh@emory.edu.
Majid Shafiq, Mayo Clinic Division of Hospital Internal Med.
James C. Lee, Emory University School of Medicine, Department of Medicine, Resident.
Susan M. Ray, Emory University School of Medicine, Division of Infectious Diseases.
Yun F. Wang, Emory University School of Medicine, Dept. Pathology & Laboratory Medicine.
Beverly Metchock, Centers for Disease Control and Prevention, Division of Tuberculosis Elimination, Mycobacteriology Laboratory Branch.
Albert M. Anderson, Emory University School of Medicine, Division of Infectious Diseases.
Minh Ly T. Nguyen, Emory University School of Medicine, Division of Infectious Diseases.
REFERENCES
- 1.Organization WH. Multidrug and extensively drug-resistant TB (M/XDR-TB): 2010 global report on surveillance and response. 2010:19.
- 2.Gandhi NR, Shah NS, Andrews JR, et al. HIV coinfection in multidrug- and extensively drug-resistant tuberculosis results in high early mortality. Am J Respir Crit Care Med. 2010 Jan 1;181(1):80–86. doi: 10.1164/rccm.200907-0989OC. [DOI] [PubMed] [Google Scholar]
- 3.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Med. 2009 Feb 10;6(2):e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998 Jun 11;393(6685):537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Vilcheze C, Jacobs WR., Jr The mechanism of isoniazid killing: clarity through the scope of genetics. Annu Rev Microbiol. 2007;61:35–50. doi: 10.1146/annurev.micro.61.111606.122346. [DOI] [PubMed] [Google Scholar]
- 6.Timmins GS, Deretic V. Mechanisms of action of isoniazid. Mol Microbiol. 2006;62(5):1220–1227. doi: 10.1111/j.1365-2958.2006.05467.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. Report of expert consultations on rapid molecular testing to detect drug-resistant tuberculosis in the United States. 2009
- 9.Hazbon MH, Brimacombe M, Bobadilla del Valle M, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006 Aug;50(8):2640–2649. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen T, Becerra MC, Murray MB. Isoniazid resistance and the future of drug-resistant tuberculosis. Microb Drug Resist. 2004 Winter;10(4):280–285. doi: 10.1089/mdr.2004.10.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalla Costa ER, Ribeiro MO, Silva MS, et al. Correlations of mutations in katG, oxyR-ahpC and inhA genes and in vitro susceptibility in Mycobacterium tuberculosis clinical strains segregated by spoligotype families from tuberculosis prevalent countries in South America. BMC Microbiol. 2009;9:39. doi: 10.1186/1471-2180-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Telenti A, Imboden P, Marchesi F, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993 Mar 13;341(8846):647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 13.Jin DJ, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988 Jul 5;202(1):45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 14.Bloch AB, Cauthen GM, Onorato IM, et al. Nationwide survey of drug-resistant tuberculosis in the United States. JAMA. 1994 Mar 2;271(9):665–671. [PubMed] [Google Scholar]
- 15.Shah NS, Lan NT, Huyen MN, et al. Validation of the line-probe assay for rapid detection of rifampicin-resistant Mycobacterium tuberculosis in Vietnam. Int J Tuberc Lung Dis. 2009 Feb;13(2):247–252. [PubMed] [Google Scholar]
- 16.Traore H, Fissette K, Bastian I, Devleeschouwer M, Portaels F. Detection of rifampicin resistance in Mycobacterium tuberculosis isolates from diverse countries by a commercial line probe assay as an initial indicator of multidrug resistance. Int J Tuberc Lung Dis. 2000 May;4(5):481–484. [PubMed] [Google Scholar]
- 17.Musser JM. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995 Oct;8(4):496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnold C, Westland L, Mowat G, Underwood A, Magee J, Gharbia S. Single-nucleotide polymorphism-based differentiation and drug resistance detection in Mycobacterium tuberculosis from isolates or directly from sputum. Clin Microbiol Infect. 2005 Feb;11(2):122–130. doi: 10.1111/j.1469-0691.2004.01034.x. [DOI] [PubMed] [Google Scholar]
- 19.CNTC. Drug-resistant tuberculosis: a survival guide for clinicians. 2nd edition. 2008. pp. 1–266. [Google Scholar]
- 20.Rusch-Gerdes S, Domehl C, Nardi G, Gismondo MR, Welscher HM, Pfyffer GE. Multicenter evaluation of the mycobacteria growth indicator tube for testing susceptibility of Mycobacterium tuberculosis to first-line drugs. J Clin Microbiol. 1999 Jan;37(1):45–48. doi: 10.1128/jcm.37.1.45-48.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bemer P, Palicova F, Rusch-Gerdes S, Drugeon HB, Pfyffer GE. Multicenter evaluation of fully automated BACTEC Mycobacteria Growth Indicator Tube 960 system for susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol. 2002 Jan;40(1):150–154. doi: 10.1128/JCM.40.1.150-154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling DI, Zwerling AA, Pai M. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur Respir J. 2008 Nov;32(5):1165–1174. doi: 10.1183/09031936.00061808. [DOI] [PubMed] [Google Scholar]
- 23.Rossau R, Traore H, De Beenhouwer H, et al. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob Agents Chemother. 1997 Oct;41(10):2093–2098. doi: 10.1128/aac.41.10.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brossier F, Veziris N, Jarlier V, Sougakoff W. Performance of MTBDR plus for detecting high/low levels of Mycobacterium tuberculosis resistance to isoniazid. Int J Tuberc Lung Dis. 2009 Feb;13(2):260–265. [PubMed] [Google Scholar]
- 25.Morgan M, Kalantri S, Flores L, Pai M. A commercial line probe assay for the rapid detection of rifampicin resistance in Mycobacterium tuberculosis: a systematic review and meta-analysis. BMC Infect Dis. 2005;5:62. doi: 10.1186/1471-2334-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makinen J, Marttila HJ, Marjamaki M, Viljanen MK, Soini H. Comparison of two commercially available DNA line probe assays for detection of multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2006 Feb;44(2):350–352. doi: 10.1128/JCM.44.2.350-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varma-Basil M, El-Hajj H, Colangeli R, et al. Rapid detection of rifampin resistance in Mycobacterium tuberculosis isolates from India and Mexico by a molecular beacon assay. J Clin Microbiol. 2004 Dec;42(12):5512–5516. doi: 10.1128/JCM.42.12.5512-5516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin SY, Probert W, Lo M, Desmond E. Rapid detection of isoniazid and rifampin resistance mutations in Mycobacterium tuberculosis complex from cultures or smear-positive sputa by use of molecular beacons. J Clin Microbiol. 2004 Sep;42(9):4204–4208. doi: 10.1128/JCM.42.9.4204-4208.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Hajj HH, Marras SA, Tyagi S, Kramer FR, Alland D. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J Clin Microbiol. 2001 Nov;39(11):4131–4137. doi: 10.1128/JCM.39.11.4131-4137.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piatek AS, Telenti A, Murray MR, et al. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob Agents Chemother. 2000 Jan;44(1):103–110. doi: 10.1128/aac.44.1.103-110.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piatek AS, Tyagi S, Pol AC, et al. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotechnol. 1998 Apr;16(4):359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 32.Gomez DI, Fisher-Hoch SP, Bordt AS, et al. Systematic interpretation of molecular beacon polymerase chain reaction for identifying rpoB mutations in Mycobacterium tuberculosis isolates with mixed resistant and susceptible bacteria. Diagn Microbiol Infect Dis. 2010 Mar 11; doi: 10.1016/j.diagmicrobio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boehme CC, Nabeta P, Hillemann D, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010 Sep 9;363(11):1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bravo LT, Tuohy MJ, Ang C, et al. Pyrosequencing for rapid detection of Mycobacterium tuberculosis resistance to rifampin, isoniazid, and fluoroquinolones. J Clin Microbiol. 2009 Dec;47(12):3985–3990. doi: 10.1128/JCM.01229-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao JR, Bai YJ, Zhang QH, Wang Y, Luo M, Yan XJ. Pyrosequencing based approach for rapid detection of rifampin-resistant Mycobacterium tuberculosis. Diagn Microbiol Infect Dis. 2005 Feb;51(2):135–137. doi: 10.1016/j.diagmicrobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Marttila HJ, Makinen J, Marjamaki M, Soini H. Prospective evaluation of pyrosequencing for the rapid detection of isoniazid and rifampin resistance in clinical Mycobacterium tuberculosis isolates. Eur J Clin Microbiol Infect Dis. 2009 Jan;28(1):33–38. doi: 10.1007/s10096-008-0584-5. [DOI] [PubMed] [Google Scholar]
- 37.Zhao JR, Bai YJ, Wang Y, Zhang QH, Luo M, Yan XJ. Development of a pyrosequencing approach for rapid screening of rifampin, isoniazid and ethambutol-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2005 Mar;9(3):328–332. [PubMed] [Google Scholar]
- 38.Jureen P, Engstrand L, Eriksson S, Alderborn A, Krabbe M, Hoffner SE. Rapid detection of rifampin resistance in Mycobacterium tuberculosis by Pyrosequencing technology. J Clin Microbiol. 2006 Jun;44(6):1925–1929. doi: 10.1128/JCM.02210-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halse TA, Edwards J, Cunningham PL, et al. Combined real-time PCR and rpoB gene pyrosequencing for rapid identification of Mycobacterium tuberculosis and determination of rifampin resistance directly in clinical specimens. J Clin Microbiol. 2010 Apr;48(4):1182–1188. doi: 10.1128/JCM.02149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Deun A, Barrera L, Bastian I, et al. Mycobacterium tuberculosis strains with highly discordant rifampin susceptibility test results. J Clin Microbiol. 2009 Nov;47(11):3501–3506. doi: 10.1128/JCM.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalokhe AS, Shafiq M, Lee JC, et al. Discordance in Mycobacterium tuberculosis Rifampin Susceptibility. Emerg Infect Dis. 2012 Mar;18(3):537–539. doi: 10.3201/eid1803.111357. [DOI] [PMC free article] [PubMed] [Google Scholar]


