Abstract
The magnocellular neurones (MCN) of the supraoptic nucleus (SON) undergo reversible changes during dehydration. We hypothesise that alterations in steady-state transcript levels might be partially responsible for this plasticity. In turn, regulation of transcript abundance might be mediated by transcription factors. We have previously used microarrays to identify changes in the expression of mRNAs encoding transcription factors in response to water deprivation. We observed down-regulation of 11 and up-regulation of 31 transcription factor transcripts, including members of the AP-1 gene family, namely c-fos, c-jun, fosl1 and junD. As JunD expression and regulation within the SON has not been previously described, we have used in situ hybridisation and quantitative RT-PCR to confirm the array results, demonstrating a significant increase in JunD mRNA levels following 24-hours and 72-hours of water deprivation. Western blot and immunohistochemistry revealed a significant increase in JunD protein expression following dehydration. Double staining fluorescence immunohistochemistry with a neurone specific marker (NeuN) demonstrated that JunD staining is predominantly neuronal. Additionally, JunD immunoreactivity is observed primarily in vasopressin containing neurones with markedly less staining seen in oxytocin containing MCNs. Furthermore, JunD is highly co-expressed with c-Fos in MCNs of the SON following dehydration. These results suggest that JunD plays a role in the regulation of gene expression within MCNs of the SON in association with other Fos and Jun family members.
Keywords: hypothalamus, osmotic regulation, plasticity, microarray, RT-PCR, in situ hybridisation
Introduction
The magnocellular neurones (MCNs) of the hypothalamic supraoptic nucleus (SON) are a major site of synthesis of two major neuropeptide hormones, vasopressin (VP) and oxytocin (OT). The antidiuretic hormone VP is crucially involved in the homeostatic control of osmotic balance and the maintenance of blood pressure [1], whilst OT primarily controls reproductive functions [2] [3], it also plays a role in the homeostatic regulation of osmotic balance through its natriuretic activity (see [4]).
VP and OT are both synthesised as parts of separate pre-propeptide precursors in the cell bodies of SON hypothalamic MCNs [1], the axons of which terminate on the blood capillaries of the posterior pituitary [1] [4]. Here, mature peptide is stored until mobilised for secretion into the circulation by electrical activities evoked by hyperosmolality [5]. While VP and OT transcripts can be detected by RT-PCR in the same MCN [6] [7], expression levels of each neuropeptide RNA differ by orders of magnitude and it is not clear whether the minority species is translated. Indeed, only a very small proportion of MCNs (~1–3%) express high, equivalent levels of both peptides [8] [9], although this proportion increases following fluid deprivation [10] and lactation [11] suggesting that transcript abundance changes under different physiological states.
Osmotic stimulation evokes VP and OT release into the systemic circulation and this is accompanied by a dramatic activity-dependent functional remodelling of the SON, a process known as function-related plasticity [12] [13] [14]. For example, alterations in the relationship between MCNs and glia, the extent of terminal contact with the basal lamina in the neurohypophysis, the type of synaptic inputs, and the extent of electrotonic coupling between MCNs, have all been documented [15] [16] [17] [18] [19] and reviewed [20]. This plasticity is governed by a complex and dynamic interplay between the intrinsic properties of the MCN, interactions between MCNs, interactions with glia, and the influences of extrinsic synaptic inputs. The response of the SON to dehydration represents a unique and tractable model for understanding the processes whereby changes in gene expression mediate neuronal plasticity [20], but the molecular mechanics of these processes remain to be elucidated.
We have hypothesised that alteration in steady-state transcript levels are associated with dehydration-induced remodelling of the SON. We have thus used microarrays to identify transcripts that have a significantly altered abundance in the SON as a consequence of 3 days of dehydration [21]. In turn, some of these alterations in RNA abundance might be mediated by transcription factors. There are many ways in which transcription factor activity might be modulated, one of which is a change in the steady-state levels of its mRNA template. We have thus examined our microarray data to identify changes in the expression of mRNAs encoding transcription factors in response to water deprivation [22]. Here, we report on one of these, JunD, a member of the activator protein (AP)-1 family, which is the subject of detailed validation studies at both the transcript and protein level described herein.
Material and Methods
All experimental procedures were approved by the University of Bristol Ethical Review Committee and carried out in accord with the Animals (Scientific Procedures) Act, 1986.
Animals
Adult male Sprague-Dawley rats (280–330 g) were obtained from commercial breeders (Harlan Sera-lab, Loughborough, UK) and housed in groups of 4 to 5 per cage at a constant temperature (22±1°C) and humidity (50–60%) under controlled light-dark cycle (10 hours light, 14 hours dark, lights on at 0700). They were given access to food (standard laboratory rat chow) and water ad libitum for a period of one week prior to experimentation. Following the one-week adaptation period, the rats were randomly assigned to one of three groups. The first group had constant access to drinking water (euhydrated control group), the second group was deprived of drinking water for 24 hours and the third group for 72 hours. All three groups had free access to food throughout the entire procedure.
Reverse transcriptase quantitative polymerase chain reaction (RT- qPCR)
Twelve animals were used for (RT-qPCR) assessment of JunD gene expression. These animals were divided into 3 groups (n=4 per group) consisting of a control group (free access to water), 24-hour water deprived group and 72-hour water deprived group. Following water deprivation, (in the case of the treated groups), the rats were stunned, decapitated using a small animal guillotine between 11:00 am – 1:00 pm, the brains were removed and tissue from the SON collected and stored at −80°C until processed.
The SON was extracted from decapitated control and dehydrated rats and homogenized in TRIzol reagent (Invitrogen Life Technologies, Paisley, UK). Total RNA was isolated according to the protocol supplied by the manufacturer and all the samples were treated with DNAse I (Invitrogen Life Technologies) to remove any genomic DNA contamination. mRNA transcripts were assessed by RT-qPCR using Sybr Green based technology allowing for continuous message quantification during amplification. The amount of cDNA in each of the samples were assessed by comparison of the number of PCR cycles required to achieve a threshold of fluorescent activity set above background occurring during the exponential phase of the reaction. Normalisation for differences in sample loading was performed by the simultaneous amplification of 18S. All RT-qPCR reactions were carried out on templates from 4 different animals for each condition run in triplicate. Prior to quantification by RT-qPCR, we performed validation experiments to ensure that the efficiency of JunD and 18 S rRNA PCRs were equal and close to 1 (data not shown). We also made certain that the signals observed resulted from RT-qPCR amplification of our target gene and not from contaminating DNA by running qPCR on all our samples minus reverse transcriptase (data not shown). We also ran a melting curve at the end of each RT-qPCR run to ensure that the desired amplicon was detected.
Specific primers for rat 18S and JunD (accession number V01270 and BC062053 respectively) were designed using Primer3 (http://frodo.wi.mit.edu/). Primers for the 18S assay (5′-ACGGAAGGGCACCACCAGGA-3′ and 5′-CACCACCACCCACGGAATCG) and JunD assay (5′AAAATCCTCCCCCCTTATCC-3′ and ACTGAAAACACAAAACCGGC-3′) were synthesised and purified by a commercial supplier (Invitrogen Life Technologies).
Reverse transcription and amplification of total RNA was carried out in a 20 μl final volume using Quantitect SYBR Green RT-PCR one step kit (Qiagen) as follows: 2 ml from a total volume of 100 ml of each mRNA sample were added to a mix of 2x Quantitect SYBR Green RT-PCR Master Mix and Quantitect RT Mix containing 1 mM for each forward and reverse primer for 18S or for JunD. Samples were chilled on ice and RT-PCR was performed using a DNA Engine Opticon RT-qPCR machine (Biorad Laboratories Inc, Waltham, MA). After a reverse transcription step at 50°C for 30 min, an initial activation step at 95°C for 15 min, each cycle consisted of a denaturation step at 94°C for 15 s, an annealing step at 58°C for 30 s, and an elongation step at 72°C for 30 s. A total of 50 cycles were performed. The fluorescent signal was acquired at the end of each elongation step. Correct PCR products were confirmed by melting curve analysis and only one peak was detected for each product (data not shown). The relative quantification of gene expression was performed using the comparative CT method [23] [24] [25], with normalisation of the target gene to the endogenous housekeeping gene 18S. The level of mRNA of each gene in the dehydrated rats was then normalised to the result obtained in the control rats using an induction factor for each gene= 2−ΔΔCT, where ΔΔCT = [CT JunD(dehydrated rat) - CT 18S(dehydrated rat) -CT JunD(control) - CT 18S(control)] and assuming that the efficiency of the PCR reaction was close to one [26]. Data are expressed as mean ± S.E.M.
In situ hybridisation histochemistry
Euhydrated (n=5) and 72-hours of water deprived rats (n=5) were decapitated (between the hours of 11:00 am and 1:00 pm). Their brains were rapidly removed and snap frozen over liquid nitrogen. Coronal sections (14 μm) of the hypothalamus were cut on a cryostat (Leica Cryocut CM3050, Germany) and thaw mounted on poly-L-lysine coated microscope slides and stored at −80°C until processed. On the day of fixation, the slide-mounted brain sections were removed from storage and allowed to equilibrate to room temperature for 15–20 minutes. Slides were placed in a glass rack and submerged in 4% paraformaldehyde (PFA; pH 7.0) for 5 min and transferred to phosphate-buffered saline (PBS) at room temperature (for 5 min). To reduce non-specific hybridisation, brain slices were acetylated in 0.1 M triethanolamine/0.9% saline buffer (pH 8.0) containing 0.25% acetic anhydride for 10 min. Sections were then dehydrated through serial ethanol dilutions at room temperature (70, 80, 95 and 100% for 2 min each). Sections were then delipidated in chloroform (5 min), and allowed to dry. A 48-mer oligonucleotide probe was used to visualize expression of JunD in the rat SON. The probe was complementary to the rat JunD gene (sequence 5′-CTC GGT GTT CTG GCT TTT GAG GGT CTT GAC TTT CTC CTC CA-3′) and the complementary sequence of the antisense probe was used as the sense probe. Aliquots (3 ml) were 3′-end labelled with [35S]dATP (1.171 mCi total activity; Perkin Elmer Life Sciences, Boston, MA) using terminal deoxynucleotidyl transferase (Boehringer Mannheim, Mannheim, Germany) to a specific activity of 559177 CPM/ml. The labelled probes were then applied to a Sephadex G25 column and spun at 2000 r.p.m. for 1 min to separate any unincorporated nucleotides from the probes. The labelled antisense oligonucleotide probes were diluted in a hybridisation buffer containing 50% de-ionized formamide, 4 x saline sodium citrate (SSC=0.6 M NaCl, 0.06 M sodium citrate, pH 7.0), 500 mg/ml sheared DNA, 250 mg yeast tRNA, 1 x Denhardts (0.02% ficoll, 0.02% polyinylpyrrolidone and 0.02% BSA) and 10% dextran sulphate. The slides were then incubated overnight in a humidified atmosphere at 37°C. On the following day, slides were rinsed in 1 x SSC, washed in 1 x SSC at 55°C for 1 hour (4 × 15 min) followed by further washes in 1 x SSC (2 × 30 min) at room temperature, given 2 quick rinses in distilled water and dried with a warm stream of air. When dry, slides were apposed to Hyperfilm (Kodak, UK) with standard 14C microscales in autoradiographic cassettes for a period of 2 weeks.
Western blot analysis
Western blot analyses of SON protein extracts were performed to ensure that the antibody labels an appropriately sized band. Nuclear proteins from pooled SON tissue samples from 10 animals from each of the respective groups were extracted based on a protocol adapted from Weinberg and Penman [27]. The samples were incubated in ice-cold hypotonic buffer of 10 mM NaCl, 10 mM Tris-HCl pH 7.4, 1.5 mM MgCl2, 0.5% NP40, protease inhibitor cocktail (Sigma, UK) for 30 minutes and homogenised using a syringe with a 23-gauge needle. The nuclei were pelleted at 1000g for 2 minutes at 4°C in a mini-centrifuge (Biofuge Fresco, Heraeus Instruments, Hanau, Germany). In order to remove the outer nuclear membranes and attached material, the nuclei were washed in the above buffer containing 1 % NP40 and 0.05% Na-Deoxycholate. The nuclei were pelleted at 1000g for 2 minutes at 4°C and re-suspended and homogenised in a buffer of 20 mM HEPES pH 7.9, 0.4 M NaCl, 1 mM EDTA, 10% Glycerol, 1 mM DTT, protease inhibitor cocktail. The homogenates were incubated in ice and vortexed occasionally for 2 hours. The homogenates were centrifuged at 15000g for 5 min at 4°C and the supernatant containing the purified nuclear proteins was stored at −20°C until used for immunoblotting studies. 10 mg of each nuclear proteins sample were solubilised in sample buffer (17.5% glycerol, 8.7% (v/v) mercaptoethanol, 5% SDS, 217 mM Tris-HCl, Blu Bromophenol) at 100°C for 10 minutes and subjected to SDS-polyacrylamide gel electrophoresis. The gel was transferred to nitrocellulose (BioRad) using a mini trans-blot electrophoretic transfer cell (BioRad) and membrane was blocked in blotting buffer (150 mM NaCl, 20 mM Tris-HCl pH 7.4, and 0.1% Tween 20) containing 5% bovine serum albumin (BSA) for 1 hour prior to incubation with the JunD antisera (1:500 dilution) for 2 hours at room temperature in blotting buffer containing 5% BSA. The membrane was washed for 30 minutes in several changes (3 × 10 minutes) of the same blotting buffer prior to incubation in a horse anti-goat IgG horseradish peroxidase-linked secondary antiserum at a dilution 1:100,000 (Vector Laboratories Inc, Burlingame, CA) for 1 hour at room temperature. Membranes were then rinsed several times in blotting buffer (3 × 10 minutes) and immunoreactive proteins were revealed with ECLplus chemiluminescence reaction (Amersham Bioscience, Bucks, UK). The JunD signal was normalised with the signal of the histone H1 protein. Briefly, the blot was stripped in stripping buffer (Tris- HCl 62.5 mM, pH 6.7, SDS 2% (v/v), mercaptoethanol 0.7% (v/v)) at 50°C for 30 minutes and washed stringently in blotting buffer. The membrane was then processed in exactly the same manner as that described above for visualisation of Histone H1 protein using a monoclonal mouse anti-histone H1 primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:200 and a peroxidase-labeled anti-mouse IgG second antibody at a dilution of 1:8000 (Vector Laboratories Inc.).
Tissue preparation for immunohistochemistry
Rats (n=5 per group) were anaesthetised with sodium pentobarbitone (100 mg/kg i.p.) and transcardially perfused with 100 mL of 0.1 M PBS (pH 7.4) at room temperature followed by 300 mL of 4% PFA in 0.1 M PBS. The brains were removed, stored and cryoprotected in fixative containing 20% sucrose overnight at 4°C. The following morning, the brains were rapidly frozen over liquid nitrogen and four sets of coronal sections (40 μm) of the entire rostro-caudal axis of the forebrain were sectioned on a cryostat. The free-floating sections were collected in 24-well tissue culture plates containing PBS prior to being processed for immunohistochemical detection of JunD.
Immunohistochemical detection of JunD
For immunohistochemical detection of JunD, we used 2 different commercially available anti-JunD antisera (Santa Cruz Biotechnology, catalogue number: sc-72 and sc-74G lot numbers: L290 and I101 respectively) directed against different sequences on the C-terminus of the JunD protein. Immunohistochemistry was performed as described previously [28]. Free-floating rat hypothalamic sections were incubated for 15 minutes in a blocking solution comprising 10% normal horse serum (NHS, Sigma) and 0.3% Triton X-100 (Sigma) in 0.1 M PBS followed by rinses in PBS (3 × 10 minutes). Sections were then incubated in either goat anti-JunD or rabbit anti-JunD primary antiserum (1:1,000 dilution for both antisera) in PBS containing 1% NHS and 0.3% Triton X-100 for 48 hours at 4°C. After the primary antibody incubation, the sections were rinsed in PBS (3 × 10 minutes) prior to a 1 hour incubation in PBS containing AlexaFluor-594 conjugated donkey anti-goat IgG or AlexaFluor-594 conjugate donkey anti-rabbit (1:500 dilution, Molecular Probes, Invitrogen), 1% NHS and 0.3% Triton X-100 at room temperature. Following rinses in PBS (3 × 10 minutes) the sections were mounted onto glass microscope slides with 0.5% gelatin and allowed to air-dry for several minutes, and coverslipped with anti-fade fluorescent mountant (Vectorshield™ Hardset; Vector Laboratories Inc.).
JunD and c-Fos double fluorescence immunohistochemistry
In order to determine whether JunD and c-Fos are co-expressed in the same neurones within the SON, we used double labelling fluorescence immunohistochemistry. The c-Fos antiserum was purchased from a commercial supplier (Santa Cruz Biotechnology, SC-52, lot number: E2903;). Brain slices containing the SON were subjected to a pre-block in 10% NHS and Triton X-100 (0.3%) in PBS for 15 minutes. After rinsing (3 × 5 min), sections were incubated with the primary antisera (goat anti-JunD; 1:1000 and rabbit anti-c-Fos; 1:1000) in PBS containing 1% NHS and Triton X-100 (0.3%) for 48 hours at 4°C. Following repeated rinses in PBS (3 × 10 min) the sections were incubated in PBS containing AlexaFluor-594 conjugated donkey anti-goat IgG (1:500; Molecular Probes, Invitrogen) and AlexaFluor 488 horse anti-rabbit IgG (1:500; Molecular Probes, Invitrogen) for 2 hours at room temperature. Following further rinses in PBS (3 × 5 min) sections were mounted and coverslipped as described above. Double staining with a variety of other markers followed a similar protocol to that described above. For double staining with VP, a mouse monoclonal antibody (PS41) directed against the VP associated neurophysin (generous gift from Dr. H. Gainer, NIH) was used. An AlexaFluor-488 conjugated donkey anti-mouse secondary antibody was used. To determine whether the JunD was expressed in neurones we used a neuronal-specific marker (mouse anti-NeuN; Chemicon International, UK; catalogue number, MAB377). A mouse anti-OT antibody (Chemicon International; Catalogue number: MAB5296) was used to determine the extent of co-localisation of JunD with OT containing neurones. An AlexaFluor-488 conjugated horse anti-mouse secondary antibody was used as the fluorochrome for all double staining described above. The concentrations of antisera were chosen based on preliminary studies where serial dilutions from 1:500 to 1:10,000 were trialled to give the optimal staining with minimal background staining.
Immunohistochemical controls
In order to control for non-specific staining, sections were also processed in the presence of a blocking peptide (JunD; sc-74P; Santa Cruz Biotechnology). Sections were also processed in the absence of either the primary or secondary antisera.
Photomicroscopy and preparation of figures
The processed sections were examined on a Leica DM IRB microscope equipped with Leica C-Plan optics and the respective filters for the detection of AlexaFluor-488 and AlexaFluor-594. Photomicrographs were taken with a Leica DC-300F digital camera using IM50 software (version 1.2, Leica, Switzerland), saved as TIFF images at 300 dpi resolution and arranged in their respective sequences in CorelDRAW (version 8.0, Corel Corp., USA) for figure production. Confocal images were acquired using a Leica TCS-NT scanning laser confocal microscope housing a Leica DM IRBE inverted epifluorescence with a two line Kr/Ar laser. Images were captured using an oil-immersion 60X objective lens, saved as TIFF images and imported into CorelDRAW for figure production.
Quantification and Statistical analysis
Quantitative densitometric analysis of the images obtained from the in situ hybridisation study was performed using ImageJ (NIH, Bethesda, MD). The density of hybridisation signal was assessed from X-ray film images by comparing the optical density of the autoradiograms to standard microscales. The SON in each section was delineated on the autoradiograms by comparison with adjacent counterstained sections (Methyl Green) and a stereotaxic atlas of the rat brain by Paxinos and Watson [29]. Results were normalised against background by subtracting the value obtained from an area outlying the SON on the same section and expressed as a ratio of control (euhydrated) levels. Four sections per rat taken at regular intervals through the SON of each rat from the respective groups were analysed. Two sections from each rat were used to establish non-specific binding.
Quantitative analysis of the immunolabelling observed was made by counting the number of immunopositive neurones located within the SON. Each section containing the SON was visualised under sufficient magnification (20X objective) to visualise all immunopositive cells at two different focal planes and the number of immunopositive neurones manually counted. As the relative sizes of JunD-like and c-Fos like immunoreactive neurones remained constant throughout the rostro-caudal extent of the SON we were confident that any error in our counts would be much smaller than those arising from biological variation. A single operative performed all of the cell counting. All values are given as mean ± SEM. The data were statistically analysed with an unpaired student’s t-test using GraphPad Prism 3.0 (GraphPad Software Inc. CA). Statistical significance was assumed when P<0.05.
Results
JunD mRNA is increased in the SON following water deprivation as determined by quantitative RT- PCR and in situ hybridisation histochemistry
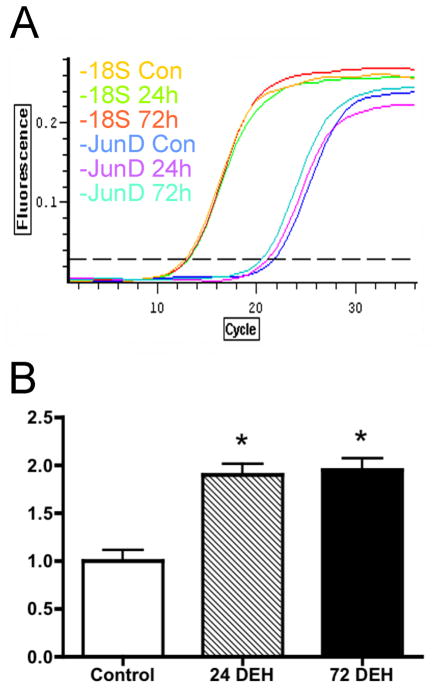
Figure 1 shows the fold change of expression of JunD mRNA induced in the SON by 24- and 72-hours of dehydration, as determined by quantitative RT-PCR. We observed a significant (one-way ANOVA, Bonferroni’s correction) increase in JunD gene expression (1.899±0.115 fold and a 1.948±0.122 fold change for CTR vs DH1 and CTR vs DH3 rats respectively). No significant difference in JunD mRNA expression was observed between rats that had undergone 24- and 72-hours of dehydration (P>0.05).
Figure 1.
JunD mRNA expression changes in the SON after 1 and 3 days of dehydration: (A) Fold change after dehydration of JunD mRNA levels were assessed relative to the internal control 18S rRNA using the Quantitect SYBR Green RT-PCR kit (Qiagen). The plots show three representative samples from one animal in each of the groups (control, 24-hours and 72-hours of water deprivation). Total mRNA was extracted from the SON of control, 24-hour and 72-hour water deprived rats (1 rat per total mRNA sample, n=4 for each condition, the PCR were run in triplicate). (B) The fold change of JunD mRNA expression was calculated as follows: Ratio (test/CTR) = 2−ΔΔCT, where ΔΔCT = [CT JunD(dehydrated rat) − CT 18S(dehydrated rat) − CT JunD(control) − CT 18S(control)]. Mean fold change ± S.E.M. *P<0.05 compared to control (Con), one-way ANOVA, bonferroni’s correction.
In situ hybridisation histochemistry revealed very little basal JunD mRNA in control rats (Fig 2A), while more intense signals were observed in the SON of 24- and 72-hour deprived groups (Fig 2B and 2C respectively). There was no significant difference in the hybridisation signals observed in the 24- and 72-hour water deprived groups (P>0.05); however, a significant difference between the control and 24-hour (P<0.05) and control and 72-hour water deprived groups (P<0.05) were observed. Interestingly, we also observed an increase in JunD mRNA expression within the PVN after 24- and 72-hours of water deprivation, particularly in the magnocelluar subdivision. Specific hybridisation signals were absent from sections hybridised with the sense probe (Fig 2D). These data are summarised in Fig 2E.
Figure 2.
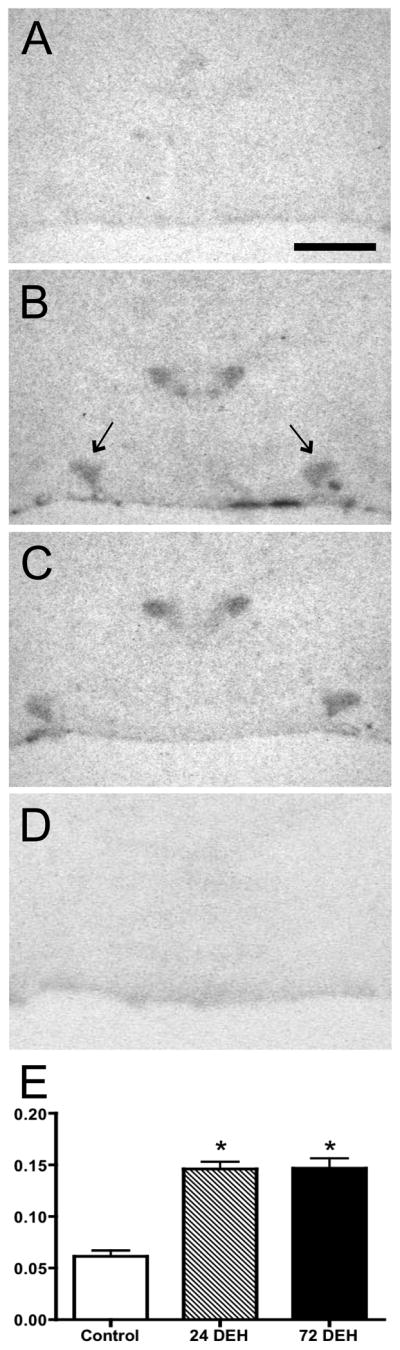
Effects of water deprivation on JunD gene expression determined by in situ hybridisation histochemistry. The signal detected in control animals (A) was extremely weak while animals that had undergone 24-hours (B) and 72-hours (C) of water deprivation showed greatly increased JunD mRNA expression. Water deprivation significantly (*P<0.001; One-way ANOVA, Bonferroni’s correction) increased JunD mRNA expression in water-deprived rats (D). Data are expressed as mean density ± S.E.M. Scale bar, 2 mm.
JunD protein is up-regulated in the SON of water deprived rats
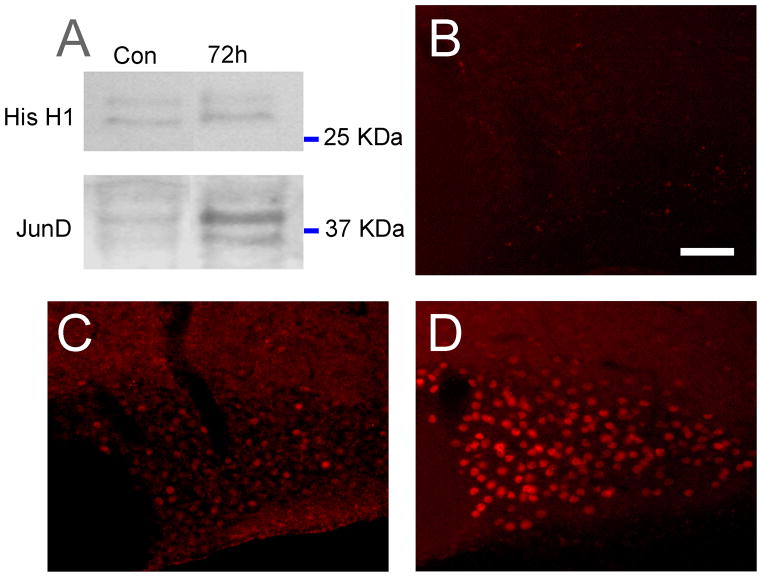
To further confirm that the anti-JunD antibody recognizes the correct protein and to assess changes in the abundance of JunD protein following water deprivation, we performed Western-blotting experiments using nuclear protein extracted from SON of control and 72-hour dehydrated rats. We detected two bands at the expected sizes of 34 and 39 kDa corresponding to both isoforms of JunD produced by alternative initiation of translation [30]. The equal loading of protein was assessed by the signal given by the nuclear protein Histone H1 in both conditions on the same blot. The normalised signal intensities of the two bands corresponding to JunD in dehydrated rats were much stronger than that of control rats (Fig 3A), suggesting an up-regulation of the protein expression in response to dehydration. To examine this further, we performed immunohistochemistry on hypothalamic sections containing the SON.
Figure 3.
An immunoblot of SON homogenates from control (CTR) and 3 day water-deprived rats (DH3). Each lane was loaded with 60 μg of protein extracted from the SON. Homogenates were probed with JunD (1:200 dilution; top) and Histone (His H1, 1:200 dilution; bottom) antisera. Immunoreactive bands were revealed with chemiluminescence ECL-plus (Amersham, USA). Two distinct bands at 34 and 39kDa were observed using the JunD antibody. A marked increase in the intensity of the bands were observed in 3 day water deprived rats (72h) compared to controls (A). Incubation of hypothalamic sections in the presence of a blocking peptide resulted in a distinct lack of staining within the SON of 72-hour water deprived rats (B). Very few JunD immunoreactive neurones were observed within the SON of control (euhydrated) rats (C); however, a striking increase in JunD immunostaining was observed following 72-hours of water deprivation (D). Scale bar, 100 μm.
Incubation of rat brain sections containing the SON with both JunD antisera revealed JunD-like immunoreactivity within areas of the brain previously reported to express JunD. Both JunD antisera gave the same pattern of expression, providing additional evidence specific staining. The pattern of JunD staining within the hypothalamus in euhydration has been reported previously [31] and is similar to the staining reported here. Omission of either the primary or secondary antiserum resulted in the absence of staining, as did incubation in the presence of the blocking peptide (Fig 3B). From these observations we are confident that the staining observed reliably represents JunD distribution within the rat SON.
In control (euhydrated) rats, JunD-like immunoreactivity was observed in the nuclei of neurones along the entire rostro-caudal extent of the SON. However, in euhydrated animals much of the staining observed was just detectable and only a small number of cells (< 20) exhibited intense staining (Fig. 3C). Following 24- and 72-hours of water deprivation a dramatic increase in JunD-like immunoreactivity was observed throughout the rostro-caudal extent of the SON (Fig. 3D). The staining observed consisted of round cell nuclei of uniform size (27.9±0.9 μm and 28.2±0.7 μm; n=100: 20 cells/rat from five 24- and 72-hour water deprived rats respectively). There was no appreciable difference in the size of JunD immunopositive SON cells of control versus 72-hour water deprived rats. Interestingly, a majority of the JunD-like immunopositive cells were located in the more ventral aspects of the SON corresponding roughly to the distribution of VP containing MCNs.
JunD immunostaining is predominantly expressed in neurones
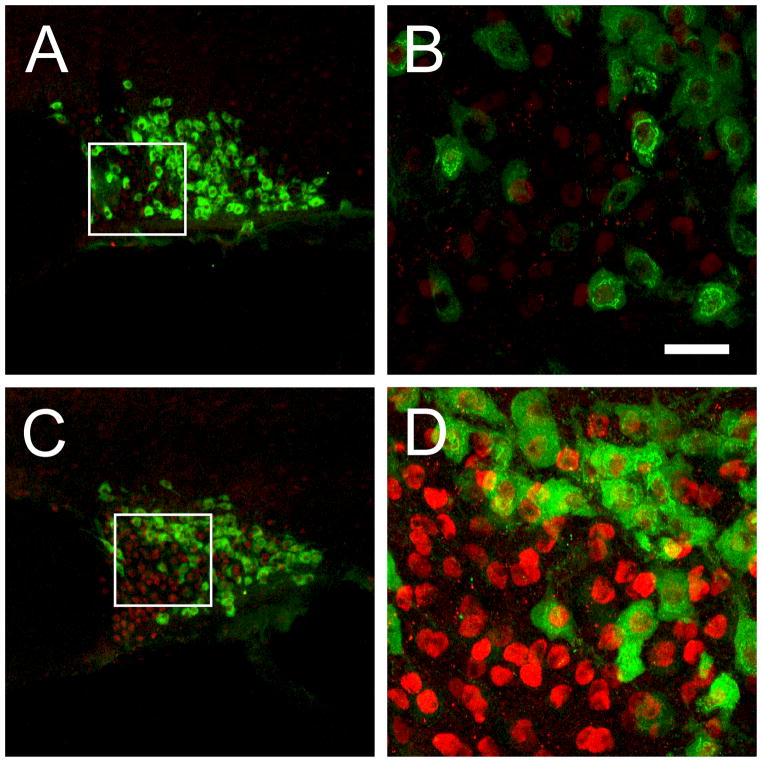
In order to elucidate whether the JunD staining was of neuronal or glia origin we performed double labelling immunohistochemistry with the neuronal marker NeuN – a vertebrate neurone-specific nuclear protein. We observed a high degree of co-localisation of JunD and NeuN positive structures (Fig 4; 94% JunD positive cells co-localised with NeuN positive cells), suggesting that a large majority of JunD expression occurs in neurones – as glia cell bodies within the SON are restricted primarily to the ventral glia limitans (an area lining the ventral borders of the SON) and send long processes dorsally amongst the MCNs. Hence, the noticeable lack of staining within this particular region of the SON immediately dorsal of the subarachnoid space suggests that JunD is primarily, if not exclusively, expressed in neurones.
Figure 4.
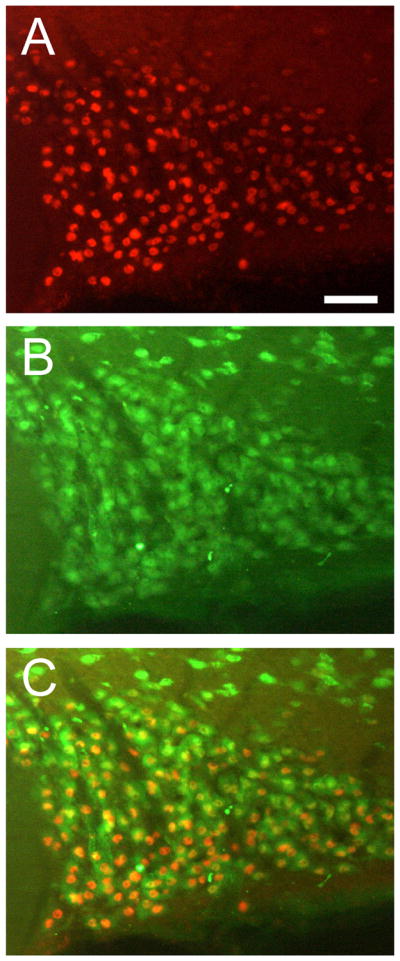
Representative photomicrographs of JunD and NeuN fluorescence immunohistochemistry in 72-hour water deprived rats. (A) and (B) are low-power photomicrographs of JunD and NeuN immunohistochemistry in a 72-hour water deprived rat respectively. In the merged picture (C) many JunD postive cells are also NeuN postitive. Scale bar, 100 μm.
JunD is preferentially expressed in VP containing MCNs
In an effort to elucidate the neurochemical phenotype of the cells that express JunD-like immunoreactivity in the SON, we performed double labelling immunohistochemistry using antibodies specific for the VP associated neurophysin (neurophysin-II) and OT. While there was little expression of JunD in control euhydrated animals, cell counts of VP-neurophysin immunopositive neurones that were also positive for JunD, in water deprived rats, revealed a significant (ANOVA, P<0.05, n=5/group) increase in the degree of co-localisation (Fig 5; 68% and 77% of VP-neurophysin positive cells co-localised with JunD after 24- and 72-hours of water deprivation respectively). Double staining with OT, however, revealed that only a small proportion of OT containing neurones increase their expression of JunD protein following 72-hours of water deprivation (Fig 6; 27% of OT positive cells co-localised with JunD).
Figure 5.
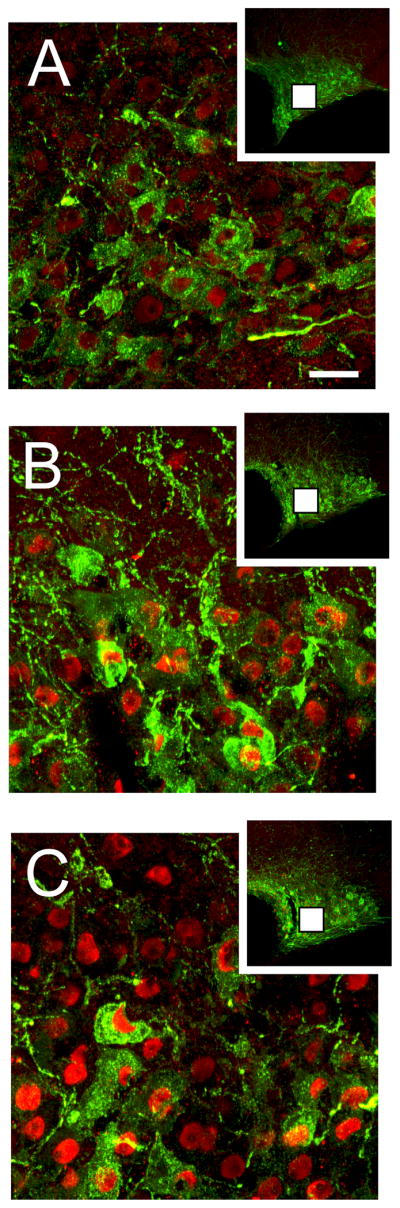
Representative confocal images of JunD and VP double labelling within the SON of control, 24-hour and 72-hour water deprived rats. Only few VP containing neurones expressed intense JunD immunoreactivity in control animals (A); however, after 24-hour (B) and 72-hours (C) of water deprivation a marked increase in JunD immunoreacitivity in VP containing neurones were observed. Inserts at the top of each panel show a photomicrograph taken at low power to show the extent of JunD labelling within the entire SON. The white boxes in the inserts show the region where the high powered photomicrographs were taken. Scale bar, 30 μm.
Figure 6.
Representative confocal images of JunD and OT double labelling immunohistochemistry within the SON of control and 72-hour water deprived rats. Low levels of JunD expression were observed in the SON of control rats (A). Panel B is an enlarged picture of the while square represented in panel A. Seventy-two hours of water deprivation induced an increase in JunD expression in OT containing neurones (C), however, there was a smaller percentage of OT containing neurones that expressed junD compared to VP and JunD double-labelled cells. Panel D is an enlarged picture of the white square represented in panel C. Scale bar, 30 μm.
JunD is co-localised with c-Fos in the SON of water-deprived rats
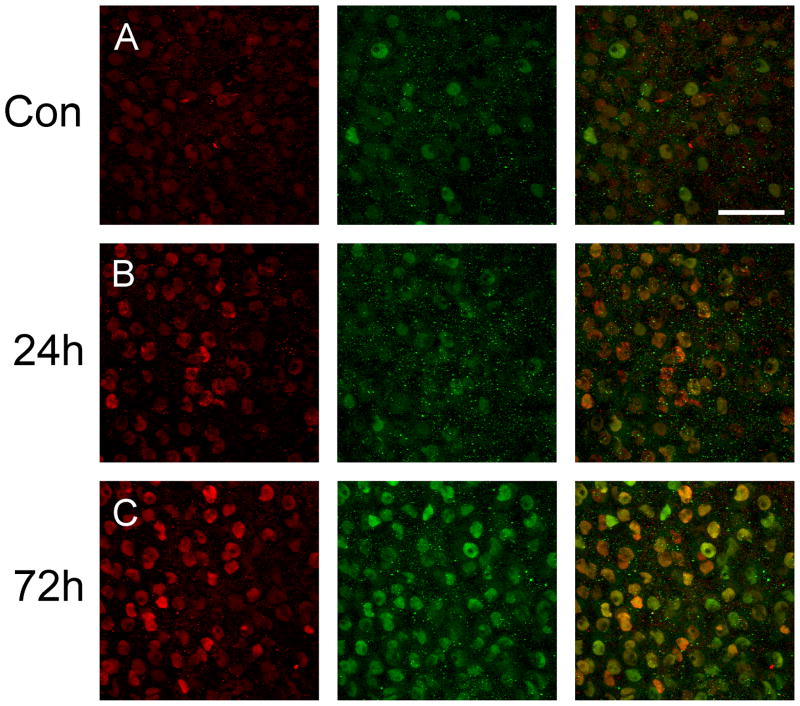
Since JunD has been shown to form heterodimers with other members of the AP-1 transcription factor family, in particular c-Fos [32], we decided to perform some JunD and c-Fos double-labelling in an effort to determine whether this might also be the case in the SON following an osmotic challenge. In control animals, basal expression of both c-Fos and JunD was sparsely scattered within the SON (Fig 7A). Twenty-four (Fig 7B) and 72- hours (Fig 7C) of water deprivation resulted in a large increase in the number of JunD immunoreactive neurones within the SON accompanied by similar increases in c-Fos immunoreactivity. In addition, we determined that approximately 76% and 87% of JunD stained cells were also c-Fos immunoreactive after 24-hours and 72-hours of water deprivation respectively.
Figure 7.
Representative confocal images of JunD and c-Fos double labelling within the SON of control and water deprived rats. While only low expression of JunD (red) and c-Fos (green) in the control animals (A), both JunD and c-Fos was markedly up-regulated after 24-hours (B) and 72-hours (C) of water deprivation. Following 72-hours of water deprivation a high percentage of co-localisation between JunD and c-Fos was observed. Scale bar, 100 μm.
Discussion
That the physiological activation of MCNs by dehydration is accompanied by a dramatic activity-dependent remodelling of the SON is well documented [12] [14] [33]. Less well understood are the molecular mechanics of these processes. As we have hypothesised that alterations in steady-state mRNA levels might be responsible, at least in part, for mediating and regulating SON functional plasticity [13] [20] [21], we have previously used microarrays to identify transcripts that are significantly altered in abundance as a consequence of 3 days of dehydration; 2543 RNA species were thus identified [22]. As these alterations in gene expression might be mediated by transcription factors, and as one of the ways in which transcription factor activity can be altered is by changing the steady-state level of its mRNA template, we analysed our microarray data to identify changes in the expression of transcription factors in response to water deprivation [22]. We identified significant changes in the abundance of transcripts encoding 42 transcription factors, 11 of which are down-regulated, whilst 31 are up-regulated [22].
A number of the putatively regulated transcription factor mRNAs identified have been shown in the past to be modulated in abundance in the SON by an osmotic challenge. Thus, while AP-1 transcription factors such as c-Fos, c-Jun and FosB have been previously studied [34] [35] [36], the observation that JunD is also regulated in the SON by dehydration is novel, and here we confirm the array results at the transcript and protein levels: quantitative RT-PCR and in situ hybridisation revealed a significant increase in JunD mRNA levels following 24-hours and 72-hours of water deprivation. Moreover, an increase in transcript levels was also observed in the PVN, a key region involved in the regulation of fluid homeostasis. Western blotting and immunocytochemistry showed that JunD protein expression is up-regulated in VP neurones following dehydration.
Originally shown to be highly abundant in numerous organs of mice, including the intestine, spleen, and lungs [37], high levels of JunD mRNA and protein have also been reported in the central nervous system (CNS) [31] [32]. JunD expression within the CNS has previously been examined in euhydrated Sprague-Dawley rats and, compared to other AP-1 transcription factors such as, c-Jun, JunB, c-Fos and FosB, was shown to be the most highly expressed in neurones [31]. In a basal state, intense immunoreactivity was reported in a number of hypothalamic structures including the paraventricular nucleus, dorsomedial nucleus and SON. In contrast, we observed very little basal expression of JunD in the SON at both the level of the mRNA and protein in control euhydrated animals. The reason for the discrepancy between our findings and those of Herdegen et al., [31] cannot be explained by differences in animal strains (as the same animal strain and anaesthetic was used in both studies). We suggest that differences in the primary antisera used may account for these discrepancies. Whereas Herdegen and colleagues [31] produced their own antisera we obtained ours from commercial source, which we validated using a number of controls and found to be highly specific for JunD. Additionally, the fact that we used two different antisera raised in two different species and obtained the same pattern of staining would suggest that the staining we observed represents genuine JunD staining.
Co-localisation studies demonstrate that JunD is primarily expressed in NeuN positive neurones. That JunD is not expressed in glia within the SON is perhaps unexpected, as several studies have reported constitutive expression of JunD in a majority of neuronal and glia nuclei in almost all CNS regions [31]; reviewed in Herdegen and Leah [32]. Within the SON almost all glia cell bodies are located within the ventral glia limitans [38] [39] and changes in glia coverage and remodelling are thought to facilitate the release of peptide hormones following an osmotic challenge [33] [39] [40] [41]. While expression of JunD has been reported in glia grown in culture [42], there is no evidence, thus far, to suggest expression of JunD in the ventral glia limitans subjacent to the SON.
JunD immunoreactivity is observed primarily in VP rather than in OT MCNs (Fig 6). We also found JunD to be preferentially up-regulated in VP containing MCNs. Whilst the number of JunD expressing OT containing MCNs slightly increased following water deprivation, this was not statistically significant. These observations beg two questions; firstly, what are the mechanisms mediating JunD expression in VP cells, and, secondly, what is the function of JunD in these neurones? A majority of the other AP-1 proteins (i.e. c-Jun, JunB, c-Fos, FosB) have been demonstrated in VP and OT expressing neurones [34] [36] [43] [44]. The observation that JunD is preferentially expressed in VP containing neurones suggest to us that JunD dimers might preferentially regulate gene transcription in VP cells, whilst non-JunD dimers mediate the responses of OT cells; hence, the molecular responses of the VP and OT cells may be different although the synaptic inputs to both cell types are thought to be similar [45] [46]. Whether this is indeed the case requires further investigation.
With regard to the regulation of JunD, it has been shown that administration of angiotensin (AngII; 100 ng) directly into the lateral ventricles does not alter basal JunD expression [47], suggesting that the increase expression of JunD expression following dehydration reported here is not due to stimulation of AngII receptors in the SON arising from efferent inputs from the subfornical organ [48]. As such, the increase in JunD might be mediated by some other efferent input or, alternatively, could be an intrinsic response of the osmosensitive MCN. For example, using an in vitro hypothalamic slice model, Meeker and Fernandes [49] showed that MCNs exposed to hypertonic culture medium increased the expression of p-CREB, a response that was unaffected in the presence of TTX, a blocker of fast sodium channels, used here to prevent synaptic transmission.
So, what is the function of JunD within VP MCNs? JunD is a member of the AP-1 family of leucine zipper transcription factors. AP-1 is a transcription factor complex formed by either homodimerisation of two Jun proteins (c-Jun, JunB, and JunD) or heterodimerisation of a Jun protein with a Fos protein (either c-Fos, FosB, Fra-1, and Fra-2) through the leucine zipper motif [50]. Upon phosphorylation, these dimers bind to AP-1 consensus sequences (5′-TGAG/CTCA-3′) to regulate the transcription of target genes [32]. We note that JunD is highly co-expressed with c-Fos in MCNs of the SON following dehydration; a figure which is comparable to that reported for c-Fos and c-Jun in rats that were osmotically stimulated using a different experimental paradigm [51]. Thus, we hypothesise that c-Fos and JunD can form heterodimers that alter the transcriptional environment of the VP MCN, altering the overall pattern of mRNA synthesis. However, a limitation of our data is not being able to predict the downstream gene changes resulting from JunD and, indeed, those genes upstream of JunD that may cause its overexpression during dehydration. Interestingly, a functional AP1 element has been identified in the rat VP gene promotor [52], which strongly suggests that the immediate-early gene products, Fos/Jun family proteins (including JunD) act as potent inducers of VP gene transcription.
To summarise, microarray analysis has revealed JunD to be a candidate transcription factor expressed in the SON and up-regulated following chronic dehydration. This hypothesis has been substantiated using independent methods that revealed that JunD mRNA and protein expression is indeed increased within the SON following water deprivation and is positively correlated with neuronal activity. Furthermore, JunD expression was observed primarily in VP and c-Fos containing cells. Hence, the expression of JunD, in conjuction with other AP-1 family members including c-Fos, may contribute to the cell-specific remodelling of the VP neurone as a consequence of an osmotic challenge.
Acknowledgments
The authors would like to thank Marie Carlucci for excellent technical assistance. This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) of the United Kingdom, grant number: 7/518346, and The National Institutes of Health, grant numbers: R-37 HL33610, RO1 HL62579 and RO1 DK57822.
References
- 1.Burbach JP, Luckman SM, Murphy D, Gainer H. Gene regulation in the magnocellular hypothalamo-neurohypophysial system. Physiol Rev. 2001;81:1197–1267. doi: 10.1152/physrev.2001.81.3.1197. [DOI] [PubMed] [Google Scholar]
- 2.Russell JA, Leng G. Sex, parturition and motherhood without oxytocin? J Endocrinol. 1998;157:343–359. doi: 10.1677/joe.0.1570343. [DOI] [PubMed] [Google Scholar]
- 3.Lipschitz DL, Crowley WR, Armstrong WE, Bealer SL. Neurochemical basis of plasticity in the magnocellular oxytocin system during gestation. Exp Neurol. 2005;196:210–223. doi: 10.1016/j.expneurol.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Antunes-Rodrigues J, de Castro M, Elias LL, Valencia MM, McCann SM. Neuroendocrine control of body fluid metabolism. Physiol Rev. 2004;84:169–208. doi: 10.1152/physrev.00017.2003. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong WE. Morphological and electrophysiological classification of hypothalamic supraoptic neurons. Prog Neurobiol. 1995;47:291–339. [PubMed] [Google Scholar]
- 6.Glasgow E, Kusano K, Chin H, Mezey E, Young WS, Gainer H. Single cell reverse transcription-polymerase chain reaction analysis of rat supraoptic magnocellular neurons: neuropeptide phenotypes and high voltage-gated calcium channel subtypes. Endocrinology. 1999;140:5391–5401. doi: 10.1210/endo.140.11.7136. [DOI] [PubMed] [Google Scholar]
- 7.Xi D, Kusano K, Gainer H. Quantitative analysis of oxytocin and vasopressin messenger ribonucleic acids in single magnocellular neurons isolated from supraoptic nucleus of rat hypothalamus. Endocrinology. 1999;140:4677–4682. doi: 10.1210/endo.140.10.7054. [DOI] [PubMed] [Google Scholar]
- 8.Kiyama H, Emson PD. Evidence for the co-expression of oxytocin and vasopressin messenger ribonucleic acids in magnocellular neurosecretory cells: simultaneous demonstration of two neurophysin messenger ribonucleic acids by hybridisation histochemistry. J Neuroendocrinol. 1990;2:257–259. doi: 10.1111/j.1365-2826.1990.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 9.Mezey E, Kiss JZ. Co-expression of vasopressin and oxytocin in hypothalamic supraoptic neurons of lactating rats. Endocrinology. 1991;129:1814–1820. doi: 10.1210/endo-129-4-1814. [DOI] [PubMed] [Google Scholar]
- 10.Telleria-Diaz A, Grinevich VV, Jirikowski GF. Colocalization of vasopressin and oxytocin in hypothalamic magnocellular neurons in water-deprived rats. Neuropeptides. 2001;35:162–167. doi: 10.1054/npep.2001.0859. [DOI] [PubMed] [Google Scholar]
- 11.Glasgow E, Kusano K, Chin H, Mezey E, Young WS, 3rd, Gainer H. Single cell reverse transcription-polymerase chain reaction analysis of rat supraoptic magnocellular neurons: neuropeptide phenotypes and high voltage-gated channel subtypes. Endocrinology. 1999;140:5391–5401. doi: 10.1210/endo.140.11.7136. [DOI] [PubMed] [Google Scholar]
- 12.Hatton GI. Function-related plasticity in hypothalamus. Ann Rev Neurosci. 1997;20:375–397. doi: 10.1146/annurev.neuro.20.1.375. [DOI] [PubMed] [Google Scholar]
- 13.Qiu J, Hindmarch CC, Yao ST, Tasker JG, Murphy D. Transcriptomic analysis of the osmotic and reproductive remodeling of the female rat supraoptic nucleus. Endocrinology. 2011;152:3483–3491. doi: 10.1210/en.2011-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theodosis DT, El Majdoubi M, Pierre K, Poulain DA. Factors governing activity-dependent structural plasticity of the hypothalamo-neurohypophysial system. Cell Mol Neurobiol. 1998;18:285–298. doi: 10.1023/A:1022577105819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka M, Cummins TR, Ishikawa K, Black JA, Ibata Y, Waxman SG. Molecular and functional remodeling of electrogenic membrane of hypothalamic neurons in response to changes in their input. Proc Natl Acad Sci (USA) 1999;96:1088–1093. doi: 10.1073/pnas.96.3.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang QZ, Hatton GI. Nitric oxide via cGMP-dependent mechanisms increases dye coupling and excitability of rat supraoptic nucleus neurons. J Neurosci. 1999;19:4270–4279. doi: 10.1523/JNEUROSCI.19-11-04270.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyata S, Takamatsu H, Maekawa S, Matsumoto N, Watanabe K, Kiyohara T. doi: 10.1002/cne.1184. [DOI] [PubMed] [Google Scholar]
- 18.Miyata S, Hatton GI. Activity-related, dynamic neuron-glial interactions in the hypothalamo-neurohypophysial system. Microsc Res Tech. 2002;56:143–157. doi: 10.1002/jemt.10012. [DOI] [PubMed] [Google Scholar]
- 19.Tasker JG, Di S, Boudaba C. Functional synaptic plasticity in hypothalamic magnocellular neurons. Prog Brain Res. 2002;139:113–119. doi: 10.1016/s0079-6123(02)39011-3. [DOI] [PubMed] [Google Scholar]
- 20.Sharman G, Ghorbel M, Leroux M, Beaucourt S, Wong LF, Murphy D. Deciphering the homeostatic plasticity within the hypothalamo-neurohypophyseal system – genomic and gene transfer studies. Prog Biophys Mol Biol. 2003;84:151–182. doi: 10.1016/j.pbiomolbio.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Hindmarch C, Yao S, Beighton G, Paton J, Murphy D. A comprehensive description of the transcriptome of the hypothalamoneurohypophyseal system in euhydrated and dehydrated rats. Proc Natl Acad Sci (USA) 2006;103:1609–1614. doi: 10.1073/pnas.0507450103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu J, Yao S, Hindmarch C, Antunes V, Paton J, Murphy D. Transcription factor expression in the hypothalamo-neurohypophyseal system of the dehydrated rat; up-regulation of Gonadotrophin inducible transcription factor 1 mRNA is mediated by cAMP-dependent protein kinase A. J Neurosci. 2007;27:2196–2203. doi: 10.1523/JNEUROSCI.5420-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fink L, Seeger W, Ermert L, Hanze J, Stahl U, Grimminger F, Kummer W, Bohle RM. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–1333. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- 24.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Ann Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Ann Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 26.Favy DA, Lafarge S, Rio P, Vissac C, Bignon YJ, Bernard-Gallon D. Real-time PCR quantification of full-length and exon 11 spliced BRCA1 transcripts in human breast cancer cell lines. Biochem Biophys Res Commun. 2000;274:73–78. doi: 10.1006/bbrc.2000.3100. [DOI] [PubMed] [Google Scholar]
- 27.Weinberg RA, Penman S. Small molecular weight monodisperse nuclear RNA. J Mol Biol. 1968;38:289–304. doi: 10.1016/0022-2836(68)90387-2. [DOI] [PubMed] [Google Scholar]
- 28.Yao ST, Gouraud S, Paton JFR, Murphy D. Water deprivation increases the expression of neuronal nitric oxide synthase (nNOS) but not Orexin-A in the lateral hypothalamic area of the rat. J Comp Neurol. 2005;490:180–193. doi: 10.1002/cne.20662. [DOI] [PubMed] [Google Scholar]
- 29.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Elsevier Academic Press; San Diego, USA: 2005. [Google Scholar]
- 30.Short JD, Pfarr CM. Translational regulation of the JunD messenger RNA. J Biol Chem. 2002;277:32697–32705. doi: 10.1074/jbc.M204553200. [DOI] [PubMed] [Google Scholar]
- 31.Herdegen T, Kovary K, Buhl A, Bravo R, Zimmermann M, Gass P. Basal expression of the inducible transcription factors c-Jun, JunB, JunD, c-Fos, FosB and Krox-24 in the adult rat brain. J Comp Neurol. 1995;354:39–56. doi: 10.1002/cne.903540105. [DOI] [PubMed] [Google Scholar]
- 32.Herdegen T, Leah JD. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res Revs. 1998;28:370–490. doi: 10.1016/s0165-0173(98)00018-6. [DOI] [PubMed] [Google Scholar]
- 33.Oliet SHR, Piet R. Anatomical remodelling of the supraoptic nucleus: changes in synaptic and extrasynaptic transmission. J Neuroendocrinol. 2004;16:303–307. doi: 10.1111/j.0953-8194.2004.01159.x. [DOI] [PubMed] [Google Scholar]
- 34.Ji LL, Fleming T, Penny ML, Toney GM, Cunningham JT. Effects of water deprivation and rehydration on c-Fos and FosB staining in the rat supraoptic nucleus and lamina terminalis region. Am J Physiol. 2005;288:R311–R321. doi: 10.1152/ajpregu.00399.2004. [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki M, Yamaguchi K, Saito J, Ozaki Y, Mera T, Hashimoto H, Fujihara H, Okimoto N, Ohnishi H, Nakamura T, Ueta Y. Expression of immediate early genes and vasopressin heteronuclear RNA in the paraventricular and supraoptic nuclei of rats after acute osmotic stimulus. J Neuroendocrinol. 2005;17:227–237. doi: 10.1111/j.1365-2826.2005.01297.x. [DOI] [PubMed] [Google Scholar]
- 36.Penny ML, Bruno SB, Cornelius J, Higgs KAN, Cunningham JT. The effect of osmotic stimulation and water availability on c-Fos and FosB staining in the supraoptic and paraventricular nuclei of the hypothalamus. Exp Neurol. 2005;194:191–202. doi: 10.1016/j.expneurol.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Hirai SI, Ryseck RP, Mechta F, Bravo R, Yaniv M. Characterization of junD: a new member of the jun proto-oncogene family. EMBO J. 1989;8:1433–1439. doi: 10.1002/j.1460-2075.1989.tb03525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawrylak N, Fleming JC, Salm AK. Dehydration and rehydration selectively and reversibly alter glial fibrillary acidic protein immunoreactivity in the rat supraoptic nucleus and subjacent glial limitans. Glia. 1991;22:260–271. doi: 10.1002/(sici)1098-1136(199803)22:3<260::aid-glia5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 39.Salm AK, Bobak JB. Dehydration-associated changes in the ventral glia limitans subjacent to the supraoptic nucleus include a reduction in the extent of the basal lamina but not astrocytic process shrinkage. Exp Neurol. 1999;160:425–432. doi: 10.1006/exnr.1999.7211. [DOI] [PubMed] [Google Scholar]
- 40.Bobak JB, Salm AK. Plasticity of astrocytes of the ventral glial limitans subjacent to the supraoptic nucleus. J Comp Neurol. 1996;376:188–197. doi: 10.1002/(SICI)1096-9861(19961209)376:2<188::AID-CNE2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 41.Theodosis DT, Piet R, Poulain DA, Oliet SH. Neuronal, glial and synaptic remodeling in the adult hypothalamus: functional consequences and role of cell surface and extracellular matrix adhesion molecules. Neurochem Int. 2004;45:491–501. doi: 10.1016/j.neuint.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Pennypacker KR, Hong JS, Mullis SB, Hudson PM, McMillian MK. Transcription factors in primary glial cultures: changes with neuronal interactions. Brain Res Mol Brain Res. 1996;37:224–230. doi: 10.1016/0169-328x(95)00318-m. [DOI] [PubMed] [Google Scholar]
- 43.Wang K, Guldenaar SEF, McCabe JT. Fos and Jun expression in rat supraoptic nucleus neurons after acute vs. repeated osmotic stimulation. Brain Res. 1997;746:117–125. doi: 10.1016/s0006-8993(96)01216-4. [DOI] [PubMed] [Google Scholar]
- 44.Moellenhoff E, Blume A, Culman J, Chatterjee B, Herdegen T, Lebrun CJ, Unger T. Effect of repetitive i.c.v. injections of ANG II on c-Fos and AT(1)-receptor expression in the rat brain. Am J Physiol. 2001;280:R1095–R1104. doi: 10.1152/ajpregu.2001.280.4.R1095. [DOI] [PubMed] [Google Scholar]
- 45.Leng G, Brown CH, Russell JA. Physiological pathways regulating the activity of magnocellular neurosecretory cells. Prog Neurobiol. 1999;57:625–655. doi: 10.1016/s0301-0082(98)00072-0. [DOI] [PubMed] [Google Scholar]
- 46.El Majdoubi M, Poulain DA, Theodosis DT. Activity-dependent morphological synaptic plasticity in an adult neurosecretory system: magnocellular oxytocin neurons of the hypothalamus. Biochem Cell Biol. 2000;78:317–327. [PubMed] [Google Scholar]
- 47.Lebrun CJ, Blume A, Herdegen T, Seifert K, Bravo R, Unger T. Angiotensin II induces a complex activation of transcription factors in the rat brain: expression of Fos, Jun and Krox proteins. Neuroscience. 1995;65:93–99. doi: 10.1016/0306-4522(94)00482-k. [DOI] [PubMed] [Google Scholar]
- 48.Shelat SG, Fluharty SJ, Flanagan-Cato LM. Adrenal steroid regulation of central angiotensin II receptor subtypes and oxytocin receptors in rat brain. Brain Res. 1998;807:135–146. doi: 10.1016/s0006-8993(98)00794-x. [DOI] [PubMed] [Google Scholar]
- 49.Meeker RB, Fernandes A. Sustained increases in activating transcription factor-2 and activator protein-2 in the rat supraoptic nucleus during water deprivation. Neuroendocrinology. 2002;76:111–120. doi: 10.1159/000064425. [DOI] [PubMed] [Google Scholar]
- 50.Curran T, Franza BR., Jr Fos and Jun: the AP-1 connection. Cell. 1988;55:395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- 51.Guldenaar SE, Wang K, McCabe JT. Double immunofluorescence staining of Fos and Jun in the hypothalamus of the rat. Cell Tissue Res. 1994;276:1–6. doi: 10.1007/BF00354777. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida M, Iwasaki Y, Asai M. Identification of a functional AP1 element in the rat vasopressin gene promotor. Endcrinology. 147:2850–2863. doi: 10.1210/en.2005-1222. [DOI] [PubMed] [Google Scholar]