Abstract
Silent mating type information regulation 1 (Sirtuin 1; SIRT1) has been reported to regulate various physiological events, such as aging and metabolism, via deacetylation of histone and nonhistone proteins. Notably, cumulative evidence supports the notion that SIRT1 has a Janus-faced role in tumorigenesis. SIRT1 contributes to anti-inflammation, genomic stability, and cancer cell death, and hence it has tumor-suppressor properties. On the other hand, SIRT1 can stimulate oncogenic signaling pathways and can create a tumor microenvironment favorable to growth and survival of cancer cells. Such dual functions of SIRT1 may be determined, at least in part, by its subcellular localization. Interestingly, SIRT1 displays differential localization in normal cells and cancer cells, which in turn may affect the substrate specificity for its deacetylase activity.
Keywords: SIRT1, cancer, subcellular localization
Introduction
Sirtuins are the mammalian orthologues of yeast silent information regulator 2 (SIR2) that have been found to extend yeast life span.1 The sirtuin protein family consists of seven isoforms, SIRT1 to SIRT7, which have specific subcellular localizations and activities.2 SIRT1, the most well-characterized member of the sirtuin family, has been known as a longevity protein in the mammals, particularly related to life-span extension induced by caloric restriction.3,4 Resveratrol, a phytoalexin found in grapes, mimics the effect of caloric restriction, which has been speculated to be mediated through activation of SIRT1.5–7 A recent study has revealed that SIRT1 is involved in the beneficial effects of resveratrol on mitochondrial function.8 However, it is still unclear whether resveratrol activates SIRT1 directly or indirectly.
SIRT1 belongs to the family of NAD+-dependent class III histone deacetylases.9,10 The deacetylation targets of SIRT1 are not limited to histones but are expanded to diverse proteins, including the tumor suppressor p53.11 Lysine acetylation and deacetylation have been recognized as crucial events for regulation of activity, stability, and subcellular localization of proteins.12 Thus, SIRT1 can modulate various cellular signaling pathways through alteration of the acetylation status of target proteins.
Deacetylation activity of SIRT1 can be modulated by multiple regulators. Two putative regulators are active regulator of SIRT1 (AROS) and deleted in breast cancer 1 (DBC1), which are positive and negative regulators of SIRT1, respectively. While the nuclear protein AROS directly binds to SIRT1, leading to enhanced deacetylase activity of the latter protein,13 DBC1 interacts with the catalytic domain of SIRT1 and negatively regulates SIRT1-dependent deacetylation.14 However, the exact roles of AROS and DBC1 as SIRT1 modulators need to be confirmed.
Furthermore, posttranslational modifications of SIRT1 affect its deacetylase activity. Sumoylation of SIRT1 at residue Lys 734 facilitates its catalytic activity, which is diminished by SENP1 desumoylase.15 Phosphorylation is also important for SIRT1 activity. Cyclin-dependent kinase 1 and c-Jun N-terminal kinase 1 have been reported to phosphorylate SIRT1, positively modulating its activity.16,17 These complex factors coordinately regulate SIRT1 activity and subsequent cellular events.
Functioning as a protein deacetylase, SIRT1 has a broad spectrum of substrates. The tumor suppressor p53 is one of the best-defined target proteins of the SIRT1 deacetylase.11 SIRT1 deacetylates and inactivates p53, thereby exerting an antiapoptotic effect.18 Moreover, SIRT1 can be involved in the DNA repair process through deacetylation of Ku70.19 SIRT1 also deacetylates and activates liver X receptor proteins, facilitating cholesterol efflux from the cell.20 Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) can modulate metabolic pathways by directly interacting with several transcription factors, such as peroxisome proliferator-activated receptor γ. SIRT1-dependent deacetylation of PGC-1α enhances its ability to cooperate with transcription factors, which, in turn, induces expression of genes involved in fatty acid oxidation and gluconeogenesis.21,22
Although the majority of investigations concern anticarcinogenic as well as antiaging effects of SIRT1, recent studies have revealed that the protein is implicated in carcinogenesis (Fig. 1). However, the exact role of SIRT1 in carcinogenesis is still controversial. This review highlights the Janus-faced role of SIRT1 in multistage carcinogenesis.
Figure 1.

The Janus face of SIRT1 in tumorigenesis. SIRT1 can inhibit inflammation and multistage carcinogenesis, acting as a tumor suppressor. Paradoxically, SIRT1 also accelerates tumorigenesis via multiple mechanisms, such as inactivation of tumor suppressors, activation of oncoproteins, and development of a microenvironment favorable for tumor survival.
SIRT1 as a putative tumor suppressor
SIRT1 inhibits aberrantly amplified proinflammatory signaling during promotion and progression of carcinogenesis
It is well known that chronic inflammation is associated with carcinogenesis, especially in the promotion and progression stages.23 SIRT1 can inhibit inflammation provoked by several proinflammatory cytokines, such as tumor necrosis factor α (TNF-α), lipopolysaccharide (LPS), and interleukins (ILs). The natural SIRT1 activator resveratrol has been shown to attenuate TNF-α–induced inflammation in mouse embryonic fibroblasts (MEFs) through upregulation of SIRT1.24 Resveratrol also ameliorates inflammation provoked during acute and restorative phases in murine liver tissues, while it failed to overcome LPS challenge in the liver of SIRT1 knockout mice.25 Moreover, SIRT1-knockdown macrophages exhibited increased inflammatory responses.26 Hepatic steatosis and inflammation were evident in SIRT1 knockout mice, lending further support to the anti-inflammatory role of SIRT1.27,28 SIRT1 expression was also found to be reduced in the lungs of patients with chronic obstructive pulmonary disease that is closely related to chronic inflammation.29
The anti-inflammatory effect of SIRT1 might be achieved by inhibition of several transcription factors related to inflammation. Nuclear factor κB (NF-κB) is a key transcription factor responsible for regulation of immune responses. SIRT1 directly interacts with the RelA/p65 subunit of NF-κB, leading to deacetylation and subsequent inactivation of NF-κB.30 However, siRNA knockdown of SIRT1 augmented acetylation of RelA/p65, as well as release of IL-8, in a monocyte–macrophage cell line.29 In the dextran sodium sulfate-induced murine colitis model, resveratrol upregulated SIRT1 expression and abrogated NF-κB activation, thus attenuating intestinal inflammation.31 In addition to NF-κB, SIRT1 inhibits other transcription factors, including those that orchestrate proinflammatory responses. Signal transducer and activator of transcription 3 (STAT3) is phosphorylated and activated in response to various proinflammatory cytokines, consequently promoting inflammation-associated carcinogenesis.32 STAT3 has been demonstrated as a binding partner as well as a substrate of SIRT1.33 Indeed, overexpression of SIRT1 inhibited acetylation, phosphorylation, and transactivation of STAT3;33 in contrast, silencing of SIRT1 potentiated IL-22–driven acetylation and phosphorylation of STAT3. These results are indicative of the existence of interplay between SIRT1 and STAT3.34 Similarly, activator protein 1 (AP-1) is inactivated through direct interaction with SIRT1, though it is unclear whether SIRT1-depdendent deacetylation is involved in this process.35 Based on the findings, one can conclude that SIRT1 inhibits proinflammatory signaling that is often inappropriately activated in transformed and cancerous cells; thus, it can be anticipated that SIRT1 inhibits inflammation-associated carcinogenesis.
SIRT1 exerts anticarcinogenic effects through multiple mechanisms
Besides inhibiting abnormally activated proinflammatory signaling and subsequently preventing inflammation-associated cancer, SIRT1 participates in suppression of multistage carcinogenesis via other mechanisms. For example, SIRT1 can counteract various genotoxic insults, including oxidative DNA damage, thereby blocking initiation of carcinogenesis. SIRT1 deacetylates and inhibits proapoptotic p53 and poly (ADP-ribose) polymerase 1 under stressful conditions, conferring adaptive cell survival.18,36 SIRT1 is also required for DNA repair processes, both nuclear excision repair and double-strand break repair, to maintain genomic stability.37,38 Furthermore, SIRT1-overexpressing MEFs showed longer telomeres, whereas telomere shortening was observed in SIRT1-deficient MEFs.39
Inappropriate overexpression of the cellular oncogene, such as c-Myc, is evident in some human malignancies; c-Myc binds to the SIRT1 promoter and induces SIRT1 expression. However, SIRT1 interacts with and deacetylates c-Myc, resulting in decreased c-Myc stability. 40 As a consequence, the transforming activity of c-Myc is compromised. The downregulation of c-Myc–mediated cellular transformation via a c-Myc–SIRT1 negative feedback loop supports a role of SIRT1 in tumor suppression.40 SIRT1 can hamper cancer cell proliferation as well. Ectopic overexpression of SIRT1 suppressed G418-resistant colony formation of human colon cancer HCT-116 cells.41 Resveratrol inhibited growth of several human cancer cell lines, which was possibly mediated by SIRT1-depedent relocalization of Werner syndrome protein (WRN) from the nucleolus to the nucleoplasm;42 in contrast, the SIRT1 inhibitor EX-527 stimulated DNA replication and proliferation of cancer cells.41 In addition to growth arrest, resveratrol triggered apoptosis of BRCA1 mutant tumor cells through SIRT1-dependent inhibition of survivin.43 SIRT1 also promoted autophagy in prostate cancer, based on the observation that overexpression of SIRT1 resulted in accumulation of autophagy-related proteins, while the SIRT1 antagonist sirtinol repressed autophagy.44
Moreover, SIRT1 can deacetylate and inactivate hypoxia-inducible factor 1α (HIF-1α), a transcription factor that can rescue solid tumors from hypoxic burden through facilitation of angiogenesis and metastasis.45 SIRT1 inhibits HIF-1α–mediated expression of oncoproteins. The tissue levels of vascular endothelial growth factor, a key stimulator of angiogenesis, were markedly reduced in xenograft tumor of SIRT1-overexpressing human sarcoma cells.45 SIRT1 negatively regulated matrix metalloproteinase-9, an enzyme responsible for cancer cell invasion and migration.46 Brouguignon et al. have shown that reveratrol acts as a chemosensitizer by repressing multidrug resistance protein 1 (MDR1) in a SIRT1-dependent manner.47
The anticarcinogenic effects of SIRT1 have been demonstrated in numerous in vivo studies. When nude mice were implanted with SIRT1-overexpressing cells, tumor growth was suppressed.40,45 In contrast, significant enlargement of xenograft tumor was observed in nude mice that had received SIRT1-knockdown colon cancer cells.41 Wang et al. have demonstrated that SIRT1+/− p53+/− double mutant mice spontaneously generate tumors at five months of age and show a higher incidence of tumors in multiple organs, compared with wild-type, SIRT1+/− and p53+/− mice.48 Moreover, SIRT1-overexpressing APCmin/+ mice developed fewer intestinal tumors compared with SIRT1 wild-type mice, further supporting the anticarcinogenic function of SIRT1.49
Oncogenic functions of SIRT1
SIRT1 is overexpressed in some tumors
On the premise that SIRT1 acts as a tumor suppressor, SIRT1 could be downregulated in many tumors. In support of this speculation, SIRT1 expression was found to be reduced in human skin tumors.37 Paradoxically, however, elevated expression of SIRT1 has been observed in various types of human malignancies. The immunostaining of SIRT1 has revealed that both the proportion of positive cells and staining intensity are significantly increased in human prostate cancer specimens.50 SIRT1 was also highly expressed in other types of human cancer tissues, such as ovary, liver, breast, stomach, and pancreas.51–55 In case of human colorectal cancer, SIRT1 overexpression was detected as well.56 However, other investigations have revealed pronounced SIRT1 expression in both normal colon and tumor tissues, even though expression is substantially reduced in some higher grade colon tumors.41,57
In spite of the conflicting observations, SIRT1 overexpression in tumors has some clinical implications. Thus, upregulation of SIRT1 in tumors was found to be associated with unsatisfactory therapeutic outcomes in some cancer patients. Kaplan–Meier analyses of survival in cancer patients with or without SIRT1 overexpression revealed that those in the SIRT1-positive group exhibited poor prognosis outcome and shorter overall survival.54,58–60 In addition to survival time, SIRT1 overexpression was correlated with the higher tumor stage and the presence of lymph node metastasis in gastric and pancreatic cancer patients.55,60 SIRT1-positive tumors showed higher expression of Ki-67, a cell proliferation marker, which may account for the poor prognosis in SIRT1-positive cancer patients.54 Interestingly, DBC1, a negative regulator of SIRT1 activity, was coordinately overexpressed together with SIRT1 in gastric cancer patients,60 which is considered compensatory expression for abnormal upregulation of SIRT1.
SIRT1 deregulates both tumor suppressors and oncoproteins
Overexpression of SIRT1 may aggravate tumorigenesis through abnormal modulation of certain proteins, particularly tumor-suppressor proteins and (proto)oncogenes. The tumor suppressor p53 is the most representative substrate of SIRT1 deacetylase. In response to genotoxic insults, p53 upregulated a potential tumor suppressor microRNA 34a (miR-34a), thus promoting apoptosis.61 However, SIRT1 might facilitate deacetylation and inactivation of p53 in cancerous cells, leading to repression of miR-34a.62 Hypermethylated in cancer 1 (HIC1) is also a tumor suppressor that cooperates with p53 to induce apoptosis in response to DNA damage.63 Primary tumors derived from HIC1+/− mice exhibited SIRT1 overexpression.63 Moreover, SIRT1 has been reported to deacetylate retinoblastoma (Rb) protein and phosphatase and tensin homologue deleted in chromosome 10 (PTEN), repressing their tumor suppressive activity.64,65
In addition to inactivation of tumor suppressors, SIRT1 overexpression is associated with deregulation of protooncogenes. Myc genes are well-known prototypic oncogenes.66 SIRT1-dependent deacetylation of c-Myc can enhance its stability, association with c-Max, and transcriptional activity.67,68 Furthermore, SIRT1 also deacetylates N-Myc, resulting in stabilization of this oncoprotein and cell proliferation.69 In turn, these Myc proteins can upregulate SIRT1 expression, consequently forming positive feedback loops between Myc proteins and SIRT1.67,69 These findings are opposite to the previously reported destabilization of c-Myc by SIRT1 as a consequence of deacetylation of c-Myc through direct interaction between two entities.40
Moreover, SIRT1 is a downstream of oncogenic BCR-ABL tyrosine kinase. The SIRT1 silencing suppressed BCR-ABL-mediated transformation of bone marrow cells and development of a chronic myelogenous leukemia (CML)-like myeloproliferative disease.70 Recently, it has been reported that SIRT1 positively regulates membrane localization and oncogenic activation of Akt via deacetylation.71 The Ras oncoproteins appear to play a role in SIRT1-associated tumorigenesis as well. The SIRT1 inhibitor sirtinol suppressed Ras activation, but it remains to be elucidated which protein is a bona fide target of SIRT1 for its oncogenic functions.72 Collectively, SIRT1 can be involved in carcinogenesis through deregulation of tumor suppressors and prototypic oncoproteins.
SIRT1 confers survival advantages to cancer cells
SIRT1 promotes cancer cell proliferation and survival. SIRT1-overexpressing hepatocellular carcinoma Hep1 cells showed enhanced proliferation, whereas there was no change in Hep1 cells transfected with a deacetylase-defective SIRT1 H363Y mutant construct.52 In contrast, SIRT1 inhibition abrogated colony formation in human breast cancer MCF-7, lung cancer H1299, and CML progenitor cells.72,73 Likewise, SIRT1 inhibitors have been reported to trigger cell death in various types of human cancer cell lines.72,74–76 Suppression of cancer cell proliferation as a consequence of SIRT1 downregulation might be attributable to telomere dysfunction and increased acetylation and subsequent activation of p53.73,77 The stimulation of cell proliferation by SIRT1 seems to be cancer specific. Ford et al. have shown that siRNA knockdown of SIRT1 leads to enhanced apoptosis of various cancer cell lines, whereas it fails to affect apoptosis or growth arrest in normal human epithelial cell lines and normal primary diploid fibroblasts.78
Epithelial to mesenchymal transition (EMT) is recognized as a plausible mechanism of tumor progression and the invasion-metastasis cascade.79 Recently, SIRT1 has emerged as a regulator of EMT-like transformation in tumors; for example, it has been demonstrated that SIRT1 is upregulated in human mammary epithelial cells during the EMT induced by tumor growth factor β.80 Conversely, SIRT1 silencing reduced expression of ZEB1, an EMT-inducing transcription factor, while restoring E-cadherin expression that is generally downregulated during the EMT process.80,81 SIRT1 thus plays a crucial role in EMT-associated signal transduction. SIRT1 silencing has also restored cell–cell adhesion, while reducing the invasiveness of cancer cells.55,81 Moreover, ectopic overexpression of SIRT1 enhanced migration of SIRT1-null MEFs, suggesting that SIRT1 directly promotes cell migration.51
SIRT1-dependent deacetylation of cell motility proteins is thought to be a feasible mechanism underlying SIRT1-promoted invasion and migration. Cortactin, an F-actin binding protein, promotes cell migration once it is acetylated.51 SIRT1 physically interacts with and deacetylates cortactin, resulting in enhanced cell migration.51 Furthermore, Dishevelled (Dvl) proteins involved in Wnt signaling also take part in SIRT1-dependent cell migration. Dvl proteins form a complex with SIRT1 and then undergo deacetylation and subsequent stabilization.82 SIRT1-dependent positive regulation of Dvl proteins seems to be crucial for Wnt-mediated cell migration. Genetic or pharmacologic inhibition of SIRT1 attenuated expression of Wnt downstream proteins and concomitantly Wnt-induced cell migration.82
Furthermore, SIRT1 contributes to acquisition of chemoresistance in several types of tumors. Various drug-resistant cancer cell lines have exhibited overexpression of SIRT1, suggesting that SIRT1 may take part in chemoresistance.83,84 In line with this notion, SIRT1 gain-of-function activity is associated with upregulated expression of MDR1, a major drug resistance molecule, in HEK293 cells.83 Conversely, inhibition of SIRT1 by use of pharmacological inhibitors or siRNA knockdown reduced MDR1 expression in cancer cells, thus enhancing chemosensitivity.74,83,85 The in vivo xenograft assay using doxorubicin-resistance MCF-7 cells also showed augmentation of doxorubicin responsiveness by a SIRT1 inhibitor amurensin G.85 In the case of CML, SIRT1 deacetylates DNA repair proteins, such as Nijmegen breakage syndrome protein 1 and Ku70, resulting in acquisition of BCR–ABL mutations and subsequent drug resistance.86 Moreover, SIRT1 renders cancer cells resistant to radiation-induced apoptosis.87 Based on these observations, it seems likely that SIRT1 overexpression confers survival advantages to cancerous or transformed cells and accelerates tumorigenesis.
Subcellular localization may account for differential roles of SIRT1 in normal versus cancer cells
As mentioned earlier, SIRT1 has dual effects on carcinogenesis (Fig. 1). SIRT1 has been reported to inhibit inflammation, transformation, tumor promotion, and progression. However, SIRT1 also exerts opposite effects, acting as a tumor promoter. Such a double-edged sword nature of SIRT1 might be potentially determined by its subcellular localization. SIRT1 has been identified as a nuclear protein at first.10 However, it has been demonstrated that SIRT1 contains at least two nuclear localization signals (NLSs) and two nuclear export signals (NESs) and hence undergoes nucleocytoplasmic shuttling.88 This implies that SIRT1 can be located in cytosol as well as nucleus. SIRT1 was found to be normally present in the cytoplasm in murine pancreatic islet cells and human embryonic kidney cells.89,90 Notably, SIRT1 was overexpressed predominantly in the cytosol of certain cancer cells, while normal epithelial cells showed nuclear localization.56,91 This phenomenon has been also observed in human colon and ovarian cancer specimens.51,56Figure 2 illustrates differential expression of SIRT1 in human colon tumor and surrounding tissues.
Figure 2.
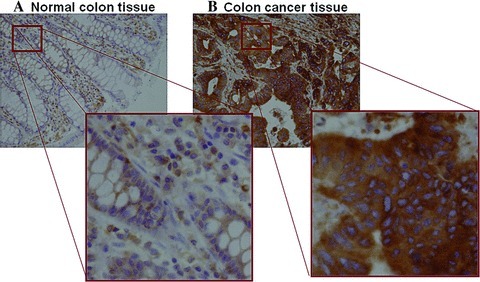
Differential subcellular localization of SIRT1 in a human colon tumor specimen and normal colonic tissues. SIRT1 is highly expressed in human colon tumor tissues (B) compared with normal counterparts (A). Notably, SIRT1 showed cytoplasmic localization in colon tumors (B), whereas the adjacent normal tissues displayed rather nuclear-concentrated expression (A).
The differential subcellular localization of SIRT1 may affect the substrate specificity in normal versus cancer cells. As illustrated in Figure 3, substrates of the SIRT1 deacetylase can be classified into two groups, cytoplasm- and nucleus-predominant, according to their subcellular localization. In the normal cells, SIRT1 seems to be present mainly in the nucleus, predominantly targeting nuclear proteins. Nuclear SIRT1 can deacetylate and inactivate transcription factors, including NF-κB, STAT3, HIF-1α, and AP-1, exerting anti-inflammatory and anticarcinogenic effects.30,33,35 Moreover, nuclear SIRT1 is also involved in maintenance of genomic stability through deacetylation of DNA repair proteins, such as PARP1, XPC, and WRN.36,37,42 SIRT1 has been reported to form both negative and positive feedback loops with c-Myc, which appear to be related to its anti- and procarcinogenic actions, respectively.40,67,68 This might be attributed to the different localization of both SIRT1 and c-Myc in normal cells and in cancer cells. Similar to SIRT1, Myc oncoproteins normally reside in the nucleus, yet cancer cells show overexpression of Myc proteins predominantly in the cytoplasm.66,92 In the nucleus of normal cells, SIRT1 deacetylates and destabilizes c-Myc, suppressing c-Myc-driven tumorigenesis.40 However, SIRT1 shows cytoplasmic localization in cancer cells, targeting cytosolic proteins as its preferred deacetylation substrates.56,91 Thus, SIRT1 can deacetylate and stabilize Myc proteins, promoting tumorigenesis.67 Cytoplasmic SIRT1 may also deacetylate and activate the Akt oncoprotein.71 Moreover, SIRT1 can deacetylate cytoplasmic proteins involved in locomotion, including cortactin and Dvl proteins, leading to enhanced cell mobility.51,82 Interestingly, p53, a well-known target of SIRT1, accumulates in the cytoplasm of various cancer cells, functioning as a proapoptotic protein; and SIRT1 might inhibit p53-dependent apoptosis in the cytoplasm of cancer cells.93,94
Figure 3.
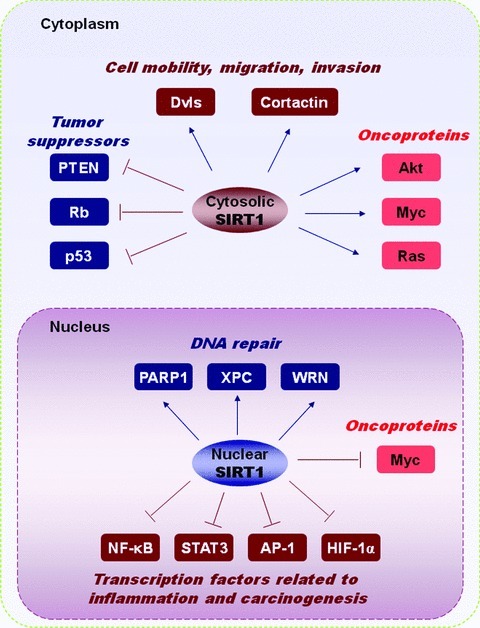
Some represantive intracellular proteins modulated by SIRT1. Nuclear and cytoplasmic SIRT1 may differentially target proteins as deacetylation substrates. Thus, SIRT1 may exert specific functions, depending on its subcellular localization.
Concluding remarks
SIRT1, a NAD+-dependent histone/protein deacetylase, has been emerging as a crucial regulator of assorted physiological events through deacetylation of various proteins related to apoptosis, DNA repair, and metabolism.10,95 SIRT1 is also involved in tumorigenesis; but it is still under much debate whether SIRT1 stimulates or suppresses carcinogenic processes (see Fig. 1). SIRT1 inhibits inflammation and activities of transcription factors that exacerbate carcinogenesis.30,33,35,49 Moreover, SIRT1 contributes to preservation of genomic stability.38,39 Thus, SIRT1 takes part in prevention, retardation, and suppression of carcinogenesis. SIRT1 is supposed to be underexpressed in tumors if it is indeed a tumor suppressor. Contrary to this supposition, substantial proportions of human cancer specimens have shown overexpression of SIRT1.51–55 SIRT1 aggravates inflammation, inactivates tumor suppressors, and, concomitantly, activates protooncogenes.63,65,71,96 In addition, SIRT1 promotes cancer cell proliferation, invasion, migration, and chemoresistance, conferring survival advantages to cancer cells.51,52,80
The subcellular localization of SIRT1 might be responsible, at least in part, for determination of its dual roles in tumorigenesis. However, there should be factors other than subcellular localization that modulate SIRT1 functions. For instance, the complex regulators of the SIRT1 activity, such as AROS and DBC1, should be considered. Moreover, SIRT1 deacetylation of c-Myc was found to modulate the stability of this oncoprotein in both positive and negative manners.40,67,68 In this case, the different subcellular localization of SIRT1 was insufficient to assign to it reciprocal regulation of c-Myc protein stability. Although the subcellular locailization of SIRT1 is not a sole determinant of functions of this deacetylase in tumorigenesis, it is evident that the nuclear and cytoplasmic SIRT1 might exert distinct functions. Cytoplasmic SIRT1 promoted neurite outgrowth in PC12 cells, which was inhibited by the nuclear SIRT1.97 Furthermore, a SIRT1 NLS mutant unable to enter the nucleus failed to suppress colony formation, whereas SIRT1-overexpressing cells showed a strong inhibitory effect.41
In conclusion, SIRT1 might modulate tumorigenesis in both positive and negative manners, partially depending on its subcellular localization. However, further investigation is required to fully clarify whether subcellular localization of SIRT1 is indeed a fate-determinant of its oncogenic versus tumor-suppressing functions. In particular, it is important to determine whether the SIRT1 NLS and/or NES contain point mutation(s) in human cancer tissues. In addition, SIRT1 mutations within the NLS and/or NES domain(s) would be useful tools for better elucidating the specific functions of nuclear and cytoplasmic SIRT1.
Acknowledgments
This work was supported by the Global Core Research Center (GCRC), Grant (No. 2012-0001184), and the World Class University (WCU) project Grant (No. R31-2012-000-10103-0) from the National Research Foundation (NRF), Ministry of Education, Science and Technology (MEST), Republic of Korea.
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Guarente L. Franklin H. Epstein Lecture: sirtuins, aging, and medicine. N. Engl. J. Med. 2011;364:2235–2244. doi: 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- 2.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 4.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 6.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen D, Guarente L. SIR2: a potential target for calorie restriction mimetics. Trends Mol. Med. 2007;13:64–71. doi: 10.1016/j.molmed.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Price NL, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imai S, et al. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 10.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 12.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim EJ, et al. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol. Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat. Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki T, et al. Phosphorylation regulates SIRT1 function. PLoS One. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasrin N, et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 19.Jeong J, et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp. Mol. Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 20.Li X, et al. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 21.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 22.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 23.Kundu JK, Surh YJ. Emerging avenues linking inflammation and cancer. Free Radic. Biol. Med. 2012;52:2013–2037. doi: 10.1016/j.freeradbiomed.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 24.Zhu X, et al. Activation of Sirt1 by resveratrol inhibits TNF-alpha induced inflammation in fibroblasts. PLoS One. 2011;6:e27081. doi: 10.1371/journal.pone.0027081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, et al. Roles of SIRT1 in the acute and restorative phases following induction of inflammation. J. Biol. Chem. 2010;285:41391–41401. doi: 10.1074/jbc.M110.174482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshizaki T, et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 2010;298:E419–E428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purushotham A, et al. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu F, et al. Lack of SIRT1 (Mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/- mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151:2504–2514. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajendrasozhan S, et al. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am. J. Respir Crit. Care Med. 2008;177:861–870. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeung F, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh UP, et al. Resveratrol (trans-3,5,4’-trihydroxystilbene) induces silent mating type information regulation-1 and down-regulates nuclear transcription factor-kappaB activation to abrogate dextran sulfate sodium-induced colitis. J. Pharmacol. Exp. Ther. 2010;332:829–839. doi: 10.1124/jpet.109.160838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie Y, et al. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat. Cell Biol. 2009;11:492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sestito R, et al. STAT3-dependent effects of IL-22 in human keratinocytes are counterregulated by sirtuin 1 through a direct inhibition of STAT3 acetylation. FASEB J. 2011;25:916–927. doi: 10.1096/fj.10-172288. [DOI] [PubMed] [Google Scholar]
- 35.Gao Z, Ye J. Inhibition of transcriptional activity of c-JUN by SIRT1. Biochem. Biophys. Res. Commun. 2008;376:793–796. doi: 10.1016/j.bbrc.2008.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajamohan SB, et al. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol. Cell Biol. 2009;29:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ming M, et al. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22623–22628. doi: 10.1073/pnas.1010377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palacios JA, et al. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J. Cell Biol. 2010;191:1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J. Cell Biol. 2009;185:203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabra N, et al. SirT1 is an inhibitor of proliferation and tumor formation in colon cancer. J. Biol. Chem. 2009;284:18210–18217. doi: 10.1074/jbc.M109.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rusin M, Zajkowicz A, Butkiewicz D. Resveratrol induces senescence-like growth inhibition of U-2 OS cells associated with the instability of telomeric DNA and upregulation of BRCA1. Mech. Ageing Dev. 2009;130:528–537. doi: 10.1016/j.mad.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Wang RH, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol. Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell MJ, et al. Disruption of a Sirt1-dependent autophagy checkpoint in the prostate results in prostatic intraepithelial neoplasia lesion formation. Cancer Res. 2011;71:964–975. doi: 10.1158/0008-5472.CAN-10-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim JH, et al. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol. Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 46.Nakamaru Y, et al. A protein deacetylase SIRT1 is a negative regulator of metalloproteinase-9. FASEB J. 2009;23:2810–2819. doi: 10.1096/fj.08-125468. [DOI] [PubMed] [Google Scholar]
- 47.Bourguignon LY, Xia W, Wong G. Hyaluronan-mediated CD44 interaction with p300 and SIRT1 regulates beta-catenin signaling and NFkappaB-specific transcription activity leading to MDR1 and Bcl-xL gene expression and chemoresistance in breast tumor cells. J. Biol. Chem. 2009;284:2657–2671. doi: 10.1074/jbc.M806708200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang RH, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Firestein R, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huffman DM, et al. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009;28:445–460. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- 52.Chen HC, et al. SIRT1 promotes tumorigenesis and resistance to chemotherapy in hepatocellular carcinoma and its expression predicts poor prognosis. Ann. Surg. Oncol.19: 2011–2019. 2012 doi: 10.1245/s10434-011-2159-4. [DOI] [PubMed] [Google Scholar]
- 53.Sung JY, et al. Balance between SIRT1 and DBC1 expression is lost in breast cancer. Cancer Sci. 2010;101:1738–1744. doi: 10.1111/j.1349-7006.2010.01573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng AN, et al. Expression of SIRT1 in gastric cardiac cancer and its clinicopathologic significance. Int. J. Surg. Pathol. 2011;19:743–750. doi: 10.1177/1066896911412181. [DOI] [PubMed] [Google Scholar]
- 55.Zhao G, et al. SIRT1 RNAi knockdown induces apoptosis and senescence, inhibits invasion and enhances chemosensitivity in pancreatic cancer cells. Gene Ther. 2011;18:920–928. doi: 10.1038/gt.2011.81. [DOI] [PubMed] [Google Scholar]
- 56.Stunkel W, et al. Function of the SIRT1 protein deacetylase in cancer. Biotechnol. J. 2007;2:1360–1368. doi: 10.1002/biot.200700087. [DOI] [PubMed] [Google Scholar]
- 57.Jang SH, et al. Loss of SIRT1 histone deacetylase expression associates with tumour progression in colorectal adenocarcinoma. J. Clin. Pathol. 2012;65:735–739. doi: 10.1136/jclinpath-2012-200685. [DOI] [PubMed] [Google Scholar]
- 58.Jang KY, et al. SIRT1 expression is associated with poor prognosis of diffuse large B-cell lymphoma. Am. J. Surg. Pathol. 2008;32:1523–1531. doi: 10.1097/PAS.0b013e31816b6478. [DOI] [PubMed] [Google Scholar]
- 59.Jang KY, et al. Expression and prognostic significance of SIRT1 in ovarian epithelial tumours. Pathology. 2009;41:366–371. doi: 10.1080/00313020902884451. [DOI] [PubMed] [Google Scholar]
- 60.Cha EJ, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin. Cancer Res. 2009;15:4453–4459. doi: 10.1158/1078-0432.CCR-08-3329. [DOI] [PubMed] [Google Scholar]
- 61.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yi J, Luo J. SIRT1 and p53, effect on cancer, senescence and beyond. Biochim. Biophys. Acta. 2010;1804:1684–1689. doi: 10.1016/j.bbapap.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen WY, et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 64.Wong S, Weber JD. Deacetylation of the retinoblastoma tumour suppressor protein by SIRT1. Biochem J. 2007;407:451–460. doi: 10.1042/BJ20070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ikenoue T, et al. PTEN acetylation modulates its interaction with PDZ domain. Cancer Res. 2008;68:6908–6912. doi: 10.1158/0008-5472.CAN-08-1107. [DOI] [PubMed] [Google Scholar]
- 66.Marcu KB, Bossone SA, Patel AJ. Myc function and regulation. Ann. Rev. Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 67.Menssen A, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc. Natl. Acad. Sci. USA. 2012;109:E187–196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mao B, et al. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. Int. J. Biochem. Cell Biol. 2011;43:1573–1581. doi: 10.1016/j.biocel.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 69.Marshall GM, et al. SIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stability. PLoS Genet. 2011;7:e1002135. doi: 10.1371/journal.pgen.1002135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan H, et al. Activation of stress response gene SIRT1 by BCR-ABL promotes leukemogenesis. Blood. 2012;119:1904–1914. doi: 10.1182/blood-2011-06-361691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sundaresan NR, et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci. Signal. 2011;4:ra46. doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- 72.Ota H, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006;25:176–185. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- 73.Li L, et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kojima K, et al. A role for SIRT1 in cell growth and chemoresistance in prostate cancer PC3 and DU145 cells. Biochem. Biophys. Res. Commun. 2008;373:423–428. doi: 10.1016/j.bbrc.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 75.Lara E, et al. Salermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene. 2009;28:781–791. doi: 10.1038/onc.2008.436. [DOI] [PubMed] [Google Scholar]
- 76.Peck B, et al. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol. Cancer Ther. 2010;9:844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- 77.Chen J, et al. Sirtuin 1 is upregulated in a subset of hepatocellular carcinomas where it is essential for telomere maintenance and tumor cell growth. Cancer Res. 2011;71:4138–4149. doi: 10.1158/0008-5472.CAN-10-4274. [DOI] [PubMed] [Google Scholar]
- 78.Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–10463. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- 79.Geiger TR, Peeper DS. Metastasis mechanisms. Biochim. Biophys. Acta. 2009;1796:293–308. doi: 10.1016/j.bbcan.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 80.Eades G, et al. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J. Biol. Chem. 2011;286:25992–26002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Byles V, et al. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene. 2012 doi: 10.1038/onc.2011.612. Jan 16. doi: 10.1038/onc.2011.612. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Holloway KR, et al. SIRT1 regulates Dishevelled proteins and promotes transient and constitutive Wnt signaling. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9216–9221. doi: 10.1073/pnas.0911325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chu F, et al. Control of multidrug resistance gene mdr1 and cancer resistance to chemotherapy by the longevity gene sirt1. Cancer Res. 2005;65:10183–10187. doi: 10.1158/0008-5472.CAN-05-2002. [DOI] [PubMed] [Google Scholar]
- 84.Liang XJ, et al. SIRT1 contributes in part to cisplatin resistance in cancer cells by altering mitochondrial metabolism. Mol. Cancer Res. 2008;6:1499–1506. doi: 10.1158/1541-7786.MCR-07-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oh WK, et al. Amurensin G, a potent natural SIRT1 inhibitor, rescues doxorubicin responsiveness via down-regulation of multidrug resistance 1. Mol. Pharmacol. 2010;78:855–864. doi: 10.1124/mol.110.065961. [DOI] [PubMed] [Google Scholar]
- 86.Wang Z, et al. SIRT1 deacetylase promotes acquisition of genetic mutations for drug resistance in CML cells. Oncogene. 2012 doi: 10.1038/onc.2012.83. Mar 12. doi: 10.1038/onc.2012.83. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun Y, et al. Downregulation of Sirt1 by antisense oligonucleotides induces apoptosis and enhances radiation sensitization in A549 lung cancer cells. Lung Cancer. 2007;58:21–29. doi: 10.1016/j.lungcan.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 88.Tanno M, et al. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 89.Moynihan KA, et al. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 90.Zhang J. The direct involvement of SirT1 in insulin-induced insulin receptor substrate-2 tyrosine phosphorylation. J. Biol. Chem. 2007;282:34356–34364. doi: 10.1074/jbc.M706644200. [DOI] [PubMed] [Google Scholar]
- 91.Byles V, et al. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. Int. J. Biol. Sci. 2010;6:599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Calcagno DQ, et al. MYC insertions in diffuse-type gastric adenocarcinoma. Anticancer Res. 2009;29:2479–2483. [PubMed] [Google Scholar]
- 93.O’Brate A, Giannakakou P. The importance of p53 location: nuclear or cytoplasmic zip code. Drug Resist. Updat. 2003;6:313–322. doi: 10.1016/j.drup.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 94.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Ann. Med. 2011;43:198–211. doi: 10.3109/07853890.2010.547211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Orecchia A, et al. Sirtinol treatment reduces inflammation in human dermal microvascular endothelial cells. PLoS One. 2011;6:e24307. doi: 10.1371/journal.pone.0024307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sugino T, et al. Protein deacetylase SIRT1 in the cytoplasm promotes nerve growth factor-induced neurite outgrowth in PC12 cells. FEBS Lett. 2010;584:2821–2826. doi: 10.1016/j.febslet.2010.04.063. [DOI] [PubMed] [Google Scholar]


