Abstract
We report the structure activity relationships of short 14-mer phosphorothioate gapmer antisense oligonucleotides (ASOs) modified with α-L-locked nucleic acid (LNA) and related modifications targeting phosphatase and tensin homologue (PTEN) messenger RNA in mice. α-L-LNA represents the α-anomer of enantio-LNA and modified oligonucleotides show LNA like binding affinity for complementary RNA. In contrast to sequence matched LNA gapmer ASOs which showed elevations in plasma alanine aminotransferase (ALT) levels indicative of hepatotoxicity, gapmer ASOs modified with α-L-LNA and related analogs in the flanks showed potent downregulation of PTEN messenger RNA in liver tissue without producing elevations in plasma ALT levels. However, the α-L-LNA ASO showed a moderate dose-dependent increase in liver and spleen weights suggesting a higher propensity for immune stimulation. Interestingly, replacing α-L-LNA nucleotides in the 3′- and 5′-flanks with R-5′-Me-α-L-LNA but not R-6′-Me- or 3′-Me-α-L-LNA nucleotides, reversed the drug induced increase in organ weights. Examination of structural models of dinucleotide units suggested that the 5′-Me group increases steric bulk in close proximity to the phosphorothioate backbone or produces subtle changes in the backbone conformation which could interfere with recognition of the ASO by putative immune receptors. Our data suggests that introducing steric bulk at the 5′-position of the sugar-phosphate backbone could be a general strategy to mitigate the immunostimulatory profile of oligonucleotide drugs. In a clinical setting, proinflammatory effects manifest themselves as injection site reactions and flu-like symptoms. Thus, a mitigation of these effects could increase patient comfort and compliance when treated with ASOs.
Keywords: gapmer, hepatotoxicity, immune stimulation, oligonucleotides, α-L-LNA
Introduction
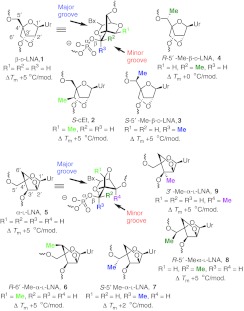
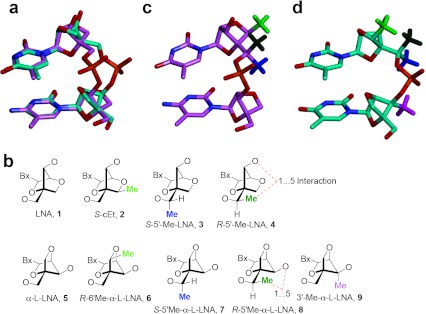
Antisense oligonucleotides (ASO) bind to their cognate mRNA in cells and modulate RNA function.1 However, unmodified oligonucleotides are highly unstable in biological media and require stabilization, either with chemical modifications or by formulation with cationic lipid based delivery vehicles, for use in animal experiments.2 Second generation ASOs, which are amongst the most advanced oligonucleotides in the clinic, best illustrate the use of chemical modifications to improve the drug-like properties of ASOs.3,4 The chemical design of a 2nd generation ASO includes a 8–14 base deoxynucleotide “gap,” flanked on either end with 2–5 2′-O-methoxyethyl RNA (MOE) nucleotides. The gap region promotes degradation of the target mRNA by RNase H-mediated cleavage while the flanking MOE nucleotides enhance affinity for cognate RNA. To examine whether further increases in affinity could improve ASO potency, we replaced MOE nucleotides in the flanks of 2nd generation ASOs with locked nucleic acid (β-D-LNA 1, referred to as LNA henceforth; also known as 2′,4′-bridged nucleic acid or BNA, Figure 1).5 This led to an increase in potency although this was sometimes accompanied by an increase in hepatotoxicity. Subsequent SAR studies showed that subtle structural changes to the LNA scaffold could change the therapeutic profile of ASOs containing these modifications. For example, introducing a methyl group in the (R) or (S) configuration at the 6′ (2′,4′-constrained-2′-O-ethyl BNA, S-cEt 2)6,7 or the 5′-position (S-5′-Me-LNA 3 or R-5′-Me-LNA 4) of LNA8 or replacing the 2′-oxygen atom in LNA with a substituted carbon atom,9,10 improved the hepatotoxicity profile of the modified ASOs while maintaining activity relative to the parent LNA benchmark. To see if more drastic changes to the LNA scaffold could impact ASO therapeutic profile while maintaining activity, we replaced LNA nucleotides in the flanks with α-L-LNA 5 nucleotides.11,12 α-L-LNA represents the α anomer of enantio-LNA (L-LNA) and also exhibits LNA-like high affinity recognition of complementary nucleic acids when incorporated into oligonucleotides.
Figure 1.
Structures and duplex stabilizing properties of LNA, S-cEt, R-5′-Me-LNA, S-5′-Me-LNA, α-L-LNA, R-6′-Me-α-L-LNA, R-5′-Me-α-L-LNA, S-5′-Me-α-L-LNA, and 3′-Me-α-L-LNA in oligonucleotide sequences used for biophysical studies. LNA, locked nucleic acid.
Wengel and co-workers first identified α-L-LNA as a high affinity RNA recognition scaffold by screening all eight stereoisomers of LNA in thermal denaturation experiments.11 Subsequent structural studies showed that α-L-LNA/DNA chimeric oligonucleotides form a seamless duplex with RNA and that the 2′,4′-bridge of α-L-LNA lies inside the major groove of the modified duplex.13,14,15 Moreover, the duplexes exhibited an intermediate character between A- and B-type helical geometries. This discovery led to some initial excitement that such duplexes could serve as substrates for RNase H although this was found not to be the case later.16 Fluiter compared the pharmacokinetic and antitumor properties of 16-mer LNA, 2′-thio-LNA, 2′-amino-LNA, and α-L-LNA modified phosphorothioate gapmer ASOs targeting H-Ras mRNA.17 The study showed that all the modified ASOs had similar Tm and activity in cell culture for reducing H-ras mRNA. In addition, the α-L-LNA ASO showed activity similar to LNA for slowing tumor growth in a xenograft model. Despite the promising antitumor activity seen with the α-L-LNA ASO, no data regarding actual knockdown of H-ras mRNA in the animal experiment was provided to ascertain if the activity was indeed related to downregulation of H-ras gene in tumor tissue. The study also did not describe the effect of ASO treatment on genes expressed in the liver where the antisense effects of phosphorothoioate gapmer ASOs are best characterized. Moreover, while the above study was carried out with 16-mer ASOs, we and others have recently shown that it is possible to improve the potency of PS gapmer ASOs in animals by further reducing ASO length down to 14- or even 12-mers.6,18,19
The interesting properties of α-L-LNA oligonucleotides spurred the synthesis of several structural analogs for use in oligonucleotide based diagnostic and therapeutic applications.20,21,22,23,24 We recently reported the synthesis and biophysical properties of oligonucleotides modified with R-6′-Me-α-L-LNA 6,25 S-5′-Me-α-L-LNA 7 and its R-5′-Me isomer 8,26 and 3′-Me-α-L-LNA 9,27 which also exhibit α-L-LNA like high affinity recognition of complementary nucleic acids but have increased steric bulk at different positions on the bicyclic scaffold (Figure 1). As seen in the LNA series, it was possible that introducing steric bulk at different locations of the α-L-LNA scaffold could modulate the biological properties of ASOs containing such modifications. In this report, we present the results from our detailed investigations of the SAR of 14-mer phosphorothioate gapmer ASOs modifed with α-L-LNA and related analogs. We show that these ASOs are capable of potent downregulation of gene expression in liver tissue without causing hepatotoxicity. However, in the sequence evaluated, α-L-LNA had a slightly higher propensity for causing increased immune stimulation which could be mitigated by replacing α-L-LNA monomers in the flanks of the ASO with R-5′-Me-α-L-LNA but not 6′-Me- or 3′-Me-α-L-LNA nucleotides.
Results
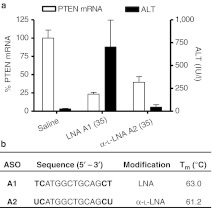
To determine whether replacing LNA with α-L-LNA in the 3′- and 5′-flanks of PS modified gapmer ASOs has an effect on the hepatotoxicity profile, we first evaluated both modifications in a 14-mer (2-10-2 design) oligonucleotide targeting mouse phosphatase and tensin homologue (PTEN). This sequence shows elevations in plasma alanine aminotransferase (ALT) levels in mice even after a single injection of ASO A1 and was previously used by us to compare the hepatotoxicity profile of LNA and related analogs (Figure 2).6,28 Mice (n = 4/group) were injected intraperitoneally with a single dose of 35 mg/kg of the LNA ASO A1 and α-L-LNA ASO A2 and PTEN mRNA in mouse liver and ALT levels were recorded post-sacrifice 72 hours after ASO administration. Both ASOs showed reductions in PTEN mRNA in liver tissue but LNA ASO A1 also showed an increase in ALT levels whereas the α-L-LNA ASO A2 did not.
Figure 2.
Evaluation of LNA and α-L-LNA antisense oligonucleotides (ASOs) for hepatotoxicity in mice. Mice (n = 4/dose group) were injected intraperitoneally (i.p.) with a single dose of 35 mg/kg of ASOs A1 and A2 formulated in saline and animals were sacrificed 72 hours after injection. (a) Phosphatase and tensin homologue (PTEN) mRNA reduction in liver normalized to saline-treated animals and plasma alanine aminotransferase (ALT) levels post-sacrifice. (b) Sequence and Tm of ASOs A1 and A2 versus complementary RNA. Modified nucleotides are indicated in bold font, all errors in ±SD. LNA, locked nucleic acid.
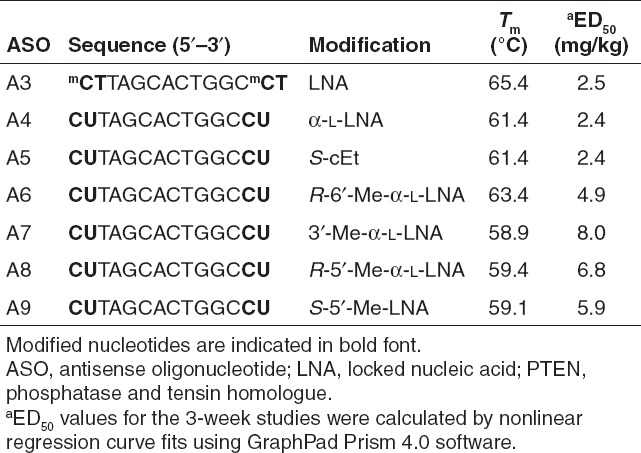
To characterize the therapeutic profile of ASOs modified with LNA 1, S-cEt 2, S-5′-Me-LNA 3, α-L-LNA 5, R-6′-Me-α-L-LNA 6, R-5′-Me-α-L-LNA 8, and 3′-Me-α-L-LNA 9, we used a different 14-mer sequence also targeting mouse PTEN (Table 1). This sequence has been used by us extensively to profile the antisense properties of ASOs modified with tcDNA and a number of BNA and HNA analogs.6,8,9,18,28,29 We first measured the Tm of ASOs A3–A9 versus complementary RNA and only observed minor differences in the ability of these modifications to stabilize oligonucleotide duplexes with complementary RNA. The LNA ASO A3 showed slightly higher Tm which can be attributed to using LNA T and 5-Me-C monomers, where each 5-Me group typically contributes +0.5 °C to overall Tm.30
Table 1. Sequence, Tm and ED50 (mg/kg) values for LNA, α-L-LNA, S-cEt, R-6′-Me-α-L-LNA, 3′-Me-α-L-LNA, R-6′-Me-α-L-LNA, S-5′-Me-LNA ASOs A3–A9, respectively, targeting mouse PTEN.
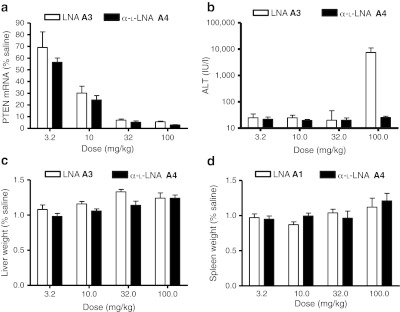
Using the second sequence, we first evaluated LNA and α-L-LNA ASOs A3 and A4, respectively, in a single dose-escalation study (Figure 3). Mice (n = 4/group) were dosed intraperitoneally with a single dose of 3.2, 10, 32, and 100 mg/kg of LNA ASO A3 and α-L-LNA ASO A4 and PTEN mRNA reduction in mouse liver and plasma ALT levels were recorded post-sacrifice 72 hours after ASO administration. In this study, both ASOs showed almost identical knockdown of PTEN mRNA at both doses tested (Figure 3a). No ALT elevations were observed for the α-L-LNA ASO A4 at all evaluated doses while the LNA ASO A3 showed dramatic elevations in transaminase levels only for the high dose group treated animals (Figure 3b). No appreciable changes in liver and spleen weights were observed for either ASO in this experiment (Figure 3c–d).
Figure 3.
Evaluation of LNA and α-L-LNA antisense oligonucleotides (ASOs) A3 and A4 for activity and hepatotoxicity in a single-dose–escalation study in mice. Mice (n = 4/dose group) were injected intraperitoneally (i.p.) with a single dose of 3.2, 10, 32, and 100 mg/kg of ASOs A3 and A4 formulated in saline and animals were sacrificed 72 hours after injection. (a) Phosphatase and tensin homologue (PTEN) mRNA reduction in liver normalized to saline-treated animals (b) plasma alanine aminotransferase (ALT) levels post-sacrifice. Percent change in (c) liver and (d) spleen weights relative to saline-treated animals. LNA, locked nucleic acid.
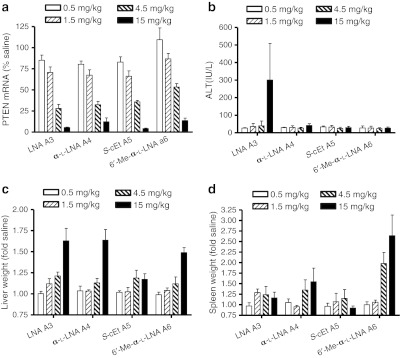
We next compared LNA ASO A3, α-L-LNA ASO A4, S-cEt ASO A5, and R-6′-Me-α-L-LNA ASO A6 using a subchronic dosing schedule (Figure 4). This dosing schedule permits evaluation of the activity and immunostimulatory profile of ASOs in a clinically more relevant manner. Mice (n = 4/group) were injected intraperitoneally with 0.5, 1.5, 4.5, and 15 mg/kg twice a week for 3 weeks and PTEN mRNA knockdown in mouse liver and plasma ALT levels were recorded post-sacrifice 48 hours after last dose. All ASOs showed very similar potency for reducing PTEN mRNA in mouse liver, except for A6 which was twofold less potent despite very similar overall Tm (Figure 4a). In this dosing regimen, all ASOs evaluated achieved near maximal PTEN mRNA knockdown at the 15 mg/kg dose but the LNA ASO A3-treated mice showed elevated ALT levels while ASOs A4, A5, and A6 did not (Figure 4b). This is consistent with previous observations where LNA gapmers produced ALT increases at doses which resulted in maximal target mRNA knockdown.5 All the ASOs except A5, showed a moderate dose-dependent increase in liver weights (Figure 4c). Interestingly, the α-L-LNA ASO A4 and to a greater degree, the R-6′-Me-α-L-LNA ASO A6 showed drug induced dose-dependent increases in spleen weight (Figure 4d) indicative of increased immune stimulation.31 In contrast, the LNA and S-cEt ASOs A3 and A5, respectively, did not show a similar dose-dependent increase in spleen weights.
Figure 4.
Evaluation of LNA A3, α-L-LNA A4, S-cEt A5, and R-6′-Me-α-L-LNA A6 for activity and hepatotoxicity in a 3-week dose-escalation study. Mice (n = 4/dose group) were injected intraperitoneally (i.p.) twice a week for 3 weeks with of 0.5, 1.5, 4.5, and 15 mg/kg of antisense oligonucleotides (ASOs) formulated in saline and animals were sacrificed 48 hours after last injection. ASOs A3, A4, and A5 were evaluated in one study while ASO A6 was evaluated in a parallel study using LNA ASO A3 as a control. Data for the LNA ASO (n = 8) reflects the average measurements from both studies while data for ASOs A4, A5, and A6 (n = 4) is from the individual studies (a) phosphatase and tensin homologue (PTEN) mRNA reduction in liver normalized to saline-treated animals (b) plasma alanine aminotransferase (ALT) levels post-sacrifice. Percent change in (c) liver and (d) spleen weights relative to saline-treated animals. All errors in ±SD.
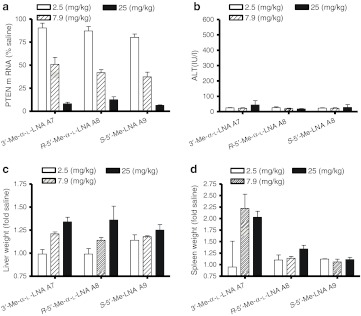
To examine whether other α-L-LNA analogs also produced similar increases in spleen weights, we evaluated the 3′-Me- and R-5′-Me-α-L-LNA modified ASOs A7 and A8, respectively in mice (Figure 5). The S-5′-Me-LNA ASO A9 was included as the control. We had previously evaluated S-5′-Me-LNA ASO A9 in animal experiments and found it to be approximately twofold less potent than LNA ASO A3.8 However, the 5′-Me group was very effective in suppressing even the modest ASO induced spleen weight increase produced by the LNA ASO A3 in that study. Mice (n = 4/group) were dosed with ASOs A7, A8, and A9 at 2.5, 7.9, and 25 mg/kg twice a week for 3 weeks and liver mRNA downregulation and plasma ALT levels were recorded post-sacrifice 48 hours after last ASO administration. In this study, the S-5′-Me-LNA ASO A9 showed the best activity for PTEN mRNA reduction in mouse liver followed by the R-5′-Me-α-L-LNA ASO A8 whereas the 3′-Me-α-LNA ASO A7 was the least active (Figure 5a) but the differences in activity were minor. The 3′-Me- and R-5′-Me-α-L-LNA ASOs A7 and A8, respectively, showed modest dose-dependent increases in liver weights but none of these ASOs produced elevations in ALT levels at all the doses evaluated (Figure 5b). However, the 3′-Me-α-L-LNA ASO A7 showed more than twofold increase in spleen weight for the 7.9 and 25 mg/kg ASO-treated groups. In contrast, the R-5′-Me-α-L-LNA ASO A8 and the S-5′-Me-LNA ASO A9 showed minimal increases in spleen weight changes (Figure 5d).
Figure 5.
Evaluation of 3′-Me-α-L-LNA A7, R-5′-Me-α-L-LNA A8, and S-5′-Me-LNA A9 for activity and hepatotoxicity in a 3-week dose-escalation study. Mice (n = 4/dose group) were injected intraperitoneally (i.p.) twice a week for 3 weeks with 2.5, 7.9, and 25 mg/kg of antisense oligonucleotides (ASOs) formulated in saline and animals were sacrificed 48 hours after last injection. (a) Phosphatase and tensin homologue (PTEN) mRNA reduction in liver normalized to saline-treated animals (b) plasma alanine aminotransferase (ALT) levels post-sacrifice. Percent change in (c) liver and (d) spleen weights relative to saline-treated animals. All errors in ±SD. LNA, locked nucleic acid.
Discussion
To help understand the relative orientations of the different methyl groups in the LNA versus the α-L-LNA series and to examine whether these differences could help explain the observed biological effects, we created structural models by overlaying dimeric units of each modification from the published NMR structures of the modified duplexes (Figure 6).15,32 We used the duplex structures to extrapolate the relative orientations of the methyl groups on the LNA and α-L-LNA scaffolds and their structural relationship with the PS backbone in single stranded ASOs. Previous studies have utilized crystal structures of dinucleoside phosphates with natural and modified sugars, as starting points to generate the double helical structures of oligonucleotide duplexes.33,34 Thus, it is reasonable to expect that the modified dinucleotide units with rigid and locked furanose rings in the flanks of ASOs A3–A9 have similar overall conformations in the single strands as those observed in the duplexes.
Figure 6.
Structural models of LNA and α-L-LNA dinucleotide units showing relative orientations of 3′-Me, 5′-Me, and 6′-Me groups. (a) Overlay of common structural units such as nucleobases, C1′ carbon and O4′ oxygen atoms of LNA (pink carbons) and α-L-LNA (teal carbons) dimers obtained from the NMR structures of modified DNA/RNA duplexes (refs. 32 and 15). Complementary RNA strand not shown for clarity. (b) Structures of the monomeric units showing relative orientations of various methyl groups. (c) Relative orientations of S-6′-Me (light green), R-5′-Me (olive green), and S-5′-Me (dark blue) groups in a LNA dinucleotide unit. (d) Relative orientations of R-6′-Me (light green), R-5′-Me (olive green), S-5′-Me (dark blue), and 3′-Me (pink) groups in an α-L-LNA dinucleotide unit. LNA, locked nucleic acid.
By overlaying the common elements such as the nucleobases, the 1′-carbon and the 4′-oxygen atoms, it becomes readily apparent that the α-L-LNA sugar-phosphate backbone is distinctly different from the LNA sugar-phosphate backbone (Figure 6a). In addition, while the 2′,4′-bridge resides in the minor groove for LNA modified duplexes, this bridge lies in the major groove for α-L-LNA modified duplexes. As a consequence, introducing substitution on the 2′,4′-bridge of LNA directs the substituent toward the minor groove while analogous substitution in the α-L-series directs the substituent into the major groove of the modified duplex. An illustration of this concept can be gained by comparing the relative orientation of the 6′-methyl groups in S-cEt 2 (S-6′-Me-LNA) versus R-6′-Me-α-L-LNA 6 (Figure 6c–d). In both cases, the methyl group (light green) points toward the 3′-hydroxyl group in the nucleoside monomer. However this group lies at the edge of the minor groove in the LNA series35 while it lies at the edge of the major groove in the α-L-series. In contrast, introducing a methyl group at the 3′-position (pink) of α-L-LNA projects this group into the minor groove of the modified duplex. The analogous substitution in LNA was not explored as substituting at this position disrupts hybridization by interfering with base stacking in the major groove.36
The relative orientation of the methyl groups at the 5′-position in LNA and α-L-LNA are more difficult to estimate. Assuming canonical orientations around torsions angles α, β, and γ, the S-configured 5′-Me group (dark blue) in LNA is most likely situated in the minor groove and occupies a position distinctly different from that of the 6′-Me group (S-cEt). We had previously shown that the R-5′-Me-LNA (olive green) analog 4 had a destabilizing effect on duplex stability relative to LNA and the S-5′-Me analog 3.8 Structural insights into this destabilizing effect were obtained by examining the crystal structures of R- and S-6′-Me substituted fluoro hexitol nucleic acid modified DNA duplexes which, like BNA modified duplexes, also exist in the A-type conformation.37 In that case, the R-6′-Me group caused an energetically unfavored 1…5 steric clash with the 3′-phosphodiester linkage resulting in duplex instability. This destabilizing interaction is intrinsic to all A-form duplexes where the sugar moieties exists in the C3′-endo sugar pucker. Interestingly, while the energetically disfavored 1…5 interaction also exists in the α-L-LNA scaffold (Figure 6b) the R-5′-Me-α-L-LNA analog 8 had a greater stabilizing effect on duplex thermal stability as compared the S-5′-Me isomer 7.26 Previous NMR studies on the R-5′-Me-α-L-LNA nucleoside monomer had indicated conformational mobility around torsion angle γ suggesting that the sugar-phosphate backbone in α-L-LNA nucleotides might be capable of adopting alternate low energy conformations in addition to the one observed in the NMR structure of the modified duplex with RNA.26
Based on the above analysis there appear to be no obvious structural reasons for the differential effects on spleen weights produced by the α-L-LNA ASO. It is also unclear if the methyl groups in different locations on the LNA and α-L-LNA scaffold are changing the binding properties of these ASOs to any putative immune receptors. However, it is known that several nucleic acid sensing proteins recognize chemically modified nucleic acids such as PS DNA, with or without functional consequences, by binding to the sugar-phosphate backbone in a sequence independent manner.38 Thus an alternate explanation for the observed effects could be that the 6′- and the 3′-methyl groups in α-L-LNA stabilize certain nucleic acid backbone conformation/s that is/are intrinsically more proinflammatory. In contrast, introducing a methyl group at the 5′-position in close proximity to the phosphorothioate linkage, shields the ASO sugar-phosphate backbone from being recognized efficiently by the immune receptors. Alternatively, the 5′-methyl group produces subtle changes in the backbone conformation such that the ASO is not recognized as efficiently by the immune receptors. Either one of these events could be responsible for the mitigation in ASO proinflammatory profile observed in our studies.
In conclusion, we report for the first time, the antisense properties of RNase H active PS gapmer ASOs modified with α-L-LNA and related analogs in animals. We find that α-L-LNA ASOs are capable of potent downregulation of gene expression in liver tissue without producing hepatoxicity. However, the α-L-LNA ASO showed a slightly increased propensity for causing immune stimulation in the oligonucleotide sequence used for evaluation in the present study. The ALT elevations seen with the LNA/DNA gapmer ASOs seen in this work and previous studies5,6,8,9,29 is in contrast to recent reports using non-gapmer LNA/DNA mixmer ASOs targeting miR12239,40 highlighting that sequence and design can significantly alter ASO therapeutic profile.
Similarly, while we only evaluated α-L-LNA in the context of two sequences, extensive profiling of this modification in several different sequences and motifs is required to arrive at general conclusions regarding any perceived benefits of this scaffold for lowering the risk of hepatotoxicity. Our results also suggest that introducing steric bulk at the 5′-position, in close proximity to the PS linkage, could be a general strategy for reducing the immunostimulatory profile of ASOs. In a clinical setting, proinflammatory effects manifest themselves as injection site reactions and flu-like symptoms. Thus, a mitigation of these effects could increase patient comfort and compliance when treated with ASOs.
Materials and Methods
Oligonucleotide synthesis and purification. LNA phosphoramidites were purchased from commercially available sources while other phosphoramidites were synthesized according to procedures described previously.7,8,12 Oligonucleotides A1–A9 were synthesized at 40 µmol scale using UnyLinker PS200 universal support, 0.2 mol/l phenylacetyldisulfide in 1:1 3-picoline:acetonitrile as a sulfur-transfer reagent and 0.7 mol/l dicyanoimidazole in acetonitrile as the activator. All phosphoramidites were used at 0.1 mol/l concentration in acetonitrile. For each of the modified analogs 4-fold excess of amidite was delivered with a 12-minute coupling time. The 5′-end dimethoxytrityl group was left on to facilitate purification. After synthesis was complete, all oligonucleotides were treated with 1:1 triethylamine:acetonitrile to remove the cyanoethyl protecting group from the phosphorothioate linkages. Subsequently, oligonucleotides were treated with concentrated aqueous NH4OH at 55 °C for 9–12 hours to cleave from support, remove heterocyclic protecting groups, and hydrolyze the UnyLinker moiety. Oligonucleotides were purified by ion-exchange chromatography using a gradient of NaBr across Source 30Q resin, with the 5′-DMT group being removed during purification using 6% (vol/vol) aqueous dichloroacetic acid. Pure fractions were desalted by binding to a C18 reverse-phase column and eluting with 50% (vol/vol) acetonitrile in water.
Tm measurements. For the Tm experiments, oligonucleotides were prepared at a concentration of 8 µmol/l in a buffer of 100 mmol/l NaCl, 10 mmol/l phosphate, 0.1 mmol/l EDTA at pH 7. The concentration of oligonucleotides was determined at 85 °C. The final oligonucleotide concentration was 4 µmol/l with mixing of equal volumes of test oligonucleotide and complementary RNA strand. Oligonucleotides were hybridized with the complementary RNA strand by heating duplex to 90 °C for 5 minutes and allowed to cool to room temperature. Using the spectrophotometer, Tm measurements were taken by heating duplex solution at a rate of 0.5 °C/minute in cuvette starting at 15 °C and heating to 85 °C. Tm values were determined using van't Hoff calculations (A260 versus temperature curve) using non-self-complementary sequences where the minimum absorbance which relates to the duplex and the maximum absorbance which relates to the non-duplex single strand are manually integrated into the program. Sequence of the RNA complement used for ASOs A1 and A2 was 5′-r(GGAAGCTGCAGCCATGATGG)-3′ and for ASOs A3–A9 was 5′-r(TCAAGGCCAGTGCTAAGAGT)-3′.
Protocols for animal experiments and RNA analysis. The Institutional Animal Care and Use Committee (IACUC) approved all procedures. Male Balb/c mice were housed 4/cage on a 12:12-hour light/dark cycle. ASO solutions were prepared in PBS and injected intraperitoneally twice a week at various concentrations for 3 weeks. Mice were sacrificed 48 hours after the last dose. Blood samples were collected by cardiac puncture, diluted 1:3 with physiological saline and plasma chemistries values were measured on the Olympus AU400 Clinical Analyzer (Beckman Coulter, CA). For the RNA analysis, liver tissues were homogenized in 4 mol/l guanidine isothiocyanate, 25 mmol/l EDTA, 50 mmol/l Tris–HCl pH 6 containing 1 mol/l β-mercaptoethanol immediately following sacrifice and homogenized. RNA was extracted using RNeasy columns (Qiagen, Valencia, CA) according to manufacturer's protocol. RNA was eluted from the columns with water. RNA samples were analyzed by fluorescence-based quantitative reverse transcription (RT)-PCR using an Applied Biosystems 7700 sequence detector (Applied Biosystems, Carlsbad, CA). Levels of target RNAs as well as those of cyclophilin A, a housekeeping gene, were determined. Target RNA levels were normalized to cyclophilin levels for each RNA sample. Primers used for determination of PTEN RNA level are as follows: FP 5′-ATGACAATCATGTTGCAGCAATTC-3′, RP 5′-CGATGCAATAAATATGCACAAATCA-3′, and PR 5′-6FAM- CTGTAAAGCTGGAAAGGGACGGACTGGT-TAMRA-3′. Primers used for determination of cyclophilin A RNA level are as follows: FP 5′-TCGCCGCTTGCTGCA-3′′, RP 5′-ATCGGCC GTGATGTCGA-3′, and PR 5′-6FAM-CCATGGTCAACCCCACC GTGTTC-TAMRA-3′.
References
- Bennett CF., and, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- Swayze EE., and, Bhat B.2007The medicinal chemistry of oligonucleotides Crooke ST.ed). Antisense Drug Technology: Principles, Strategies, and Applications2nd edn. CRC Press: Boca Raton; 143–182. [Google Scholar]
- Altmann KH, Martin P, Dean NM., and, Monia BP. Second generation antisense oligonucleotides – inhibition of pkc-alpha and c-raf kinase expression by chimeric oligonucleotides incorporating 6'-substituted carbocyclic nucleosides and 2'-O-ethylene glycol substituted ribonucleosides. Nucleosides, Nucleotides and Nucleic Acids. 1997;16:917–926. [Google Scholar]
- Raal FJ, Santos RD, Blom DJ, Marais AD, Charng MJ, Cromwell WC.et al. (2010Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial Lancet 375998–1006. [DOI] [PubMed] [Google Scholar]
- Swayze EE, Siwkowski AM, Wancewicz EV, Migawa MT, Wyrzykiewicz TK, Hung G.et al. (2007Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals Nucleic Acids Res 35687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth PP, Siwkowski A, Allerson CR, Vasquez G, Lee S, Prakash TP.et al. (2009Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals J Med Chem 5210–13. [DOI] [PubMed] [Google Scholar]
- Seth PP, Vasquez G, Allerson CA, Berdeja A, Gaus H, Kinberger GA.et al. (2010Synthesis and biophysical evaluation of 2′,4′-constrained 2′O-methoxyethyl and 2′,4′-constrained 2′O-ethyl nucleic acid analogues J Org Chem 751569–1581. [DOI] [PubMed] [Google Scholar]
- Seth PP, Allerson CR, Siwkowski A, Vasquez G, Berdeja A, Migawa MT.et al. (2010Configuration of the 5′-Methyl Group Modulates the Biophysical and Biological Properties of Locked Nucleic Acid (LNA) Oligonucleotides J Med Chem 538309–8318. [DOI] [PubMed] [Google Scholar]
- Seth PP, Allerson CR, Berdeja A, Siwkowski A, Pallan PS, Gaus H.et al. (2010An exocyclic methylene group acts as a bioisostere of the 2′-oxygen atom in LNA J Am Chem Soc 13214942–14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., and, Chattopadhyaya J. Intramolecular Free-Radical Cyclization Reactions on Pentose Sugars for the Synthesis of Carba-LNA and Carba-ENA and the Application of Their Modified Oligonucleotides as Potential RNA Targeted Therapeutics. Chem Rev. 2012;112:3808–3832. doi: 10.1021/cr100306q. [DOI] [PubMed] [Google Scholar]
- Rajwanshi VK, Håkansson AE, Sørensen MD, Pitsch S, Singh SK, Kumar R.et al. (2000The Eight Stereoisomers of LNA (Locked Nucleic Acid): A Remarkable Family of Strong RNA Binding Molecules We acknowledge the Danish Natural Science Research Council, the Danish Technical Research Council, and Exiqon A/S for financial support. Ms Britta M. Dahl is thanked for oligonucleotide synthesis, Dr. Carl E. Olsen for MALDI-MS analysis, and Ms. Karen Jørgensen for recording CD spectra Angew Chem Int Ed Engl 391656–1659. [DOI] [PubMed] [Google Scholar]
- Sørensen MD, Kvaernø L, Bryld T, Håkansson AE, Verbeure B, Gaubert G.et al. (2002α-L-ribo-configured locked nucleic acid (α-L-LNA): synthesis and properties J Am Chem Soc 1242164–2176. [DOI] [PubMed] [Google Scholar]
- Petersen M, Håkansson AE, Wengel J., and, Jacobsen JP. α-L-LNA (α-L-ribo configured locked nucleic acid) recognition of RNA. A study by NMR spectroscopy and molecular dynamics simulations. J Am Chem Soc. 2001;123:7431–7432. doi: 10.1021/ja010557u. [DOI] [PubMed] [Google Scholar]
- Nielsen KM, Petersen M, Håkansson AE, Wengel J., and, Jacobsen JP. α-L-LNA (α-L-ribo configured locked nucleic acid) recognition of DNA: an NMR spectroscopic study. Chemistry. 2002;8:3001–3009. doi: 10.1002/1521-3765(20020703)8:13<3001::AID-CHEM3001>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Nielsen JT, Stein PC., and, Petersen M. NMR structure of an α-L-LNA:RNA hybrid: structural implications for RNase H recognition. Nucleic Acids Res. 2003;31:5858–5867. doi: 10.1093/nar/gkg800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondensgaard K, Petersen M, Singh SK, Rajwanshi VK, Kumar R, Wengel J.et al. (2000Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity Chemistry 62687–2695. [DOI] [PubMed] [Google Scholar]
- Fluiter K, Frieden M, Vreijling J, Rosenbohm C, De Wissel MB, Christensen SM.et al. (2005On the in vitro and in vivo properties of four locked nucleic acid nucleotides incorporated into an anti-H-Ras antisense oligonucleotide Chembiochem 61104–1109. [DOI] [PubMed] [Google Scholar]
- Murray S, Ittig D, Koller E, Berdeja A, Chappell A, Prakash TP.et al. (2012TricycloDNA-modified oligo-2'-deoxyribonucleotides reduce scavenger receptor B1 mRNA in hepatic and extra-hepatic tissues–a comparative study of oligonucleotide length, design and chemistry Nucleic Acids Res [DOI] [PMC free article] [PubMed]
- Straarup EM, Fisker N, Hedtjärn M, Lindholm MW, Rosenbohm C, Aarup V.et al. (2010Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates Nucleic Acids Res 387100–7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TS, Wengel J., and, Hrdlicka PJ. 2′-N-(pyren-1-yl)acetyl-2′-amino-α-L-LNA: synthesis and detection of single nucleotide mismatches in DNA and RNA targets. Chembiochem. 2007;8:1122–1125. doi: 10.1002/cbic.200700144. [DOI] [PubMed] [Google Scholar]
- Kumar TS, Madsen AS, Østergaard ME, Wengel J., and, Hrdlicka PJ. Nucleic acid structural engineering using pyrene-functionalized 2′-amino-α-L-LNA monomers and abasic sites. J Org Chem. 2008;73:7060–7066. doi: 10.1021/jo800551j. [DOI] [PubMed] [Google Scholar]
- Kumar TS, Madsen AS, Østergaard ME, Sau SP, Wengel J., and, Hrdlicka PJ. Functionalized 2′-amino-α-L-LNA: directed positioning of intercalators for DNA targeting. J Org Chem. 2009;74:1070–1081. doi: 10.1021/jo802037v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yuan F, Zhou C, Plashkevych O., and, Chattopadhyaya J. Free-radical ring closure to conformationally locked α-L-carba-LNAs and synthesis of their oligos: nuclease stability, target RNA specificity, and elicitation of RNase H. J Org Chem. 2010;75:6122–6140. doi: 10.1021/jo100900v. [DOI] [PubMed] [Google Scholar]
- Seth PP, Allerson CR, Berdeja A., and, Swayze EE. Replacing the 2′-oxygen with an exocyclic methylene group reverses the stabilization effects of α-L-LNA. Bioorg Med Chem Lett. 2011;21:588–591. doi: 10.1016/j.bmcl.2010.10.025. [DOI] [PubMed] [Google Scholar]
- Seth PP, Yu J, Allerson CR, Berdeja A., and, Swayze EE. Synthesis and biophysical characterization of R-6′-Me-α-L-LNA modified oligonucleotides. Bioorg Med Chem Lett. 2011;21:1122–1125. doi: 10.1016/j.bmcl.2010.12.119. [DOI] [PubMed] [Google Scholar]
- Seth PP, Allerson CR, Ostergaard ME., and, Swayze EE. Structural requirements for hybridization at the 5′-position are different in α-L-LNA as compared to β-D-LNA. Bioorg Med Chem Lett. 2012;22:296–299. doi: 10.1016/j.bmcl.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Seth PP, Allerson CA, Østergaard ME., and, Swayze EE. Synthesis and biophysical evaluation of 3′-Me-α-L-LNA – Substitution in the minor groove of α-L-LNA duplexes. Bioorg Med Chem Lett. 2011;21:4690–4694. doi: 10.1016/j.bmcl.2011.06.104. [DOI] [PubMed] [Google Scholar]
- Prakash TP, Siwkowski A, Allerson CR, Migawa MT, Lee S, Gaus HJ.et al. (2010Antisense oligonucleotides containing conformationally constrained 2′,4′-(N-methoxy)aminomethylene and 2′,4′-aminooxymethylene and 2′-O,4′-C-aminomethylene bridged nucleoside analogues show improved potency in animal models J Med Chem 531636–1650. [DOI] [PubMed] [Google Scholar]
- Egli M, Pallan PS, Allerson CR, Prakash TP, Berdeja A, Yu J.et al. (2011Synthesis, improved antisense activity and structural rationale for the divergent RNA affinities of 3′-fluoro hexitol nucleic acid (FHNA and Ara-FHNA) modified oligonucleotides J Am Chem Soc 13316642–16649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier SM., and, Altmann KH. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S, Stecker K, Brooks D, Monteith D, Conklin B., and, Bennett CF. Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J Pharmacol Exp Ther. 2000;292:468–479. [PubMed] [Google Scholar]
- Petersen M, Bondensgaard K, Wengel J., and, Jacobsen JP. Locked nucleic acid (LNA) recognition of RNA: NMR solution structures of LNA:RNA hybrids. J Am Chem Soc. 2002;124:5974–5982. doi: 10.1021/ja012288d. [DOI] [PubMed] [Google Scholar]
- Rosenberg JM, Seeman NC, Day RO., and, Rich A. RNA double helices generated from crystal structures of double helical dinucleoside phosphates. Biochem Biophys Res Commun. 1976;69:979–987. doi: 10.1016/0006-291x(76)90469-1. [DOI] [PubMed] [Google Scholar]
- Egli M. Structural aspects of nucleic acid analogs and antisense oligonucleotides. Angew Chem Int Ed. 1996;35:1894–1909. [Google Scholar]
- Pallan PS, Allerson CR, Berdeja A, Seth PP, Swayze EE, Prakash TP.et al. (2012Structure and nuclease resistance of 2′,4′-constrained 2′-O-methoxyethyl (cMOE) and 2′-O-ethyl (cEt) modified DNAs Chem Commun (Camb) 488195–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldgaard M, Hansen FG., and, Wengel J. 3′-C-Branched LNA-type nucleosides locked in an N-type furanose ring conformation: synthesis, incorporation into oligodeoxynucleotides, and hybridization studies. J Org Chem. 2004;69:6310–6322. doi: 10.1021/jo049159a. [DOI] [PubMed] [Google Scholar]
- Pallan PS, Yu J, Allerson CR, Swayze EE, Seth P., and, Egli M. Insights from crystal structures into the opposite effects on RNA affinity caused by the S- and R-6′-methyl backbone modifications of 3′-fluoro hexitol nucleic acid. Biochemistry. 2012;51:7–9. doi: 10.1021/bi201810r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H, Chiba S, Ban T, Nakaima Y, Onoe T, Honda K.et al. (2011Suppression of immune responses by nonimmunogenic oligodeoxynucleotides with high affinity for high-mobility group box proteins (HMGBs) Proc Natl Acad Sci USA 10811542–11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME.et al. (2010Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection Science 327198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S.et al. (2008LNA-mediated microRNA silencing in non-human primates Nature 452896–899. [DOI] [PubMed] [Google Scholar]