Abstract
Oligonucleotide aptamer-mediated in vivo cell targeting of small interfering RNAs (siRNAs) is emerging as a useful approach to enhance the efficacy and reduce the adverse effects resulting from siRNA-mediated genetic interference. A current main impediment in aptamer-mediated siRNA targeting is that the activity of the siRNA is often compromised when conjugated to an aptamer, often requiring labor intensive and time consuming design and testing of multiple configurations to identify a conjugate in which the siRNA activity has not been significantly reduced. Here, we show that the thermal stability of the siRNA is an important parameter of siRNA activity in its conjugated form, and that siRNAs with lower melting temperature (Tm) are not or are minimally affected when conjugated to the 3′ end of 2′F-pyrimidine-modified aptamers. In addition, the configuration of the aptamer-siRNA conjugate retains activity comparable with the free siRNA duplex when the passenger strand is co-transcribed with the aptamer and 3′ overhangs on the passenger strand are removed. The approach described in this paper significantly reduces the time and effort necessary to screening siRNA sequences that retain biological activity upon aptamer conjugation, facilitating the process of identifying candidate aptamer-siRNA conjugates suitable for in vivo testing.
Keywords: aptamer, aptamer-siRNA conjugates, aptamer targeting, siRNA
Introduction
The limited specificity of drugs, notably that of cytotoxic drugs used in cancer therapy, is often a limiting factor in their clinical utility. A general approach to enhance therapeutic index of a drug, the differential between its beneficial and tolerable toxic dose, is to target the drug to the appropriate cells in the body, for example to target chemotherapeutic drugs to the tumor lesions of the cancer patients.
Cell targeting requires identification of products expressed specifically on the surface of targeted cell and the development of corresponding ligands to which the drug can be conjugated. To date, antibodies afford the most versatile and commonly used platform for generating targeting ligands with the necessary specificity and avidity.1,2 Nonetheless, since antibodies must be produced in cell culture systems their development and clinical manufacture is challenging and expensive. In addition, antibody–drug conjugation chemistries are complex, requiring extensive purification and quality control steps that may affect the drug's activity and reduce conjugate yield.
Oligonucleotide-based aptamers (aptamers) offer an alternative platform technology for generating ligands that can bind their targets with specificity and avidity comparable with antibodies.3 Unlike antibodies however, aptamers can be synthesized in a relatively simple and cost-effective cell-free chemical process. Aptamers have been recently used as ligands for targeting small interfering RNAs (siRNAs) to specific subsets of cells in mice, such as tumor cells,4,5,6 immune cells7 or virally-infected cells,8 that in each instance was accompanied by significant biological and therapeutic effects. Conjugation of the aptamer to its siRNA cargo was achieved by co-transcribing the aptamer and one of the two siRNA strands from a double-stranded DNA template, followed by hybridization to the complementary siRNA strand.4,6,7,8 In one instance both siRNA strands were co-transcribed with the aptamer as a short-hairpin RNA using a loop separating the two strands to allow for their intramolecular hybridization.5 Therefore, unlike antibody–drug conjugation, conjugation of aptamers to their oligonucleotide cargo is significantly simpler.
A current main impediment in aptamer-mediated siRNA targeting is that the activity of the siRNA is often compromised when conjugated to an aptamer, limiting the ability to use previously validated siRNA sequences (ref. 4 and A. Berezhnoy, R. Brenneman and E. Gilboa, unpublished data). Identification of active aptamer-siRNA conjugates often requires laborious synthesis and screening of multiple candidate conjugates to identify one in which the siRNA activity has not been significantly inhibited. It is not clear why siRNA action is negatively affected when conjugated to an aptamer, nor are there guiding rules how to design active conjugates. For example, the configurations in which the antisense (guide) strand of the siRNA fused to the 3′ end to the aptamer was used in some studies,4 the opposite configuration in which the sense (passenger) strand was fused downstream to the aptamer was used in other studies,7,8 and in yet another study a configuration where both siRNA strands were fused downstream to the aptamer was used.5
Given that aptamer-mediated siRNA targeting is emerging as a potentially useful approach for in vivo drug targeting it is desirable to understand what parameters affect the function of the conjugated siRNA and establish guiding rules for the design of functional aptamer-siRNA conjugates. In this study, we show that the thermal stability of the siRNA is an important parameter of siRNA activity in its conjugated form, and that siRNAs with lower melting temperature (Tm) are not or minimally affected when conjugated to aptamer compared to those with higher Tm. The methodology described in this paper significantly reduces the time and effort necessary to identify siRNA sequences that retain biological activity upon aptamer conjugation and may encourage increased application of this technology for in vivo studies.
Results
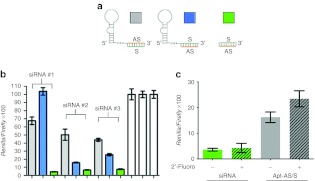
We generated multiple aptamer-siRNA conjugates where one strand of the siRNA duplex is contiguous with the 3′ end of the aptamer. Aptamer-siRNA conjugates were transcribed from corresponding double-stranded DNA template PCR products using a modified T7 polymerase to incorporate 2′-fluoro-modified pyrimidines in the RNA backbone. Two configurations were generated in which either of the two strands of the siRNA, the antisense (AS)/guide (gray) or sense (S)/passenger strand (blue), were co-linearly transcribed with the aptamer, and the chemically synthesized complementary strand hybridized to the fusion transcript (Figure 1a). siRNA activity was tested in transiently transfected HEK293T cells using the siCHECK assay measuring the normalized reporter activity of Renilla luciferase encoding the siRNA target sense sequence in its 3′-untranslated region. As shown in Figure 1b, the conjugated siRNA in the configuration in which the S strand was co-transcribed with the aptamer (Apt-S/AS; blue bars) was more active than the alternative configuration (Apt-AS/S; gray bars). However, even in the Apt-S/AS configuration siRNA activity appeared to be significantly reduced as compared with that of the nonconjugated siRNA duplex (green bars).
Figure 1.
Effect of conjugation on small interfering RNA (siRNA) inhibition. (a) Two configurations of aptamer-siRNA conjugates and unconjugated, unmodified siRNA that did not contain 5′-fluoro-modified pyrimidines were transfected into HEK293T cells and target inhibition assayed. (b) Cells were co-transfected with siRNA duplex or 4-1BB aptamer-siRNA conjugates and reporter plasmid containing short sequences corresponding to the murine TGFβRII siRNA targets cloned into the 3′ untranslated region (3′-untranslated region) of Renilla luciferase. After 48 hours, the normalized Renilla luciferase activity was measured in the siCHECK assay as described in Methods. White bars: conjugated and unconjugated control siRNA and untreated respectively. Conditions transfected in triplicate and data representative of at least two independent experiments. (c) Unmodified sense strand of raptor siRNA #23 (Table 1) was hybridized to unmodified or 2′-fluoropyrimidine modified antisense strand, or to unmodified aptamer-antisense fusion or 2′-fluoropyrimidine modifed aptamer-antisense fusion. Silencing activity was determined in the siCHECK system.
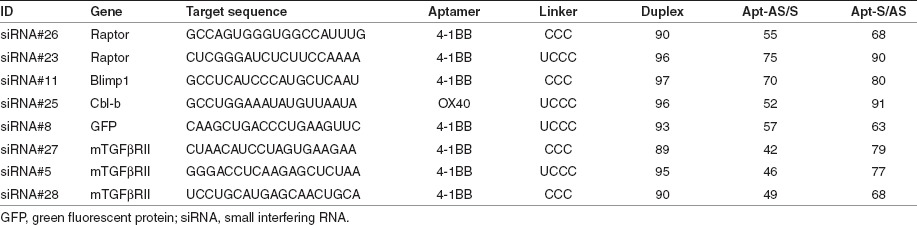
Table 1 shows a compilation of data from several experiments in which eight siRNAs corresponding to multiple targets were assessed for their inhibitory activity when conjugated to 4-1BB or OX40 binding aptamers using either of the two configurations depicted in Figure 1a. These experiments suggest that, in general, conjugation of siRNAs to aptamer in either configuration negatively affects its inhibitory activity but that the Apt-S/AS configuration was less detrimental than the Apt-AS/S configuration. To test whether the diminished silencing activity of Apt-AS/S conjugates is due to the incorporation of 2′-fluoro-modified pyrimidines in the transcribed AS strand, or its conjugation to the 3′ end of the aptamer sequence, we compared the silencing activities of free duplex siRNA and Apt-AS/S conjugate whereby the AS strand either contained or did not contain 2′-fluoromodified pyrimidines. As shown in Figure 1c silencing activity was diminished primarily as a result of aptamer conjugation and to a lesser extent by incorporation of 2-fluoro modified pyrimidines.
Table 1. siRNAs generated against murine targets were conjugated to 4-1BB-binding aptamer using either of the two configurations shown in Figure 1a, and tested for siRNA inhibition using the siCHECK assay. Values shown correspond to the percentage reduction in normalized Renilla expression. The data was compiled from multiple independent experiments with each condition transfected in triplicate and normalized to activity of a control aptamer-siRNA conjugate that did not recognize a sense target in the siCHECK Renilla 3′-untranslated region.
Several studies have shown that reducing the thermal stability at the 5′ end of the AS strand by introducing a mutation in the S (wobble) enhances its inhibitory activity.4,9 As shown in Figure 2, consistent with these observations, introducing a wobble by changing C→U in the 5′-end region of the S strand (pink bars) led to a small improvement in the inhibitory activity of the conjugated siRNA, though it did not fully restore the inhibitory activity of the unconjugated, native siRNA (green bars) that did not contain a wobble. This observation, however, raised the possibility that the reduced inhibitory activity of the aptamer-conjugated siRNA might be affected by the overall thermal stability of the siRNA.
Figure 2.
Introducing a C→U wobble enhances the inhibitory activity of the conjugated small interfering RNA (siRNA). A wobble was generated by replacing C with U in the 3′ region of the siRNA sense strand. The unmodified and wobble-containing 4-1BB aptamer-S transcripts were conjugated to an AS siRNA strand in the Apt-S/AS configuration (TGFβRII (#4, #5, and #6), Blimp-1 (#7), and green fluorescent protein (#8) siRNAs). The inhibitory activities of the aptamer-conjugated siRNAs and siRNA duplexes were tested in the siCHECK assay. White bar, aptamer-conjugated control siRNAs and control siRNAs duplex. Conditions transfected in triplicate and data representative of at least two independent experiments.
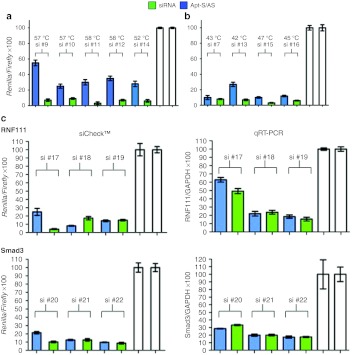
In a retrospective analysis we determined whether stratifying candidate sequences by siRNA Tm was predictive of the impact of conjugation on its activity. As shown in Table 2, there was a striking, but not absolute, correlation between the Tm and the conjugated siRNA activity. A majority of siRNAs with reduced overall thermal stability, calculated Tm <50 °C (see Methods section), retained high levels of inhibitory activity comparable with that of unconjugated siRNA, whereas siRNAs exhibiting higher thermal stability were less active in their conjugated form. We were, however, unable to identify specific motifs within the siRNA sequence that predict the impact of aptamer conjugation on siRNA function. Nonetheless, by selecting siRNA candidates with lower Tm, we were able to readily identify aptamer-siRNA conjugates with minimal-to-no loss in siRNA activity (Figure 3).
Table 2. The calculated Tm of siRNAs and the impact of conjugation on their activity.

Figure 3.
Small interfering RNA (siRNA) activity in conjugates prescreened for siRNAs with reduced melting temperature (Tm). Candidate siRNAs were predicted by the algorithms described in Methods and confirmed to have high efficiency knockdown of respective targets—murine Blimp-1 (#7, #9, #10, and #11) and T-bet (#12, #13, #14, and #15) in siCHECK assay. (a) Highest scoring candidates, all which had a relative Tm of 57–58 °C, were conjugated to a 4-1BB-binding aptamer using the more favorable Apt-S/AS configuration and tested for target inhibition using the siCHECK assay. White bars, control siRNA as free duplex or conjugate. (b) Same as a except that siRNAs with reduced Tm were used. (c) Candidate siRNAs targeted to murine Smad3 and Rnf111 genes were conjugated to 4-1BB binding aptamer (Apt-S/AS configuration) and tested for target inhibition using the ψcheck assay (left panels) or downregulation of endogenous transcripts measured by quantitative reverse-transcription-PCR (right panels). White bar, aptamer-conjugated control siRNAs and control siRNAs duplex. Conditions transfected in triplicate and data representative of at least two independent experiments.
Using the siCHECK assay, we screened numerous unconjugated siRNAs versus two genes Tbx21 and PRDM1 for silencing efficiency above 90% against their respective sense targets. From that pool, in the experiment shown in Figure 3a, we initially chose siRNAs that scored the highest in the HPC Dispatcher and Thermo/Dharmacon algorithms without paying attention to their Tm that happened to be about 57–58 °C. Consistent with Table 2, siRNA activity of all four Apt-S/AS conjugates (blue bars) was significantly compromised relative to the corresponding free duplexes (green bars). Subsequently, we generated a second set of conjugates, this time choosing siRNAs with lower Tm below 50 °C, and analyzed them for siRNA inhibition in the siCHECK assay. As shown in Figure 3b, three of five conjugates (blue bars) exhibited significantly improved siRNA activity comparable with that of the unconjugated, native siRNA duplex (green bars). Figure 3c shows two additional examples in which 4-1BB aptamer-mTGFβRII siRNA and 4-1BB aptamer-mRnf111 siRNA conjugates were generated using low Tm siRNAs, and analyzed for siRNA inhibition in both the siCHECK assay as well as knockdown of the endogenous mRNA target in transfected HEPA1-6 cells as measured by quantitative reverse-transcription-PCR. Despite some discordance between the two assays, in the majority of conjugates tested siRNA activity was not or minimally affected as compared with the free duplex siRNA; positive predictions by siCHECK against a short synthetic targets were supported by siRNA activity against the endogenous target in the quantitative reverse-transcription-PCR assay.
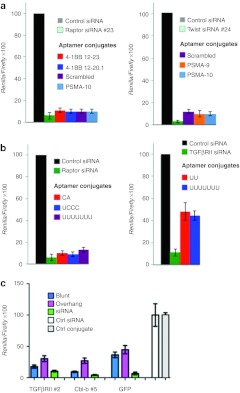
One question is whether the nucleotide composition of the aptamer and/or the linker sequence between the aptamer and siRNA affects the inhibitory activity of the conjugated siRNA. To this end we conjugated two different low Tm siRNAs to various aptamers and measured their activity in both the unconjugated and conjugated forms. As shown in Figure 4a, there was no aptamer-specific effect observed on siRNA function. This observation, therefore, suggests that the inhibitory activity of conjugated siRNA is by and large a property of the siRNA. No significant effect was seen using several linker sequences shown in Figure 4b, whereas the addition of a UU 3′ overhang invariably reduced the inhibitory activity of the aptamer-conjugated siRNA (Figure 4c).
Figure 4.
Effect of aptamer sequence, linker sequence and sense strand 3′ overhang, on small interfering RNA (siRNA) activity. (a) Either of two siRNAs were conjugated to two distinct prostate specific membrane antigen (PSMA)-binding or 4-1BB-binding aptamers, or to a scrambled aptamer using the Apt-S/AS configuration, and tested for siRNA inhibition in the siCHECK assay. (b) PSMA aptamer-raptor siRNA or 4-1BB aptamer-TGFβRII siRNA conjugated using varying linker sequences between the aptamer and siRNA were tested for siRNA inhibition in the ψcheck assay. (c) 4-1BB aptamer-TGFβRII siRNA, OX40 aptamer-Cbl-b siRNA and 4-1BB aptamer-GFP siRNAs with or without a template-encoded UU overhang at the 3′ end of the sense sequence were tested in the ψcheck assay. Conditions transfected in triplicate and data representative of at least two independent experiments. GFP, green fluorescent protein.
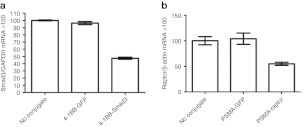
In the experiments described above the aptamer-siRNA conjugates were transfected into cells in an aptamer-independent process using a transfection reagent. To determine whether the aptamer-siRNA conjugates can inhibit their targets in an aptamer-targeted manner, Apt-S/AS configured conjugates harboring low Tm siRNA that have retained activity in the conjugated form were incubated in the absence of transfection reagents with cells expressing the product to which the aptamer binds, and siRNA inhibition was determined 48 hours later by quantitative reverse-transcription-PCR. As shown in Figure 5, aptamer-targeted delivery of a Smad3 (Tm = 48.9 °C; Figure 5a) or raptor siRNA (Tm = 48.9 °C; Figure 5b) to 4-1BB- or prostate specific membrane antigen expressing cells, respectively, led to the downregulation of the corresponding RNA transcripts in the cell. By contrast, a 4-1BB aptamer fused to a high Tm raptor siRNA (Tm = 59.2 °C), which was equally potent as free duplex by less potent when transfected as conjugate, was ineffective in this setting (data not shown).
Figure 5.
Aptamer-dependent small interfering RNA (siRNA) inhibition using Apt-S/AS conjugates and low Tm siRNAs. (a) 4-1BB-aptamer-Smad3 #3siRNA conjugate shown in Figure 3 or 4-1BB aptamer conjugated to a green fluorescent protein (GFP) siRNA were incubated with antigen-activated ovalbumin-specific transgenic CD8+ OT-I cells in the absence of a transfection reagent, and 48 hours later relative Smad3 and GAPDH mRNA transcript levels were assessed by quantitative reverse-transcription-PCR. Assay was performed in triplicate and data representative of at least two independent experiments. (b) Same as a except for using prostate specific membrane antigen (PSMA)-aptamer conjugated to GFP siRNA or to raptor siRNA and measuring relative raptor and GAPDH mRNA transcripts. Experimental data shown is representative of triplicate incubations and at least two independent experiments.
Discussion
In this study, we describe the design of aptamer-siRNA conjugates that minimizes the negative impact of conjugation on siRNA activity. We first show that the activity of siRNA is generally, but not always, better preserved when the sense strand is fused downstream to the aptamer (Apt-S/AS, Figure 1).
Although other configurations were able to yield reasonably effective conjugates as tested in vitro, the success rate of Apt-S/AS conjugates was significantly higher (Table 1). Since the aptamer-siRNA fusion is generated by co-transcription in the presence of 2′-fluoro-modified pyrimidines, a combination of an extended sequence and transcription errors of T7 polymerase with base modifications13 seemed to have an additive, albeit negative effect on siRNA activity when the AS strand is transcribed. These same considerations, however, are less critical in Apt-S/AS configuration when the AS is an unmodified, synthetic oligonucleotide. Not withstanding, the siRNA activity in the Apt-S/AS configuration appeared to be often reduced as compared with that of unconjugated, unmodified siRNA.
We also found that in the Apt-S/AS configuration the relative thermal stability of the siRNA (measured as Tm) exerted a significant influence on its activity when conjugated to the aptamer. As shown in Table 2 and Figure 3, with some exceptions (e.g., si #13), siRNAs with calculated Tm below 50 °C exhibited minimal-to-no loss of function in their conjugated form. It should be noted that the calculated Tm did not take into account the incremental stability conferred by the 2′-fluoro-modified nucleotides present in the sense strand or physiological salt conditions and, therefore, provides a comparative rather than absolute measure of the thermal stability of the siRNAs. We have so far failed to identify specific motifs in the siRNA sequence that correlated with preservation of activity in their conjugated form. For example, there was no apparent position effect of the 2′-fluoro-modified pyrimidines in the siRNA sense or antisense strands. Given that the activity of the conjugated siRNA appears to be largely independent of the upstream aptamer sequence, as shown in Figure 4, it appears that thermal stability of the siRNA is the main if not sole determinant of whether and to what extents its activity is compromised when conjugated to an aptamer.
These findings offer useful guidance for identifying siRNAs amenable to incorporation into aptamer-targeted siRNA conjugates generated by in vitro transcription for preclinical testing. Candidate siRNAs are first stratified by prediction scores from >1 siRNA selection algorithm, then those with reduced thermal stability (lower Tm) are chosen and the S strand incorporated into the aptamer of interest at the 3′ end of the transcription template. Using these criteria, in our experience two to three of four siRNA candidates for a given target exhibited no to little reduction when conjugated to an aptamer as compared with unconjugated native siRNA (which unlike the conjugated siRNA are not 2′-fluoro pyrimidine modified). This compares favorably to our prior experience when siRNAs were chosen solely based on their inhibitory activity as free duplex, in which case only 1/5 to 1/20 siRNAs retained significant activity as conjugates.
The precise biochemical reasons underlying the observation that low Tm siRNAs are more amenable to aptamer incorporation are not known. Since low and high Tm siRNAs demonstrate >80–90% activity when transfected as free duplex the loss of such activity when high Tm siRNAs are used as aptamer-siRNA conjugates must be at least partially related to a failure of one of the processing steps requisite for loading the guide strand into the RNA-induced silencing complex. A plausible explanation why the activity of siRNA when conjugated to an aptamer becomes dependent on its thermal stability was provided by the work of Gu et al.10 MicroRNA and short-hairpin RNAs loading into mammalian Argonaute (Ago) proteins and formation of the RNA-induced silencing complex is a two-step process: physical association followed by activation. Activation involves the displacement of the passenger strand, and is the rate-limiting step in RNA-induced silencing complex formation. Displacement (unwinding) of the passenger strand can occur via either cleavage or noncleavage mechanisms that are mediated by distinct Ago proteins. Gu et al.10 demonstrated that whereas cleavage-based passenger strand displacement mediated by Ago2 is thermoresistant, noncleavage-based displacement of the passenger strand is thermosensitive.10 Given the known structure of Ago2-siRNA complex, it is conceivable that the presence of a bulky structure like an aptamer at the end of the siRNA, the end that corresponds to the 5′ end of the passenger strand that is conjugated to the aptamer in the Apt-S/AS configuration as shown in Figure 1a, would interfere with its action, leaving as the only option noncleavage-based, and thereby thermosensitive, strand displacement and RNA-induced silencing complex formation. Further biochemical studies, however, are warranted to probe the mechanistic explanations behind the intracellular processing differences between high and low Tm aptamer-siRNA conjugates.
Materials and Methods
siRNA candidates selection. To identify candidate siRNA sequences three open-access siRNA selection algorithms were used: (i) HPC Dispatcher (http://infosci.coh.org/hpcdispatcher//), (ii) Thermo/Dharmacon siDesign Center (http://www.dharmacon.com/designcenter/designcenterpage.aspx), and (iii) siRNA Scales (http://gesteland.genetics.utah.edu/siRNA_scales/. The top candidates from each program were cross-referenced and sequences mostly predicted by all programs were further considered for in vitro testing. Candidate siRNAs were tested in silico for their ability to be incorporated at the 3′ end of the 4-1BB aptamer without compromising its secondary structure using RNAStructure 5.1 (Supplementary Data online).11 The relative Tm of the unmodified passenger and guide RNA strands was estimated using OligoAnalyzer 2.1 using the default conditions under the RNA setting (IDT; http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/).
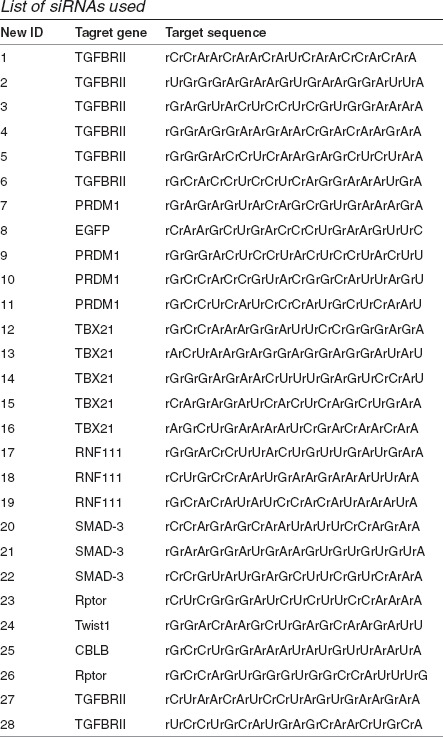
List of siRNAs used
Aptamer-siRNA conjugates. For 4-1BB aptamer-siRNA screening studies, a modified version of an RNA aptamer binding murine 4-1BB: 5′-GGGGGAATTCTAATACGACTCACTA TAGGGCGGGAGAGAGGAAGAGGGATGGGCGACCGAA CGTGCCCTTCAAAGCCGTTCACTAACCAGTGGCA TAACCCAGAGGTCGATAGTACTGGATCCCGCCCTCCC-3′ was used as a double-stranded DNA template for PCR to generate a double-stranded DNA transcription template encoding the 4-1BB aptamer monomer with a 5′ T7 promoter (bold) and 3′ siRNA extension. 4-1BB-siRNA transcripts were generated using the Durascribe T7 Transcription kit (Illumnia/Epicentre, Madison, WI) with a modified T7 RNA polymerase (Y693F) that incorporates 2′-F-modified RNA pyrimidines. Aptamer transcripts were purified by 10% denaturing PAGE as previously described (McNamara et al., 2008). To produce aptamer-siRNA chimeras, purified transcripts (1 µmol/l) in Dulbecco-modified phosphate buffered saline (Ca2+/Mg2+) were heated at 85 °C for 5 minutes in a heat block, cooled to room temperature then heated to 65 °C for 5 minutes and the complementary siRNA strand (2 µmol/l) added and cooled to room temperature by removal from the heat block. Unannealed siRNA was removed from the conjugation reaction by successive washes with Dulbecco-modified phosphate buffered saline +/+ and the volume reduced by centrifugation using Amicon Ultracel-30K columns (Milipore, Billerica, MA) and quantified using a Nanodrop spectrophotometer (Thermo, Asheville, NC). Conjugate annealing was verified using 3% agarose-EtBr gels run at 40 V for 3 hours to visualize electrophoretic mobility shifts as compared with the unannealed transcript without denaturing conjugates. For in vitro receptor-mediated delivery studies using 4-1BB-siRNA conjugates a 4-1BB dimer-siRNA was generated using the following double-stranded DNA template: 5′-GGGGGAATTCTAATACGACTCACTATAGGGCGGG AGAGAGGAAGAGGGATGGGCGACCGAACGTGCCCTT CAAAGCCGTTCACTAACCAGTGGCATAACCCAGAGGTC GATAGTACTGGATCCCGCCCTCCTGCGGCCGAGAGAG GAAGAGGGATGGGCGACCGAACGTGCCCTTCAAAGC CGTTCACTAACCAGTGGCATAACCCAGAGGTCGATAG TACTGGATCGGCCGCTCCC-3′ where underlined sequences represent 4-1BB monomer binding sequences and italics represent a single-stranded 2′-F-pyrimidine linker sequence. prostate specific membrane antigen-siRNA conjugates were generated in a similar fashion as described in reference.12
Cell culture. HEK293T, HEPA1-6, and B16-F10.9 were obtained from ATCC (Manassas, VA) and cultured at 37 °C in 5% CO2 in Dulbecco's modified Eagle medium (Gibco, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 IU/ml), and streptomycin (100 μg/ml; all from Invitrogen, Carlsbad, CA). Cells were passaged when 80% confluent using 0.05% trypsin/EDTA (Gibco, Grand Island, NY) and plated in complete Dulbecco's modified Eagle medium without antibiotics to prepare for transfection. For primary OT-I cell cultures, splenocytes were prepared from whole spleens from OT-I/Ly5.1 mice and cultured in complete RPMI-1640 (Invitrogen, Carlsbad, CA) at 37 °C in 5% CO2.
OT-I T cell assay. For receptor-mediated siRNA delivery freshly prepared OT-I total splenocytes were activated overnight with 100 pMSIINFEKL peptide (Anaspec, Fremont, CA) in complete RPMI-1640 (Invitrogen, Carlsbad, CA). After activation, an aliquot of cells was used to determine CD8+ OT-I cell percentage and to confirm 4-1BB expression by staining with anti-mCD8a and anti-m41bb antibody incubated with prostate specific membrane antigen expressing 4T1 cells (eBioscience, San Diego, CA) and flow cytometry. 48 hours post-activation, cells were washed and seeded at 100,000 cells/well into a 96 well plate (NUNC Rochester, NY) in complete RPMI-1640. Cells were pulsed every 8 hours with 4-1BB aptamer-siRNA conjugates (800 nmol/l/pulse) three times by direct addition to the culture medium without transfection reagents. One day after the last treatment cells were harvested by centrifugation, lysed and mRNA isolated using the RNEasy Kit (Qiagen,Valencia, CA).
siCHECK assay. Synthetic 30 nucleotide mRNA sense sequences for target genes were ligated with T4 DNA ligase (New England Biolabs, Beverly, MA) as annealed DNA oligonucleotides with XhoI 5′ and NotI 3′ overhangs into the 3′ end of the gene for Renilla luciferase in the siCHECK-2 vector (Promega, Madison, WI) previously digested with XhoI and NotI restriction enzymes and dephosphorylated with Antarctic phosphatase (New England Biolabs). HEK293T cells were plated in 24 well plates (NUNC, Rochester, NY) and transfected in triplicate with siCHECK containing the candidate target sense sequences (100 ng) and siRNA or conjugates (5 pmol) using Lipofectamine 2000 reagent (Invitrogen, Grand Island, NY). Transfections were halted after 6 hours by addition of 1 ml complete Dulbecco's modified Eagle medium. 24–48 hours post-transfection Renilla and Firefly luciferase activity were measured using the Dual-Glo Luciferase Kit (Promega) according to the manufacturer's instructions. Luminescence values were recorded using a Wallac VICTOR2 1420 multilabel counter (Perkin Elmer, Waltham, MA). The relative Renilla activity was calculated by normalizing each condition to the control Firefly luciferase activity. Candidate siRNA activity was normalized to a corresponding aptamer-control siRNA or a control siRNA where the control was either a green fluorescent protein or pGL3 siRNA that does not have significant recognition of sense sites within the siCHECK-2 target plasmid.
Quantitative reverse-transcription-PCR. HEPA1-6 cells were plated in 6 well plates in Dulbecco's modified Eagle medium without antibiotics and 24 hours later transfected with 50 pmol of siRNA or aptamer-siRNA conjugates using Lipofectamine RNAiMAX (Invitrogen, Grand Island, NY). 24–48 hours after transfection cells were washed twice with Dulbecco-modified phosphate buffered saline −/−, RNA isolated using the RNEasy Kit (Qiagen) and converted to cDNA using the High Capacity cDNA kit (Applied Biosystems, Carlsbad, CA). Predesigned Taqman probe sets for specific genes of interest were used to analyze gene expression by qPCR on a StepOne qPCR device (Applied Biosystems) with β-actin and/or GAPDH or the 18S ribosomal subunit genes used as internal reference controls where indicated. Samples were analyzed using the comparative threshold (Ct) method of analysis with each experimental sample normalized as percent expression of the control groups.
Statistical analysis. GraphPad Prism v5.06 (GraphPad Software, La Jolla, CA) was used to analyze and plot the data and is shown as the median value ± SEM. Unpaired Student's t-tests with two-tailed P values were used to determine statistical power of the results with significance given as P ≤ 0.05.
SUPPLEMENTARY MATERIAL Supplementary Data.
Acknowledgments
The authors declare no commercial affiliation, consulting arrangements, stock, equity or any other potential financial conflict of interest. Funding was provided by a bequest from the Dodson Estate andthe Sylvester Comprehensive Cancer Center (Miller School of Medicine, University of Miami), and a grant (KG090348) from the Susan G. Komen for the Cure of Breast Cancer Foundation.
Supplementary Material
References
- Campoli M, Ferris R, Ferrone S., and, Wang X. Immunotherapy of malignant disease with tumor antigen-specific monoclonal antibodies. Clin Cancer Res. 2010;16:11–20. doi: 10.1158/1078-0432.CCR-09-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastan I, Hassan R, FitzGerald DJ., and, Kreitman RJ. Immunotoxin treatment of cancer. Annu Rev Med. 2007;58:221–237. doi: 10.1146/annurev.med.58.070605.115320. [DOI] [PubMed] [Google Scholar]
- Keefe AD, Pai S., and, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR.et al. (2009Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors Nat Biotechnol 27839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X, Zhang Y, Ribas J, Chowdhury WH, Castanares M, Zhang Z, Laiho M, Deweese TL., and, Lupold SE. Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts. J Clin Invest 121: 2383–2390. 2011. [DOI] [PMC free article] [PubMed]
- Pastor F, Kolonias D, Giangrande PH., and, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–230. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler LA, Trifonova R, Vrbanac V, Basar E, McKernan S, Xu Z.et al. (2011Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras J Clin Invest 1212401–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H.et al. (2011An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice Sci Transl Med 366ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck K, Volper EM, Spengler RM, Long DD, Chan CY, Ding Y.et al. (2009Rational design leads to more potent RNA interference against hepatitis B virus: factors effecting silencing efficiency Mol Ther 17538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Huang Y, Grimm D, Rossi JJ.et al. (2011Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes Proc Natl Acad Sci USA 1089208–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS., and, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO., 2nd, , Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E.et al. (2006Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras Nat Biotechnol 241005–1015. [DOI] [PubMed] [Google Scholar]
- Blidner RA, Hammer RP, Lopez MJ, Robinson SO., and, Monroe WT. Fully 2'-deoxy-2'-fluoro substituted nucleic acids induce RNA interference in mammalian cell culture. Chem Biol Drug Des. 2007;70:113–122. doi: 10.1111/j.1747-0285.2007.00542.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.