Abstract
Haplogroup G, together with J2 clades, has been associated with the spread of agriculture, especially in the European context. However, interpretations based on simple haplogroup frequency clines do not recognize underlying patterns of genetic diversification. Although progress has been recently made in resolving the haplogroup G phylogeny, a comprehensive survey of the geographic distribution patterns of the significant sub-clades of this haplogroup has not been conducted yet. Here we present the haplogroup frequency distribution and STR variation of 16 informative G sub-clades by evaluating 1472 haplogroup G chromosomes belonging to 98 populations ranging from Europe to Pakistan. Although no basal G-M201* chromosomes were detected in our data set, the homeland of this haplogroup has been estimated to be somewhere nearby eastern Anatolia, Armenia or western Iran, the only areas characterized by the co-presence of deep basal branches as well as the occurrence of high sub-haplogroup diversity. The P303 SNP defines the most frequent and widespread G sub-haplogroup. However, its sub-clades have more localized distribution with the U1-defined branch largely restricted to Near/Middle Eastern and the Caucasus, whereas L497 lineages essentially occur in Europe where they likely originated. In contrast, the only U1 representative in Europe is the G-M527 lineage whose distribution pattern is consistent with regions of Greek colonization. No clinal patterns were detected suggesting that the distributions are rather indicative of isolation by distance and demographic complexities.
Keywords: Y-chromosome, haplogroup G, human evolution, population genetics
Introduction
The Y-chromosomal haplogroup G (hg G) is currently defined as one of the 20 standard haplogroups comprising the global Y-chromosome phylogeny.1 The phylogeographic demarcation zone of hg G is largely restricted to populations of the Caucasus and the Near/Middle East and southern Europe. Hg G is most common in the Caucasus with a maximum frequency exceeding 70% in North Ossetians,2, 3 decreasing to 13% in Iran4 and then rapidly dissipating further eastward. Hg G also occurs at frequencies ranging from 5 to 15% in both the rest of Near/Middle East and southern European countries (especially Italy and Greece), with a decreasing frequency gradient towards the Balkans and northern Europe. The presence of hg G was first reported in Europe and Georgia5 and later described in additional populations of the Caucasus.6 Subsequently, several data sets containing hg G-related lineages have been presented in studies of different European populations7, 8, 9, 10 and so on, as well as studies involving several Middle Eastern and South Asian populations.4, 11, 12, 13
Hg G, together with J2 clades, has been associated with the spread of agriculture,5 especially in the European context. However, interpretations based on coarse haplogroup resolution frequency clines are unsophisticated and do not recognize underlying patterns of genetic diversification. The complexity is apparent in both the phylogenetic resolution and geographic patterning within hgs G and J2a. These patterns have been related to different migratory events and demographic processes.2, 10, 11, 14, 15, 16
Although the phylogenetic resolution within hg G has progressed,1, 17 a comprehensive survey of the geographic distribution patterns of significant hg G sub-clades has not been conducted. Here we address this issue with a phylogeographic overview of the distribution of informative G sub-clades from South/Mediterranean Europe, Near/Middle East, the Caucasus and Central/South Asia. The new phylogenetic and phylogeographic information provides additional insights into the demographic history and migratory events in Eurasia involving hg G.
Materials and methods
The present study comprises data from 98 populations totaling 17 577 individuals, of which 1472 were members of hg G. The haplogroup frequency data are presented in Supplementary Table S1. The hg G individuals in Supplementary Table S1 were either first genotyped for this study or updated to present phylogenetic resolution from earlier studies.2, 4, 10, 11, 13, 16, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27 All hg G (M201-derived) samples were genotyped in a hierarchical manner for the following binary markers: M285, P20, P287, P15, L91 P16, M286, P303, U1, L497, M406, Page19, M287 and M377. Specifications for most markers have been previously reported,1, 17, 28 ISOGG 2011 (http://www.isogg.org/tree/). In addition, we introduce five new markers: M426, M461, M485, M527 and M547 (Supplementary Table S2). Furthermore, markers Page94, U5, U8 and L30 were typed in contextually appropriate samples to establish the position of the five new markers within the phylogeny. We genotyped binary markers following PCR amplification, by either Denaturing High Performance Liquid Chromatography, RFLP analysis, Taqman assay (Applied Biosystems, Foster City, CA, USA) or direct Sanger sequencing methodology.
A subset of 693 samples was typed for short tandem repeats of Y-chromosome (Y-STRs) using the 17 STR markers in the Applied Biosystems AmpFlSTR Yfiler Kit according to manufacturer recommendations. Two additional markers, DYS38829, 30 and DYS46131 were typed separately. The fragments were run on the ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems). The results were analyzed using the ABI PRISM program GeneMapper 4.0 (Applied Biosystems). Y-STR haplotypes were used to construct phylogenetic networks for haplogroups G-P303, G-P16 and G-M377, using the program Network 4.6.0.0 (Fluxus-Engineering, Suffolk, England, UK) and applying the median-joining algorithm. The G-P303 phylogenetic network was constructed using 248 G2a3b-P303-derived 19-locus haplotypes from populations representing Europe, Middle/Near East, South/Central Asia and the Caucasus and belonging to five sub-clades P303*, U1, M527, M426 and L497. Similarly, G-P16 and G-M377 networks were created using 104 P16-derived 19-locus haplotypes and 61G-M377-derived 9-locus haplotypes, with both groups representing European, Near/Middle Eastern and central/west Asian populations. The identities of the specific 19 loci that define the STR haplotypes are reported in Supplementary Table S3 and Figure 4 legend.
The coalescent times (Td) of various haplogroups were estimated using the ASDo methodology described by Zhivotovsky et al,32 modified according to Sengupta et al.13 We used the evolutionary effective mutation rate of 6.9 × 10–4 per 25 years, as pedigree rates are arguably only pertinent to shallow rooted familial pedigrees,33 as they do not consider the evolutionary consequences of population dynamics including the rapid extinction of newly appearing microsatellite alleles. Moreover, the accuracy and validity of the evolutionary rate has been independently confirmed in several deep-rooted Hutterite pedigrees.34 Furthermore pedigree rate-based estimates cannot be substantiated, as they are often inconsistent with dateable archeological knowledge, for example, as clearly illustrated regarding the peopling of the Americas.35 Coalescent times based on 10 STR loci (DYS19, DYS388, DYS389I, DYS389b, DYS390, DYS391, DYS392, DYS393, DYS439, DYS461-TAGA counts) and the median haplotypes of specific hg G sub-haplogroups are presented in Supplementary Table S4. For the multi-copy STR DYS389I,II the DYS389b value was DYS389I subtracted from DYS389II. Also for P15* and L91 lineages Td estimates, DYS19 was excluded owing to duplications in these lineages.36
The formula for the coalescence calculations is as follows: Age=25/1000 × ASD0/0.00069.
‘ASD0' is the average squared difference in the number of repeats between all current chromosomes of a sample and the founder haplotype, which is estimated as the median of current haplotypes. ‘25' and ‘0.00069' denote the assumed average generation time in years and the effective mutation rate, respectively, and ‘1000' is used to convert the result of the equation (into thousands of years). SD was also calculated for the age estimates according to the following formula: 25/1000 × √(ASD0 variance)/0.00069.
Such temporal estimates must be viewed with caution owing to differences in individual STR locus mutation rates, sensitivity to rare outlier STR alleles and complexities related to multiple potential founders during a demographic event. Nonetheless, coalescent times provide a valuable/informative relative metric for estimating the time of lineage formation. Spatial frequency maps for hg G sub-clades that attained 10% frequency in at least one population were obtained by applying the haplogroup frequencies from Supplementary Table S1. The frequency data were converted into isofrequency maps using the Surfer software (version 8, Golden Software, Inc., Golden, CO, USA), following the kriging algorithm using advanced options to use bodies of waters as breaklines. Artefactual values below 0% values were not depicted. Spatial autocorrelation analysis was carried out to assess the presence/absence of clines regarding informative G sub-haplogroups. The Moran's I coefficient was calculated using the PASSAGE software v.1.1 (Phoenix, AZ, USA) with binary weight matrix, nine distance classes and random distribution assumption. To accommodate for variability in sample sizes and hg G content, haplogroup diversity was calculated using the method of Nei37 only in the 52 instances when total population sample size exceeded 50 individuals and ≥5 hg G chromosomes were observed.
Principal component analysis based on G sub-haplogroup frequencies was performed using the freeware POPSTR program (http://harpending.humanevo.utah.edu/popstr/).
Results and Discussion
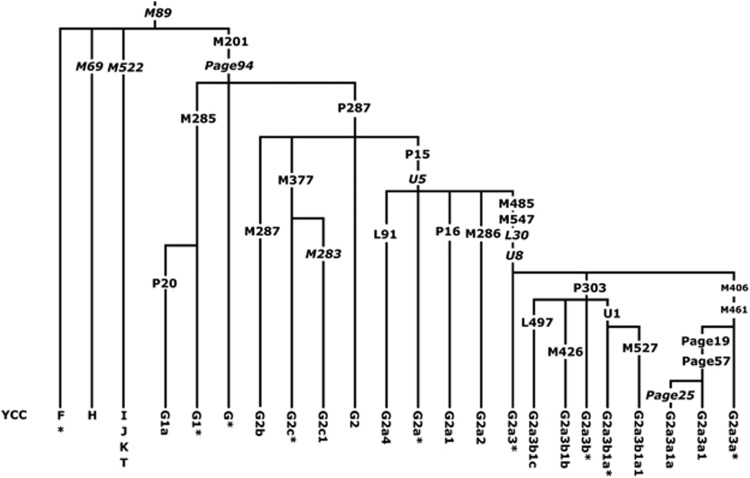
The phylogenetic relationships of the various sub-haplogroups investigated are shown in Figure 1. Notably no basal G-M201*, Page94*(xM285, P287) chromosomes were detected in our data set.
Figure 1.
Phylogenetic relationships of studied binary markers within haplogroup G in wider context of M89-defined clade. The naming of sub-clades is according to YCC nomenclature principles.
We emphasize that our assessments are based solely on contemporary DNA distributions rather than actual prehistoric patterns. Thus inferences regarding migratory histories must be viewed cautiously, as diversities may have changed over the time spans discussed. Nonetheless, our approach using high-resolution phylogenetic relationships as well as their phylogeography to infer the possible origin of a genetic variant provides a more plausible deduction than simply the region of highest frequency. We attempted to localize the potential geographic origin of haplogroup G-M201 by considering those locations containing both G1-M285- and G2-P287-related lineages as well as the co-occurrence of high sub-haplogroup diversity. Specifically, we intersected these criteria by applying the following filters. First, we calculated haplogroup diversity using data in Supplementary Table S1 for the 52 instances when total population sample size exceeded 50 individuals and ≥5 hg G chromosomes were observed. Then we applied a 10% overall hg G frequency threshold and the additional specification that both haplogroup G1 and G2 lineages also be present. In the ten remaining populations, haplogroup diversity spanned from a low of 0.21 in Adyghes, to highs of 0.88 in Azeris (Iran) and 0.89 in eastern Anatolia and 0.90 in Armenia. We estimate that the geographic origin of hg G plausibly locates somewhere nearby eastern Anatolia, Armenia or western Iran. The general frequency pattern of hg G overall (Figure 2a) shows that the spread of hg G extends over an area from southern Europe to the Near/Middle East and the Caucasus, but then decreases rapidly toward southern and Central Asia.
Figure 2.
(a)–(f) Spatial frequency maps of haplogroup G (hg G) and its sub-clades with frequencies over 10%. In the case of the general frequency pattern of hg G, panel (a) was obtained by applying the frequencies from Supplementary Table S1 together with data taken from the literature, concerning 569 individuals representing 7 populations comprising Algerians,47 Oromo and Amhara Ethiopians,48 and Berbers, Arabs and Saharawis from Morocco.49 Dots on the map (a) indicate the approximate locations of the sampled populations. Spatial frequency maps for sub-clades (panels b–f) were obtained by applying the frequencies from Supplementary Table S1 using the Surfer software (version 8, Golden Software, Inc.), following the kriging algorithm with option to use bodies of water as breaklines.
Although not exceeding 3% frequency overall, haplogroup G1-M285 reflects a branching event that is phylogenetically equivalent to the more widespread companion G2-P287 branch in the sense that both branches coalesce directly to the root of G-M201. Although the low frequency of hg G1-M285 makes it impractical to justify displaying a spatial frequency map, it is found (Supplementary Table S1) in the Near/Middle East including Anatolia, the Arabian Peninsula and Persian Gulf region, as well as Iran and the South Caucasus (mostly Armenians). Although hg G1 frequency distribution, overall, extends further eastward as far as Central Asian Kazakhs (present even among Altaian Kazakhs38 with identical STR haplotypes compared with the main Kazakh population), it is virtually absent in Europe. Although the present-day frequency of G1 is low across its spread zone, the expansion time estimate (Supplementary Table S4) of 19 271±6158 years attests to considerable antiquity.
In contrast to G1, the absolute majority of hg G samples belonged to G2-P287-related sub-clades, with the vast majority of them being associated with G2a-P15-related lineages. Using Y-STR data, the Td expansion time for all combined P15-affiliated chromosomes was estimated to be 15 082±2217 years ago. Important caveats to consider include the fact that Td is sensitive to authentic rare outlier alleles and that multiple founders during population formation will inflate the age estimate of the event. Thus, these estimates should be viewed as the upper bounds of dispersal times. Considering these issues, we acknowledge that the variance of the age estimates may be underestimated. While neither knowledge of paleo-climate, archeology or genetic evidence from a single locus using modern populations provides an unimpeachable microcosm of pre-historical expansions, considering them together cautiously provides a contextual framework for discussion.
The suggested relevant pre-historical climatic and archeological periods specified in conjunction with lineage-specific estimated expansion times are specified in the summary portion of Supplementary Table S4.
The G2 clade consists of one widespread but relatively infrequent collection of P287*, M377, M286 and M287 chromosomes versus a more abundant assemblage consisting of G2a-related P15*, P16 and M485-related lineages. A network of 61 G2c-M377 lineages from Europe, the Near/Middle East and Central and South Asia reveals founder lineages (one pronounced founder in Ashkenazi Jews and a far distant one among South Asian individuals) and diverged lineages (Supplementary Figure S1). The corresponding coalescent estimate for M377 is 5600 years ago (Supplementary Table S4). Unresolved G2a-P15* lineages occur across a wide area extending from the Near/Middle East to the Balkans and Western Europe in the west, the Caucasus (especially the South Caucasus) in the north and Pakistan in the east. Although both broadly distributed, G2a-P15* and its downstream L91 sub-lineage have low frequencies, with the exception of Sardinia and Corsica. It is notable that Ötzi the 5300-year-old Alpine mummy was derived for the L91 SNP and his autosomal affinity was nearest to modern Sardinians.28
The G2a2-M286 lineage is very rare, so far detected only in some individuals in Anatolia and the South Caucasus. On the other hand, G2a3-M485-associated lineages, or more precisely its G2a3b-P303-derived branch, represent the most common assemblage, whereas the paraphyletic G2a3-M485* lineages display overall low occurrence in the Near/Middle East, Europe and the Caucasus. Interestingly, the L30 SNP, phylogenetically equivalent to M485, M547 and U8, was detected in an approximately 7000-year-old Neolithic specimen from Germany, although this ancient DNA sample was not resolved further to additional sub-clade levels.39
Geographic spread patterns of the P303-derived groups defined by L497, U1 and P15(xP303)-derived P16 and M406 lineages, all of which achieve a peak frequency of at least 10%, are presented in Figures 2b–f, respectively. These five major sub-clades of the G2 branch show distinct distribution patterns over the whole area of their spread. However, no clinal patterns were detected in the spatial autocorrelation analysis of the five sub-haplogroup frequencies with distance, suggesting that the distributions are not clinal but rather indicative of isolation by distance and demographic complexities. This is not surprising, as clines are not expected in cases of sharp changes in haplogroup frequency over a relatively small distance such as those observed for hg G, for instance between the Caucasus and Eastern Europe.
The overall coalescent age estimate (Supplementary Table S4) for P303 is 12 600 years ago. Although compared with G1-M285, the phylogenetic level of P303 (Figure 1) is shallower but its geographic spread zone covers the whole hg G distribution area (Figure 2b). The highest frequency values for P303 are detected in populations from Caucasus region, being especially high among South Caucasian Abkhazians (24%) and among Northwest (NW) Caucasian Adyghe and Cherkessians—39.7% and 36.5%, respectively. In the Near/Middle East, the highest P303 frequency is detected among Palestinians (17.8%), whereas in Europe the frequency does not exceed 6%.
Another frequent sub-clade of the G2a3-M485 lineage is G2a3a-M406 (Figure 2e). In contrast to its widely dispersed sister clade defined by P303, hg G-M406 has a peak frequency in Cappadocia, Mediterranean Anatolia and Central Anatolia (6–7%) and it is not detected in most other regions with considerable P303 frequency. The expansion time of G-M406 in Anatolia is 12 800 years ago, which corresponds to climatic improvement at the beginning of the Holocene and the commencement of sedentary hunter-forager settlements at locations, such as Gobekli Tepi in Southeast Anatolia, thought to be critical for the domestication of crops (wheat and barley) that propelled the development of the Neolithic. G2a3a-M406 has a modest presence in Thessaly and the Peloponnese (4%),10 areas of the initial Greek Neolithic settlements. More distantly, G2a3a-M406 occurs in Italy (3%) with a Td of 8100 years ago, consistent with the model of maritime Neolithic colonization of the Italian peninsula from coastal Anatolia and/or the Levant. Finally, to the east, G2a3a-M406 has an expansion time of 8800 years ago in Iran, a time horizon that corresponds to the first Neolithic settlements of the Zagros Mountains of Iran. Thus, G2a3a-M406, along with other lineages, such as J2a3b1-M92 and J2a4h2-DYS445=616, may track the expansion of the Neolithic from Central/Mediterranean Anatolia to Greece/Italy and Iran.
Concerning the presence of hg G in the Caucasus, one of its distinguishing features is lower haplogroup diversity in numerous populations (Supplementary Table S1) compared with Anatolia and Armenia, implying that hg G is intrusive in the Caucasus rather than autochthonous. Another notable feature is its uneven distribution. Hg G is very frequent in NW Caucasus and South Caucasus, covering about 45% of the paternal lineages in both regions2 in this study. Conversely, hg G is present in Northeast Caucasus only at an average frequency of 5% (range 0–19%). Interestingly, the decrease of hg G frequency towards the eastern European populations inhabiting the area adjacent to NW Caucasus, such as southern Russians and Ukrainians,18, 40 is very rapid and the borderline very sharp, indicating that gene flow from the Caucasus in the northern direction has been negligible. Moreover, these general frequencies mostly consist of two notable lineages. First, the G2a1-P16 lineage is effectively Caucasus specific and accounts for about one-third of the Caucasian male gene pool (Figure 2f). G-P16 has a high frequency in South and NW Caucasus, with the highest frequency among North Ossetians—63.6%. G-P16 is also occasionally present in Northeast Caucasus at lower frequencies (Supplementary Table S1), consistent with a previous report.3 Outside the Caucasus, hg G-P16 occurs at ≥1% frequency only in Anatolia, Armenia, Russia and Spain, while being essentially absent elsewhere. A network analysis of representative hg G-P16 Y-STR haplotypes reveals a diffuse cluster (Supplementary Figure S2). The coalescence age estimate of 9400 years for P16 coincides with the early Holocene (Supplementary Table S4). The second common hg G lineage in the Caucasus is U1, which has its highest frequencies in the South (22.8% in Abkhazians) and NW Caucasus (about 39.7% in Adyghe and 36.5% in Cherkessians), but also reaches the Near/Middle East with the highest frequency in Palestinians (16.7%) and, shows extremely low frequency in Eastern Europe.
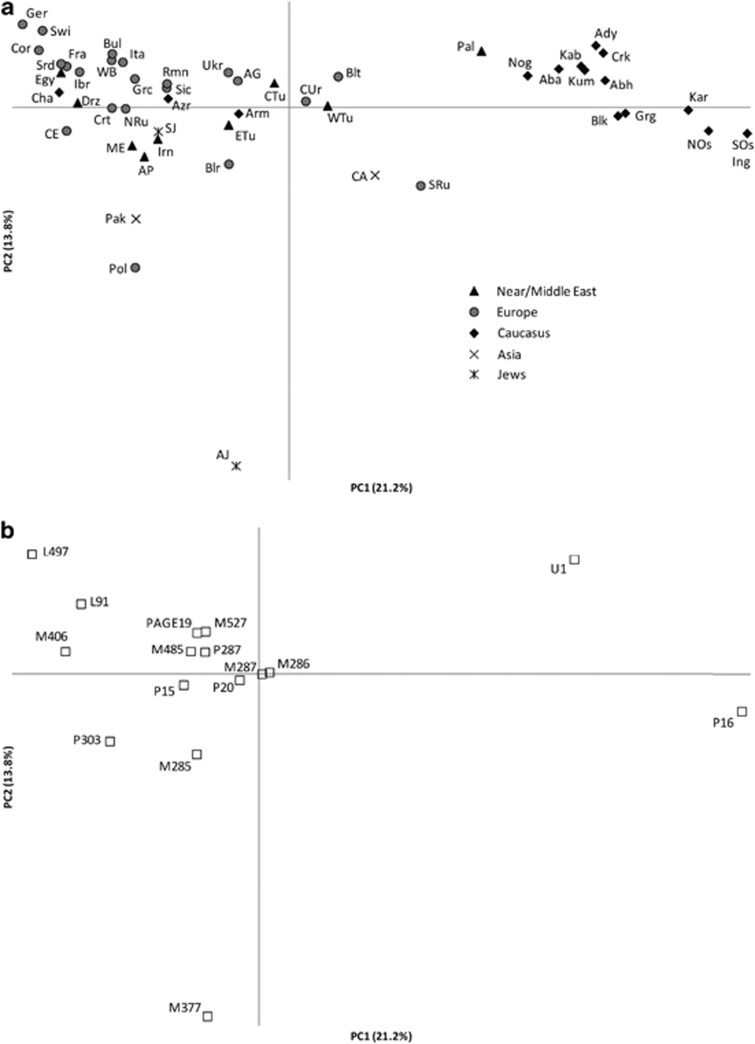
We performed principal component analysis to determine the affinities of various hg G fractions with respect to total M201 among different populations, using the frequency distributions of the following sub-clades: M285, P20, M377, M287, P287, P15*, P16, M286, M485, P303*, L497, U1*, M527, M406 and Page19. The first principal component separates the populations of the Caucasus from those of Europe, with the Near/Middle Eastern populations being intermediate (Figure 3a). The second component, influenced by the relatively high presence of M377, separates Ashkenazi Jews from other populations (Figure 3a). A plot of the sub-clades included in the principal component analysis (Figure 3b) indicates that the clustering of the populations from NW Caucasus is due to their U1* frequency, whereas L497 lineages account for the separation of central Europeans. Furthermore, the U1-specific sub-clade M527 is most pronounced among Ukrainians and Anatolian Greeks.
Figure 3.
(a) Principal component analysis by population. The 96 populations were collapsed into 50 regionally defined populations by excluding populations where the total G count was less than n=5. Population codes: Baltics (Blt), Belarusians (Blr), Poles (Pol), Ukrainians (Ukr), northern Russians (NRu), southern and central Russians (SRu), Circum-Uralic (CUr), Germans (Ger), Central Europeans (CE), Iberians (Ibr), French (Fra), Sardinians (Srd), Corsica (Cor), Sicilians (Sic), Italians (Ita), Switzerlands (Swi), Western Balkans (WB), Romanians (Rmn), Bulgarians (Bul), Crete (Crt), Greeks (Grc), Anatolian Greeks (AG), Egyptians (Egy), Near/Middle Easterners (ME), Ashkenazi Jews (AJ), Sephardic Jews (SJ), Arabian Peninsula (AP), Palestinians (Pal), Druze (Drz), Western Turks (WTu), Central Turks (CTu), Eastern Turks (ETu), Iranians (Irn), Abkhazians (Abh), Armenians (Arm), Georgians (Grg), South Ossetians (SOs), Iranian Azeris (Azr), Abazins (Aba), Adyghes (Ady), Balkars (Blk), Cherkessians (Crk), Kabardins (Kab), Karachays (Kar), Kuban Nogays (Nog), North Ossetians (NOs), Chamalals (Cha), Ingushes (Ing), Kumyks (Kum), Central Asians (CA), Pakistani (Pak). (b) Principal component analysis by hg G sub-clades: (A) M285, P20, P287, P15, L92 P16, M286, M485, P303, U1, L497, M527, M406, Page19, M287 and M377 sub-haplogroups with respect to total M201.
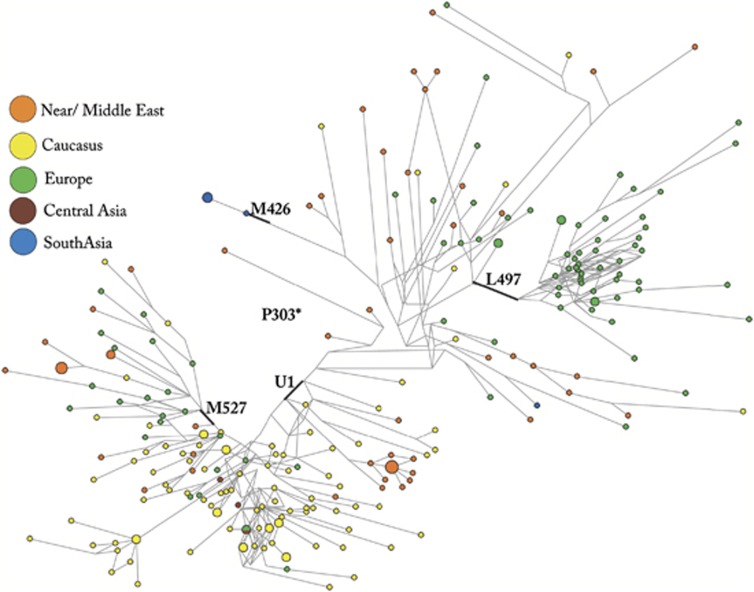
In the G2a3b-P303 network (Figure 4), there are several region-specific clusters, indicating a considerable history for this SNP. Taken as a collective group, P303-derived chromosomes are the most widespread of all hg G lineages (Supplementary Table S1 and Figure 2b) and clearly display differential geographic partitioning between L497 (Figure 2c) and U1 (xM527) (Figure 2d). Looking still more closely at the distribution of P303 sub-clades, some distinct patterns emerge in the network (Figure 4). The non-clustering paraphyletic, hg G sub-group P303* residuals consist of samples from Near/Middle Eastern, Caucasian and European populations. Its estimated Td of 12 095±3000 years ago suggests considerable antiquity allowing time to accumulate STR diversity and also to disperse relatively widely. The hg G-U1 subclade is characterized by several sub-clusters of haplotypes, including a more diverse cluster mostly represented by Caucasus populations. A more compact cluster of Near/Middle Eastern samples is also resolved in the network. The M527-defined sub-clade is unusual in that it reflects the presence of hg G-U1 that is otherwise rare in Europe. Although M527 frequency (Supplementary Table S1) is relatively low (1–6%), its phylogeographic distribution in regions such as southern Italy, Ukraine and the Levant (Druze and Palestinians) often coincides with areas associated with the Neolithic and post-Neolithic expansions into the Greek Aegean beginning approximately 7000 years ago.41 The expansion time (Td) of M527 is 7100±2300 years ago and is consistent with a Middle to Late Neolithic expansion of M527 in the Aegean. The presence of M527 in Provence, southern Italy and Ukraine may reflect subsequent Greek maritime Iron Age colonization events16 and perhaps, given its appearance among the Druze and Palestinians, even episodes associated with the enigmatic marauding Sea Peoples.42
Figure 4.
Network of 248 samples P303 derived from Supplementary Table S3. The network was obtained using the biallelic markers P303, M426, L497, U1, M527 and 19 STR loci (DYS19, DYS388, DYS389I, DYS389b, DYS390, DYS391, DYS392, DYS393, DYS439, DYS461 (TAGA counts), DYS385a,b, DYS437, DYS438, DYS448, DYS456, DYS458, DYS635, YGATAH4). The Network 4.6.0.0 (Fluxus-Engineering) program was used (median-joining algorithm and the post-processing option). Circles represent microsatellite haplotypes, the areas of the circles and sectors are proportional to haplotype frequency (smallest circle corresponds to one individual) and the geographic area is indicated by color.
The hg G2a3b1c-L497 sub-cluster, on the other hand, has so far been found essentially in European populations and therefore is probably autochthonous to Europe. While acknowledging that the inference of the age and geographic source of dispersals of Y chromosome haplogroups from the frequency and STR diversity data can be approximate at best, we speculate that this lineage could potentially be associated with the Linearbandkeramik (LBK) culture of Central Europe, as its highest frequency (3.4–5.1%) and Td estimate (Supplementary Table S4) of 10 870±3029 years ago occur there. Whereas the presence of Mideastern mtDNA in Tuscany43 supports the model of early Iron Age migrants from Anatolia (putative Etruscans) colonizing Central Italy,44 the occurrence of the G2a3b1c-L497 lineage in Italy is most likely associated to migratory flows from the north. An assessment of the Y-chromosome phylogeography-based proposal that the spread of G2a-L497 chromosomes originated from Central Europe could be achieved by typing this SNP in the Holocene period human remains from Germany31 as well as those from France and Spain.45, 46 Certainly, Y chromosome represents only a small part of human genome and any population-level interpretation of gene flow in this region would have to be supported by genome-wide evidence.
Acknowledgments
JD and JC were supported by ANR program AFGHAPOP No BLAN07-9_222301'. RV and DMB thank the European Commission, Directorate-General for Research for FP7 Ecogene grant 205419. RV thanks the European Union Regional Development Fund for support through the Centre of Excellence in Genomics, the Estonian Ministry of Education and Research for the Basic Research grant SF 0270177As08. SR thanks the Estonian Science Foundation for grant 7445 and M Metspalu for grant 8973. EKK thanks the Russian Academy of Sciences Program for Fundamental Research ‘Biodiversity and dynamics of gene pools', the Ministry of Education and Science of the Russian Federation for state contracts P-325 and 02.740.11.07.01, and the Russian Foundation for Basic Research for grants 04-04-48678-а and 07-04-01016-а. IK thanks the Russian Foundation for Basic Research for grant 08-06-97011 and the Grant of the President of the Russian Federation of state support for young Russian scientists MK-488.2006.4. OS thanks the Italian Ministry of the University: Progetti Ricerca Interesse Nazionale 2009 and FIRB-Futuro in Ricerca 2008 and Fondazione Alma Mater Ticinensins. AAL thanks the Sorenson Molecular Genealogy Foundation. PAU thanks Professor Carlos D Bustamante. MH and MHS are thankful to the ‘National Institute for Genetic Engineering and Biotechnology', Tehran, Iran, and the ‘National Research Institute for Science policy', Tehran, Iran, for providing the samples.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunusbayev B, Metspalu M, Järve M, et al. The Caucasus as an asymmetric semipermeable barrier to ancient human migrations. Mol Biol Evol. 2011;29:359–365. doi: 10.1093/molbev/msr221. [DOI] [PubMed] [Google Scholar]
- Balanovsky O, Dibirova K, Dybo A, et al. Parallel evolution of genes and languages in the Caucasus region. Mol Biol Evol. 2011;28:2905–2920. doi: 10.1093/molbev/msr126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regueiro M, Cadenas AM, Gayden T, Underhill PA, Herrera RJ. Iran: tricontinental nexus for Y-chromosome driven migration. Hum Hered. 2006;61:132–143. doi: 10.1159/000093774. [DOI] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, et al. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- Nasidze I, Quinque D, Dupanloup I, et al. Genetic evidence concerning the origins of South and North Ossetians. Ann Hum Genet. 2004;68:588–599. doi: 10.1046/j.1529-8817.2004.00131.x. [DOI] [PubMed] [Google Scholar]
- Goncalves R, Freitas A, Branco M, et al. Y-chromosome lineages from Portugal, Madeira and Acores record elements of Sephardim and Berber ancestry. Ann Hum Genet. 2005;69:443–454. doi: 10.1111/j.1529-8817.2005.00161.x. [DOI] [PubMed] [Google Scholar]
- Capelli C, Brisighelli F, Scarnicci F, et al. Y chromosome genetic variation in the Italian peninsula is clinal and supports an admixture model for the Mesolithic-Neolithic encounter. Mol Phylogenet Evol. 2007;44:228–239. doi: 10.1016/j.ympev.2006.11.030. [DOI] [PubMed] [Google Scholar]
- Battaglia V, Fornarino S, Al-Zahery N, et al. Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur J Hum Genet. 2009;17:820–830. doi: 10.1038/ejhg.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RJ, Ozcan SS, Carter T, et al. Differential Y-chromosome Anatolian influences on the Greek and Cretan Neolithic. Ann Hum Genet. 2008;72:205–214. doi: 10.1111/j.1469-1809.2007.00414.x. [DOI] [PubMed] [Google Scholar]
- Cinnioglu C, King R, Kivisild T, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- Hammer MF, Behar DM, Karafet TM, et al. Extended Y chromosome haplotypes resolve multiple and unique lineages of the Jewish priesthood. Hum Genet. 2009;126:707–717. doi: 10.1007/s00439-009-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, et al. Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Magri C, Benuzzi G, et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet. 2004;74:1023–1034. doi: 10.1086/386295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalloua PA, Xue Y, Khalife J, et al. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am J Hum Genet. 2008;82:873–882. doi: 10.1016/j.ajhg.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RJ, DiCristofaro J, Kouvatsi A, et al. The coming of the Greeks to Provence and Corsica: Y-chromosome models of archaic Greek colonization of the western Mediterranean. BMC Evol Biol. 2011;11:69. doi: 10.1186/1471-2148-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims LM, Garvey D, Ballantyne J. Improved resolution haplogroup G phylogeny in the Y chromosome, revealed by a set of newly characterized SNPs. PLoS One. 2009;4:e5792. doi: 10.1371/journal.pone.0005792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanovsky O, Rootsi S, Pshenichnov A, et al. Two sources of the Russian patrilineal heritage in their Eurasian context. Am J Hum Genet. 2008;82:236–250. doi: 10.1016/j.ajhg.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores C, Maca-Meyer N, Gonzalez AM, et al. Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography. Eur J Hum Genet. 2004;12:855–863. doi: 10.1038/sj.ejhg.5201225. [DOI] [PubMed] [Google Scholar]
- Barac L, Pericic M, Klaric IM, et al. Y chromosomal heritage of Croatian population and its island isolates. Eur J Hum Genet. 2003;11:535–542. doi: 10.1038/sj.ejhg.5200992. [DOI] [PubMed] [Google Scholar]
- Pericic M, Lauc LB, Klaric IM, Janicijevic B, Rudan P. Review of croatian genetic heritage as revealed by mitochondrial DNA and Y chromosomal lineages. Croat Med J. 2005;46:502–513. [PubMed] [Google Scholar]
- Martinez L, Underhill PA, Zhivotovsky LA, et al. Paleolithic Y-haplogroup heritage predominates in a Cretan highland plateau. Eur J Hum Genet. 2007;15:485–493. doi: 10.1038/sj.ejhg.5201769. [DOI] [PubMed] [Google Scholar]
- Luis JR, Rowold DJ, Regueiro M, et al. The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet. 2004;74:788–788. doi: 10.1086/382286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Yunusbayev B, Metspalu M, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- Cadenas AM, Zhivotovsky LA, Cavalli-Sforza LL, Underhill PA, Herrera RJ. Y-chromosome diversity characterizes the Gulf of Oman. Eur J Hum Genet. 2008;16:374–386. doi: 10.1038/sj.ejhg.5201934. [DOI] [PubMed] [Google Scholar]
- Chiaroni J, King RJ, Myres NM, et al. The emergence of Y-chromosome haplogroup J1e among Arabic-speaking populations. Eur J Hum Genet. 2010;18:348–353. doi: 10.1038/ejhg.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, et al. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet. 2003;72:313–332. doi: 10.1086/346068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Graefen A, Ball M, et al. New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 2012. p. 3. [DOI] [PubMed]
- de Knijff P, Kayser M, Caglia A, et al. Chromosome Y microsatellites: population genetic and evolutionary aspects. Int J Legal Med. 1997;110:134–149. doi: 10.1007/s004140050052. [DOI] [PubMed] [Google Scholar]
- Kayser M, Caglia A, Corach D, et al. Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med. 1997;110:141–149. doi: 10.1007/s004140050051. [DOI] [PubMed] [Google Scholar]
- White PS, Tatum OL, Deaven LL, Longmire JL. New, male-specific microsatellite markers from the human Y chromosome. Genomics. 1999;57:433–437. doi: 10.1006/geno.1999.5782. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioglu C, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Feldman MW. Difference between evolutionarily effective and germ line mutation rate due to stochastically varying haplogroup size. Mol Biol Evol. 2006;23:2268–2270. doi: 10.1093/molbev/msl105. [DOI] [PubMed] [Google Scholar]
- Pichler I, Fuchsberger C, Platzer C, et al. Drawing the history of the Hutterite population on a genetic landscape: inference from Y-chromosome and mtDNA genotypes. Eur J Hum Genet. 2010;18:463–470. doi: 10.1038/ejhg.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulik MC, Zhadanov SI, Osipova LP, et al. Mitochondrial DNA and Y Chromosome Variation Provides Evidence for a Recent Common Ancestry between Native Americans and Indigenous Altaians. Am J Hum Genet. 2012;90:573. doi: 10.1016/j.ajhg.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli C, Brisighelli F, Scarnicci F, Blanco-Verea A, Brion M, Pascali VL. Phylogenetic evidence for multiple independent duplication events at the DYS19 locus. Forensic Sci Int-Gen. 2007;1:287–290. doi: 10.1016/j.fsigen.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Dulik MC, Osipova LP, Schurr TG. Y-chromosome variation in Altaian Kazakhs reveals a common paternal gene pool for Kazakhs and the influence of Mongolian expansions. PLoS One. 2011;6:e17548. doi: 10.1371/journal.pone.0017548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W, Balanovsky O, Sanchez JJ, et al. Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS Biol. 2010;8:e1000536. doi: 10.1371/journal.pbio.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharkov VN, Stepanov VA, Borinskaya SA, et al. Gene pool structure of Eastern Ukrainians as inferred from the Y-chromosome haplogroups. Russ J Genet. 2004;40:326–331. [Google Scholar]
- Cavalli-Sforza L, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton: Princeton University Press; 1994. [Google Scholar]
- Kaniewski D, Van Campo E, Van Lerberghe K, et al. The Sea Peoples, from cuneiform tablets to carbon dating. PLoS One. 2011;6:e20232. doi: 10.1371/journal.pone.0020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilli A, Olivieri A, Pala M, et al. Mitochondrial DNA variation of modern Tuscans supports the near eastern origin of Etruscans. Am J Hum Genet. 2007;80:759–768. doi: 10.1086/512822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernesi C, Caramelli D, Dupanloup I, et al. The Etruscans: a population-genetic study. Am J Hum Genet. 2004;74:694–704. doi: 10.1086/383284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacan M, Keyser C, Ricaut FX, et al. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proc Natl Acad Sci USA. 2011;108:9788–9791. doi: 10.1073/pnas.1100723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacan M, Keyser C, Ricaut FX, et al. Ancient DNA suggests the leading role played by men in the Neolithic dissemination. Proc Natl Acad Sci USA. 2011;108:18255–18259. doi: 10.1073/pnas.1113061108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet. 2000;67:1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J. High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet. 2001;68:1019–1029. doi: 10.1086/319521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.