Abstract
Protein-o-mannosyl transferase 1 (POMT1) is a glycosyltransferase involved in α-dystroglycan (α-DG) glycosylation. Clinical phenotype in POMT1-mutated patients ranges from congenital muscular dystrophy (CMD) with structural brain abnormalities, to limb-girdle muscular dystrophy (LGMD) with microcephaly and mental retardation, to mild LGMD. No cardiac involvement has until now been reported in POMT1-mutated patients. We report three patients who harbored compound heterozygous POMT1 mutations and showed left ventricular (LV) dilation and/or decrease in myocardial contractile force: two had a LGMD phenotype with a normal or close-to-normal cognitive profile and one had CMD with mental retardation and normal brain MRI. Reduced or absent α-DG immunolabeling in muscle biopsies were identified in all three patients. Bioinformatic tools were used to study the potential effect of POMT1-detected mutations. All the detected POMT1 mutations were predicted in silico to interfere with protein folding and/or glycosyltransferase function. The report on the patients described here has widened the clinical spectrum associated with POMT1 mutations to include cardiomyopathy. The functional impact of known and novel POMT1 mutations was predicted with a bioinformatics approach, and results were compared with previous in vitro studies of protein-o-mannosylase function.
Keywords: POMT1, LGMD, CMD, cardiomyopathy, α-dystroglycan glycosylation
Introduction
Protein-o-mannosyl transferase 1 (POMT 1, OMIM *607423), together with its homolog POMT2 (OMIM *607439), is part of a heteromeric complex involved in the initiation of o-mannosyl glycan synthesis in the endoplasmic reticulum,1, 2 which is the first step in the attachment of o-mannose-linked glycan moieties to α-dystroglycan (α-DG).3 α-DG has a structural role in muscle fiber integrity, connecting the dystrophin–glycoprotein complex to the extracellular matrix.4 POMT1 mutations result in a reduction in α-DG glycosylation in skeletal muscles5, 6, 7 and the clinical spectrum of the phenotype ranges from severe Walker–Warburg syndrome (WWS, OMIM 236670)5 to milder forms of congenital muscular dystrophy (CMD) with microcephaly and mental retardation without eye abnormalities (CMD-MR, OMIM 613155),6 and to limb-girdle muscular dystrophy (LGMD) with normal brain structure and mental retardation (LGMD2K, OMIM 609308).7, 8
Five other genes – POMT2, protein o-mannose β-1, 2-N-acetylglucosaminyltransferase (POMGnT1, OMIM *606822), fukutin (FKTN, OMIM *607440), FKTN-related protein (FKRP, OMIM *606596) and like-glycosyltransferase (LARGE, OMIM *608840) – are involved in α-DG glycosylation and their mutations lead to heterogeneous phenotypes characterized by combinations of muscular, cerebral and ophthalmic involvement.9 Cardiac involvement has until now been reported only in patients with FKRP10, 11, 12, 13 and FKTN14 gene mutations. Furthermore, autosomal recessive mutations in genes previously involved in multisystemic congenital (N-) glycosylation disorders (CGDs) have been recently linked to impaired o-mannosylation of dystroglycan, with a cardiomyopathic phenotype, associated to muscular dystrophy (dolichol–phosphate–mannose synthase complex, OMIM *605951)15 or without significant muscular involvement (dolichol kinase, OMIM *610746).16
Overall, cardiomyopathy is increasingly being described in the clinical spectrum of glycosyltransferase mutations. The present is a report on three unrelated patients affected by cardiomyopathy in association with compound heterozygous POMT1 mutations, with different neuromuscular phenotypes.
Materials and methods
Patients
We screened archived muscle biopsies of 247 patients affected by LGMD, CMD, muscle weakness or creatine kinase (CK) elevation of unknown cause for α-DG glycosylation defect by immunohistochemistry. Dystrophin, α-sarcoglycan, calpain and dysferlin were normal by immunohistochemistry and/or immunoblotting in patients' biopsy. A mild to complete reduction of immunolabeling for a glycosylated epitope of α-DG was found in 107 patients, who were subsequently screened for mutations in glycosyltransferase genes. POMT1 mutations were found in nine patients, distributed by phenotype as follows: three LGMD, four CMD with mental retardation and normal brain MRI and two WWS. All these patients routinely undergo a periodic screening for cardiological abnormalities by EKG end echocardiography; three patients showing signs of cardiomyopathy were selected for the present study (Table 1).
Table 1. Patients' clinical and molecular features.
| POMT1 mutations | |||||||
|---|---|---|---|---|---|---|---|
| Patient | Disease onset (years) | Current age | Phenotype | Mentation/brain MRI | Cardiomyopathy onset (years) | Nucleotide change | Amino-acid change |
| 1 | Birth | 17 | CMD-MR | MR/ microcephaly | 17 | c.2005G>A c.1241+1G>A | p.Ala699Thr p.His384_Thr414del |
| 2 | 3 | 20 | LGMD-MR | Slight MR/ normal | 12 | c.430A>G c.1241C>T | p.Asn144Asp p.Thr414Met |
| 3 | 33 | 34 | LGMD-NORM | Normal/normal | 34 | c.1864C>T r(=,1052_1053insGTAAG) | p.Arg622* =, p.Ile351Metfs15 |
Abbreviations: CMD-MR, congenital muscular dystrophy with mental retardation; LGMD-MR, limb-girdle muscular dystrophy with mental retardation; LGMD-NOMR, LGMD with no mental retardation; MR, mental retardation; MRI, magnetic resonance imaging; POMT1, protein-o-mannosyltransferase 1.
Patient 1 is a 17-year-old boy described in a previous report.17 Hypotonic at birth, the patient acquired stable head control at 8 months and the ability to sit unsupported at 15 months, but never learned to walk. He had severe mental retardation and autistic features. A brain MRI carried out at 6 years of age was normal. On neurological examination, diffuse muscle wasting, muscle weakness, mild calf hypertrophy, severe scoliosis with rigid spine and microcephaly were present. Tendon reflexes were normal. Serum CK was 6000 U/l. A muscle biopsy, taken when the patient was 12 years old, revealed dystrophic features and reduced immunolabeling of α-DG and dystrophin. Dystrophin gene analysis did not identify any mutations and the observed slight decreased of dystrophin immunostaining was probably secondary to nonspecific proteolysis.
At the age of 16, the patient was admitted to the hospital for respiratory distress. Nocturnal noninvasive ventilation was begun and a gastrostomy was carried out because of severe swallowing disturbances. An electrocardiogram (EKG) and an echocardiography carried out at that time were normal. After 1 year, the patient developed acute respiratory distress, prompting a complete cardiac evaluation. An echocardiography showed a moderate LV dysfunction (LV end diastolic volume index (LVEDVi) 50 ml/m2: n.v.<70 ml/m2; LV ejection fraction (LVEF) 40%: n.v.≥50%) but a poor acoustic window due to scoliosis did not permit assessment of right ventricular (RV) function. Diuretic therapy was begun and cardiac ultrasound performed 6 months later demonstrated stable parameters (LVEDV index 56 ml/ m2, LVEF 44%).
Patient 2 is a 20-year-old man who showed normal psychomotor development, who had come to medical attention at the age of 3 because of the occasional finding of elevated CK levels (∼10,000 U/l). At the age of 5 years a muscle biopsy showed mild myopathic alterations and perimysial fibrosis. Immunohistochemical analysis of dystrophin, α-, β- and γ-sarcoglycan and β-dystroglycan was normal. Dystrophin gene analysis did not identify any mutations.
At the age of 12 years the patient, until then asymptomatic, underwent a routine echocardiography, which documented a diffuse LV wall hypokinesia with normal LVEDVi (69 ml/m2) and LVEF (50%). He presented at the age of 17 years with shortness of breath, cough, easy fatigability and abdominal pain. An electrocardiogram showed LV hypertrophy (voltage criteria Sokolow-Lyon index=38 mm, n.v.⩽35 mm), and an echocardiography showed a moderate LV dilation (LVEDVi 81 ml/m2) with moderate–severe systolic dysfunction (LVEF 36%) as well as moderate RV dilation (RVEDVi 88 ml/m2, n.v.⩽60 ml/m2). The patient responded to β-blockers and angiotensin receptor 1 blocker (sartanics) therapy. A cardiac echo carried out when the patient was 20 years of age showed a LVEDVi of 92 ml/m2 with an EF of 47% and mild hypokinesia of LV walls. The RV was moderately dilated (RVEDi 98 ml/m2) and kinesis was normal (RVEF 70%). Conventional spirometry showed mild obstruction and a normal forced vital capacity.
Currently, the patient has no difficult rising from the floor or climbing stairs. A neurological examination showed calf and thigh hypertrophy, relative wasting of the scapulo-humeral girdle, and a mild symmetrical weakness of proximal muscles. A brain MRI was normal, but neuropsychological evaluation showed executive dysfunctions (categorization ability, set-shifting and planning) and significant visuo-spatial learning impairment. The patient's IQ was in the normal range (82).
Patient 3 is a 34-year-old man who was well until the age of 33 years, when he began to complain muscle weakness in the lower limbs and myalgias in the shoulder girdle. Serum CK was 981 U/l and a muscle biopsy was consistent with a severe myopathy with type I fiber predominance (90%), central nuclei and cores in the majority of fibers. A neurological examination revealed calf hypertrophy and moderate weakness of bilateral triceps brachii. He had no difficulty rising from the floor, walking long distances or climbing stairs.
A diagnosis of an initial biventricular dilatation was made when the patient was 34 years old on the basis of a cardiac echo, which showed a LVEDVi of 78 ml/m2, and a LVEF of 67% the RV was moderately dilated, the ejection fraction was normal (RVEDi 74 ml/m2, RVEF 59%) as were the kinesis indexes. A conventional spirometry was normal.
α-DG glycosylation and laminin α2 studies
α-DG glycosylation was studied on 8-μm thick cryosections of frozen muscle tissue, using an antibody directed against an o-glycosylated epitope of α-dystroglycan (IIH-6; Upstate Biotechnology, Lake Placid, NY, USA); laminin α2 was studied using an antibody directed against the carboxyl-terminus of the protein (mAb 1922, 80 kDa, Chemicon, Temecula, CA, USA; 1:1,000).
Gene mutation studies
DNA was extracted from peripheral blood. The complete coding regions, including intron/exon boundaries of POMT1, POMT2, POMGnT1, FKTN and LARGE, were screened for mutations by PCR/single stranded conformation polymorphism/sequencing or by direct sequencing (primers are available upon request). Restriction fragment length polymorphism analysis was used to confirm gene variants, to verify segregation in the family and to assess frequency in 110 control chromosomes.
Bioinformatics
An integrative bioinformatic approach was used to elucidate the sequence–structure–function relationship of POMT1. The human POMT1 sequence was downloaded from UniProt18 with an accession number Q9Y6A1. PSI-BLAST19 was used with standard parameters for a single iteration on the UniProt sequence database to search for homologous sequences. InterPro20 and ELM21 were used to search for known domains and interacting motifs, respectively. The secondary structure was analyzed with the consensus method,22 while disordered regions were searched with SPRITZ23 and transmembrane helices predicted with TOPCONS.24 The structure of the MIR domains (found in Mannosyltransferases, Inositol triphosphate receptors and Ryanodin receptors) was modeled with HOMER (URL: http://protein.bio.unipd.it/homer/) from the template structure with PDB code 1T9F previously identified with PSI-BLAST, with loops positioned using a fast divide and conquer approach25 and the final model being evaluated with FRST.26 The structure was visualized using PyMol (DeLano Scientific, URL: http://www.pymol.org/). The I-Mutant,27 Mupro28 and SNPs3D29 servers were used to estimate effects of the mutations in terms of protein stability. Other two predictor, SNAP30 and PhD-SNP,31 were used to classify variants as disease-related or as neutral polymorphisms.
Results
α-DG glycosylation and laminin α2 studies
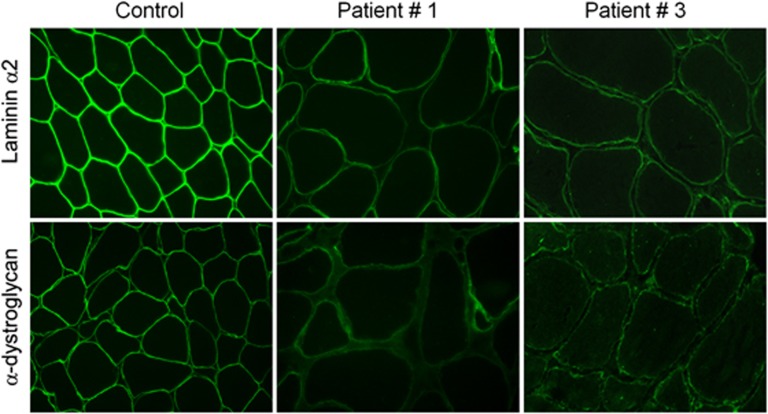
Immunofluorescence analysis of muscle biopsies revealed severe reduction of α-DG glycosylation in patient 1 and 2 and moderate reduction in patient 3 with respect to control (Figure 1). Few α-DG-negative fibers were observed in patients 1 and 3. Laminin α2 expression was slightly reduced in the patient muscle biopsies compared with control.
Figure 1.
Reduced α-DG glycosylation in POMT1-mutated patients. α-DG immunostaining using an antibody directed against a glycosylated epitope shows a normal labeling at the periphery of each fiber in the control's muscle, in comparison with patients 1 and 3 where the majority of myofibers show a faint immunoreaction and variability of the intensity of the labeling. Laminin α2 immunostaining using an antibody directed against the 80-kDa carboxyl-terminus shows a subtle reduction of the labeling in the patients.
Gene mutation studies
Patients' molecular features are summarized in Table 1. The patient 1 was compound heterozygous for two POMT1 mutations: a missense mutation, c.2005G>A, p.Ala669Thr and a donor splice site mutation in intron 12, POMT1 c.1241+1G>A.17 cDNA analysis showed that the c.1241+1G>A14 results in the in-frame skipping of exon 12 (p.His384_Thr414del; Supplementary Figure 1). Two novel POMT1 missense mutations were identified in patient 2: c.430A>G, p.Asn144Asp and c.1241C>T, p.Thr414Met.
The patient 3 was found to be heterozygous for the nonsense mutation c.1864C>T, predicting a premature stop codon (p.Arg622*). In addition, sequencing of patient's cDNA identified a splice defect that incorporates five bases at the junction exon 10–exon 11 r.1052_1053insGTAAG (Supplementary Figure 2). Full sequencing of genomic DNA identified a number of variations in intron 10–exon 11 c.1052+49 g>a (Hom), c.1052+184 g>a (Hom), c.1052+246 g>a (Hom), c.1052+276 t>c (Hom), c.1053–172 c>t (Hom), c.1053–113 c>g (Hom), c.1053–102 g>a (Het) and c.1113T>C D371D (Hom). All these have unknown significance and none predicted a cryptic splice site compatible with the aberrant transcript observed. The likely scenario is a leaking splicing defect leading to two different transcripts: one alternative transcript resulting in an out-of-frame insertion of five base pairs and a normally spliced transcript consistent with the production of a normal, but reduced protein product, and thereby consistent with partial α-DG glycosylation defect. Another possible scenario is that this patient harbors a large deletion on one allele or a deep intronic mutation that alters splicing, leading to a large deletion undetected by genomic DNA sequencing.
No FKRP, POMT2, POMGnT1, FKTN and LARGE mutations were detected in any of the patients. Identified mutations were not detected in 110 control chromosomes.
In silico prediction of mutation effects
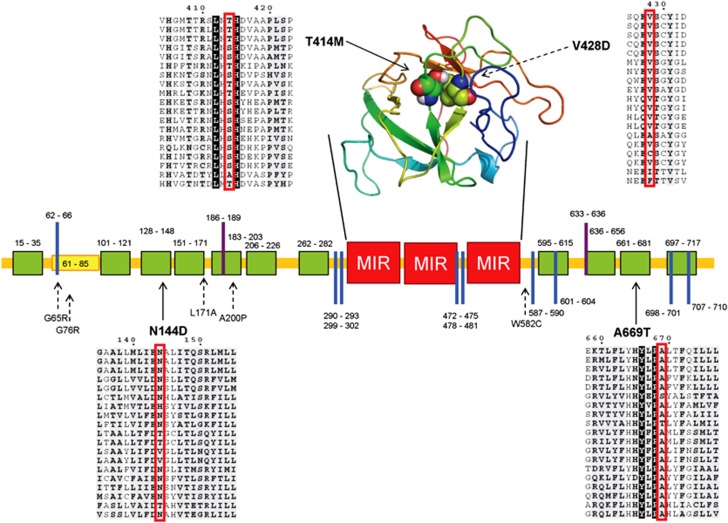
The sequence of human POMT1 (NG_008896) was analyzed with several bioinformatics methods to characterize mutation sites. As expected, several transmembrane helices were predicted and the known MIR domains detected. A consensus approach was used to delimit the single transmembrane helices, as various methods provided slightly different predictions, especially for the second and last helices. The structure of the MIR domain was predicted by homology modeling from a template structure with 31% sequence identity. Secondary structure and disorder predictions were used in combination with ELM to identify possible functional motifs locations. Figure 2 summarizes the analysis of the POMT1 sequence and the positions of disease-associated mutations.
Figure 2.
Schematic overview of POMT1. The sequence of human POMT1 is shown as a horizontal line, with transmembrane helices (green), disordered regions (yellow) and MIR domains (red) shown as boxes. The modeled structure of the three MIR domains is shown above the sequence, colored from N- to C-terminus in blue to red. C-mannosylation and glycosaminoglycan attachment sites predicted by ELM are shown as purple and blue bars, respectively. Disease-associated mutations are shown with arrows pointing to the relevant position in the sequence together with the local sequence context from a multiple sequence alignment. Previously known mutations are shown with dotted lines. Both mutations falling into the MIR domain are shown with their residues as spheres in the structure.
The mutations were analyzed using several prediction methods to determine possible pathogenicity and compared with known mutations with experimentally measured enzyme activity.2, 32, 33 All substitutions occur at conserved positions (ConSeq score of 7–9), except for G76R, which presents a medium score of 5. However, at this position charged residues like arginine are never present in homologous sequences. Furthermore, all amino-acid substitutions are predicted to be destabilizing or pathogenic by most of the used prediction methods (Table 2). Two of the identified missense mutations (N144D and A669T) are located in transmembrane helices and have similar predicted effects as previously identified mutations. The two substitutions introduce, respectively, a negatively charged and a polar residue that appear to have a destabilizing effect on protein folding (Table 2). The T414M mutation is a part of the modeled MIR domain (Figure 2) and it is in close proximity to the WWS-associated V428D mutation,5 and probably destabilizes the protein by a similar mechanism.
Table 2. Summary of missense POMT1 mutation effects.
| Mutation | Phenotype | Enzymatic activity | I-Mutant (ΔΔG) | SNPs3D (SVM score) | Mupro (C score) | SNAP (RI, EA) | PhD-SNP (RI) | ConSeq (conservation level) | AAs in alignment |
|---|---|---|---|---|---|---|---|---|---|
| N144Da | LGMD-MR | ? | Decrease (−0.41) | Deleterious (−1.73) | Increase (0.3) | Non-neutral (3, 78%) | Disease (5) | 9 | N |
| T414Ma | LGMD-MR | ? | Decrease (−0.33) | Deleterious (−2.17) | Decrease (−0.66) | Non-neutral (4, 82%) | Neutral (1) | 8 | A, S, T |
| A669T |
CMD-MR |
0.004 pmol/h/mg proteins32c |
Decrease (−1.37) |
Deleterious (−2.39) |
Decrease (−0.92) |
Non-neutral (1, 63%) |
Disease (5) |
9 |
A, S |
| G65R |
LGMD-MR |
0.002 pmol/h/mg proteins32b,c |
Decrease (−1.31) |
Deleterious (−2.66) |
Decrease (−0.69) |
Non-neutral (1, 63%) |
Disease (7) |
9 |
H, G |
| G76R | WWS | None2 10%35 | Decrease (−0.57) | Deleterious (−0.90) | Increase (+0.69) | Non-neutral (1, 63%) | Disease (5) | 5 | A, T, M, I, G, V |
| L171A | LGMD-MR | 40%35 | Decrease (−2.22) | Non-deleterious (+0.51) | Decrease (−1) | Neutral (0, 53%) | Disease (8) | 8 | I,L,V |
| A200P | LGMD-MR | None32 | Decrease (−1.05) | Deleterious (−0.89) | Decrease (−0.1) | Non-neutral (3, 78%) | Disease (7) | 7 | A, T, D, G, V |
| V428D | WWS | None2 | Decrease (−1.11) | Deleterious (−3.26) | Decrease (−0.87) | Non-neutral (6, 93%) | Disease (9) | 8 | F, I, V |
| W582C | LGMD-MR | 0.002 pmol/h/mg proteins32b,c | Decrease (−1.53) | Non-deleterious (+0.94) | Decrease (−1) | Non-neutral (3, 78%) | Disease (6) | 7 | F, W |
Abbreviations: AAs, amino acids; C score, confidence score; CMD-MR, congenital muscular dystrophy with mental retardation; EA, expected accuracy; LGMD-MR, limb-girdle muscular dystrophy with mental retardation; RI, reliability index; SVM, support vector machine; WWS, Walker–Warburg syndrome.
The mutations are listed with their associated phenotype, enzymatic activity and several in silico predictions. I-Mutant, MUpro and SNPs3D predict changes in protein stability in terms of ΔΔG, whereas SNAP and PhD-SNP predict a variant as disease-related or as neutral polymorphism. In the table we report reliability parameters for each prediction in parentheses. I-Mutant calculates the free energy change value (ΔΔG), where a ΔΔG<0 indicates decrease of stability. SNPs3D uses a SVM to find the separation pattern between a set of disease and non-deleterious SNPs. A positive score indicates variants classified as non-deleterious. MUpro predictions were reported with the confidence score (C score). A negative score indicates the mutation decreases protein stability, where lower scores imply higher confidence. In SNAP, variations are listed as ‘neutral' or ‘non-neutral' with reliability indices (RI; range 0–9) and EA (range 1–100%) indicative of confidence in prediction. Higher RI correlates strongly with higher prediction accuracy. EA is a number of correctly predicted neutral or non-neutral samples (at a given reliability index) in the SNAP testing set. SNAP only reports predictions that are made with at least 50% accuracy. PhD-SNP classifies a mutation as disease-related or as neutral polymorphism. As with SNAP, the RI indicates the confidence of predictions. ConSeqscores the sequence conservation from 0 to 9, with 9 being highly conserved and 0 being highly unconserved (ie, variable). The last column shows the residue types present in that position of the multiple sequence alignment.
Novel mutation.
Measured in lymphoblasts from a compound heterozygous carrier of p.G65R and p.W582C.
Average range of POMT activity in controls is 0.041±0.013 pmol/h/mg proteins.32
Discussion
Although the clinical spectrum of dystroglycanopathies is broad, cardiac involvement has been reported only in patients with FKRP10, 11, 12, 13 and FKTN14 mutations, or in rare forms allelic to CGDs.15, 16 Similar to other known glycosyltransferases, POMT1 is expressed ubiquitously in all human tissues. Skeletal and cardiac muscles, in particular, show above-average levels of expression.3 It is thus quite surprising that no signs of cardiomyopathy have been described in the ∼40 previously reported patients carrying POMT1 mutations (Leiden muscular dystrophies pages at http://www.dmd.nl).
The patients in our series all presented with dilation and/or decreased LV contractility, variable RV involvement, and all had a good response to pharmacological therapy.
Muscle and CNS involvement in the patients was variable ranging from the mild to the severe ends of the POMT1 clinical spectrum. Patient 1, whose clinical immunohistochemical and genetic features were documented before the development of cardiomyopathy,17 had CMD with severe mental retardation and a normal brain MRI, a phenotype known to be associated with POMT1 mutations.33 Patients 2 and 3, conversely, differ from the classical LGMD2K, which usually includes overt mental retardation.7, 8
It has been hypothesized that mutations, which completely disrupt mannosyltransferase activity, are associated with more severe phenotypes (WWS), while those allowing residual enzyme activity are linked to milder ones (CMD-MR/LGMD2K).4, 7 Recent findings suggest that this correlation is weaker with regard to putative glycosyltransferase genes, such as FKTN or FKRP, but stronger for genes with a known enzyme product, such as POMT1.34 In fact, studies that measure POMT1 activity in Sf9 cell lines coexpressing mutated POMT1 with wild-type POMT22 or in immortalized lymphoblasts from patients carrying POMT1 mutations32 have demonstrated a marked reduction in POMT activity in the mutations/patients studied, but were unable to precisely predict phenotype severity. On the other hand, measurement of POMT activity using dermal fibroblasts from POMT1-mutated patients showed that clinical phenotype severity is inversely correlated with POMT1 activity.35
A direct correlation between mannosyltransferase activity and clinical severity, however, does not seem to apply to heart involvement, which in our patients appears possible with very different degrees of neuromuscular severity, and with both complete and partial glycosylation defects (detected in the skeletal muscle).
It remains to be established if the development of cardiomyopathy in our patients can be mutation-dependent and if specific POMT1 mutations can predispose to cardiac deterioration. Indeed, all the identified mutations in our patients seem to indicate some degree of functional relevance. Some of the identified mutations, such as stop-codon mutations or the in-frame skipping of exon 12, which codes for a portion of the catalytic MIR domain, have an easily predictable deleterious effect on enzyme activity. Novel missense mutations, on the other hand, need further studies to better assess pathogenicity. In silico predictions of protein structure, summarized in Table 2, have localized these mutations into transmembrane helices, probably interfering with protein folding and stability, or into the MIR domain, in close proximity with previously described WWS-associated mutations, which completely impair catalytic function, and thus probably alter the protein by means of a similar mechanism.5 We did not, however, expect our patients to have a complete defect of POMT1 enzymatic function, especially in those cases in which the phenotype was relatively mild and/or there was residual α-DG immunolabeling with antibodies against glycosylated epitopes. This may explain why some identified mutations have a predicted benign or slightly damaging effect on enzyme structure with some of the bioinformatic models that have been used, suggesting that they allow for the expression of a partially viable and functioning enzyme.
The mechanism of both cardiomyocyte and muscle fiber damage in dystroglycanopathies is probably loss of dystroglycan function due to insufficient glycosylation and subsequent accumulation of membrane damage in response to exercise-induced stress, as suggested by animal models.36 In our patients, the myocardium may have been particularly stressed by specific conditions, such as respiratory failure (patient 1) or several years of relatively strenuous exercise in adults with a globally preserved motor function (patients 2 and 3).
We speculate that all or most of patients with severe WWS phenotypes would probably develop cardiomyopathy if their lifespan was longer, while in patients with more residual enzymatic activity and milder phenotypes, different mutations may determine a different pattern and timing of multisystemic involvement, explaining the absence of cardiomyopathy in most CMD and LGMD patients.
Bioinformatic prediction of the effect of missense mutations upon protein folding and function appears to yield results, which are consistent with in vitro enzymatic assay findings found in the literature for identical or similar mutations (Table 2), and may prove useful especially in those laboratories in which these assays are unavailable. Further studies on α-DG glycosylation in myocardial tissue will help in clarifying genotype–phenotype correlations and the mechanisms by which POMT1 mutations and dystroglycanopathies in general selectively involve the myocardium.
Our report expands the phenotypical spectrum of POMT1 mutations, adding cardiomyopathy to LGMD forms with slight cognitive impairment and to CMD-MR. In the light of these findings, we would recommend that clinicians monitor patients with POMT1 mutations closely, regardless of their neuromuscular phenotype, to detect precocious signs of cardiac dysfunction. Gadolinium-enhanced cardiac MRI studies in these patients may help to detect subclinical heart involvement, making timely therapeutic interventions possible.
Acknowledgments
This study is funded by the Italian Telethon (GUP06004 and GTF02009), EuroBioBank and University of Padova (CPDA098382/09).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Manya H, Chiba A, Yoshida A, et al. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc Natl Acad Sci USA. 2004;101:500–505. doi: 10.1073/pnas.0307228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaka-Manya K, Manya H, Nakajima A, Kawakita M, Endo T. Physical and functional association of human protein O-mannosyltransferases 1 and 2. J Biol Chem. 2006;281:19339–19345. doi: 10.1074/jbc.M601091200. [DOI] [PubMed] [Google Scholar]
- Perez Jurado LA, Coloma A, Cruces J. Identification of a human homolog of the Drosophila rotated abdomen gene (POMT1) encoding a putative protein O-mannosyl-transferase, and assignment to human chromosome 9q34.1. Genomics. 1999;58:171–180. doi: 10.1006/geno.1999.5819. [DOI] [PubMed] [Google Scholar]
- Barresi R, Campbell KP. Dystroglycan: from biosynthesis to pathogenesis of human disease. J Cell Sci. 2006;119:199–207. doi: 10.1242/jcs.02814. [DOI] [PubMed] [Google Scholar]
- Beltran-Valero De Bernabe D, Currier S, Steinbrecher A, et al. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am J Hum Genet. 2007;71:1033–1043. doi: 10.1086/342975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Reeuwijk J, Maugenre S, van den Elzen C, et al. The expanding phenotype of POMT1 mutations: from Walker-Warburg syndrome to congenital muscular dystrophy, microcephaly, and mental retardation. Hum Mutat. 2006;27:453–459. doi: 10.1002/humu.20313. [DOI] [PubMed] [Google Scholar]
- Balci B, Uyanik G, Dincer P, et al. An autosomal recessive limb girdle muscular dystrophy (LGMD2) with mental retardation is allelic to Walker-Warburg sindrome (WWS) caused by a mutation in the POMT1 gene. Neuromuscul Disord. 2005;15:271–275. doi: 10.1016/j.nmd.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Godfrey C, Clement E, Mein R, et al. Refining genotype phenotype correlations in muscular dystrophies with defective glycosylation of dystroglycan. Brain. 2007;130:2725–2735. doi: 10.1093/brain/awm212. [DOI] [PubMed] [Google Scholar]
- Muntoni F, Torelli S, Brockington M. Muscular dystrophies due to glycosylation defects. Neurotherapeutics. 2008;5:627–632. doi: 10.1016/j.nurt.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe M, Bourke J, Eagle M, et al. Cardiac and respiratory failure in limb girdle muscular dystrophy 2I. Ann Neurol. 2004;56:738–741. doi: 10.1002/ana.20283. [DOI] [PubMed] [Google Scholar]
- Bourteel H, Vermersch P, Cuisset JM, et al. Clinical and mutational spectrum of limb-girdle muscular dystrophy type 2I in 11 French patients. J Neurol Neurosurg Psychiatry. 2009;80:1405–1408. doi: 10.1136/jnnp.2007.141804. [DOI] [PubMed] [Google Scholar]
- Boito C, Melacini P, Vianello A, et al. Clinical and molecular characterization of patients with limb-girdle muscular dystrophy type 2I. Arch Neurol. 2005;62:1894–1899. doi: 10.1001/archneur.62.12.1894. [DOI] [PubMed] [Google Scholar]
- Margeta M, Connolly AM, Winder TL, et al. Cardiac pathology exceeds skeletal muscle pathology in two cases of limb-girdle muscular dystrophy type 2I. Muscle Nerve. 2009;40:883–889. doi: 10.1002/mus.21432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Hayashi YK, Noguchi S, et al. Fukutin gene mutations cause dilated cardiomyopathy with minimal muscle weakness. Ann Neurol. 2006;60:597–602. doi: 10.1002/ana.20973. [DOI] [PubMed] [Google Scholar]
- Lefeber DJ, Schönberger J, Morava E, et al. Deficiency of Dol-P-Man synthase subunit DPM3 bridges the congenital disorders of glycosylation with the dystroglycanopathies. Am J Hum Genet. 2009;85:76–86. doi: 10.1016/j.ajhg.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeber DJ, Brouwer AP, Morava E, et al. Autosomal recessive dilated cardiomyopathy due to DOLK mutations results from abnormal dystroglycan O-mannosylation. PLoS Genet. 2011;7:e1002427. doi: 10.1371/journal.pgen.1002427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico A, Tessa A, Bruno C, et al. Expanding the clinical spectrum of POMT1 phenotype. Neurology. 2006;66:1564–1567. doi: 10.1212/01.wnl.0000216145.66476.36. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, et al. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll P, Linding R, Gemund C, et al. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M, Tosatto SC, Lengauer T, Valle G. Simple consensus procedures are effective and sufficient in secondary structure prediction. Protein Eng. 2003;16:459–462. doi: 10.1093/protein/gzg063. [DOI] [PubMed] [Google Scholar]
- Vullo A, Bortolami O, Pollastri G, Tosatto SC. Spritz: a server for the prediction of intrinsically disordered regions in protein sequences using kernel machines. Nucleic Acids Res. 2006;34:164–168. doi: 10.1093/nar/gkl166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsel A, Viklund H, Falk J, Lindahl E, von Heijne G, Elofsson A. Prediction of membrane-protein topology from first principles. Proc Natl Acad Sci USA. 2008;105:7177–7181. doi: 10.1073/pnas.0711151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosatto SC, Bindewald E, Hesser J, Manner R. A divide and conquer approach to fast loop modeling. Protein Eng. 2002;15:279–286. doi: 10.1093/protein/15.4.279. [DOI] [PubMed] [Google Scholar]
- Tosatto SC. The Victor/FRST Function for Model Quality Estimation. J Comput Biol. 2005;12:1316–1327. doi: 10.1089/cmb.2005.12.1316. [DOI] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, Casadio R. I-Mutant20: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Randall A, Baldi P. Prediction of Protein Stability Changes for Single-Site Mutations Using Support Vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–2734. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- Manya H, Bouchet C, Yanagisawa A, et al. Protein O-mannosyltransferase activities in lymphoblasts from patients with α-dystroglycanopathies. Neuromusc Disord. 2008;18:45–51. doi: 10.1016/j.nmd.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Jimenez-Mallebrera C, Torelli S, Feng L, et al. A comparative study of alpha dystroglycan glycosylation in dystroglycanopathies suggests that the hypoglycosylation of alpha-dystroglycan does not consistently correlate with clinical severity. Brain Pathol. 2009;19:596–611. doi: 10.1111/j.1750-3639.2008.00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercuri E, Messina S, Bruno C, et al. Congenital muscular dystrophies with defective glycosylation of dystroglycan: a population study. Neurology. 2009;26:1802–1809. doi: 10.1212/01.wnl.0000346518.68110.60. [DOI] [PubMed] [Google Scholar]
- Lommel M, Cirak S, Willer T, et al. Correlation of enzyme activity and clinical phenotype in POMT1-associated dystroglycanopathies. Neurology. 2010;74:157–164. doi: 10.1212/WNL.0b013e3181c919d6. [DOI] [PubMed] [Google Scholar]
- Michele DE, Kabaeva Z, Davis SL, Weiss RM, Campbell KP. Dystroglycan matrix receptor function in cardiac myocytes is important for limiting activity induced myocardial damage. Circ Res. 2009;105:984–993. doi: 10.1161/CIRCRESAHA.109.199489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.