Abstract
Only 20 patients with deletions of 18q12.2 have been reported in the literature and the associated phenotype includes borderline intellectual disability, behavioral problems, seizures, obesity, and eye manifestations. Here, we report a male patient with a de novo translocation involving chromosomes 12 and 18, with borderline IQ, developmental and behavioral disorders, myopia, obesity, and febrile seizures in childhood. We characterized the rearrangement with Affymetrix SNP 6.0 Array analysis and next-generation mate pair sequencing and found truncation of CELF4 at 18q12.2. This second report of a patient with a neurodevelopmental phenotype and a translocation involving CELF4 supports that CELF4 is responsible for the phenotype associated with deletion of 18q12.2. Our study illustrates the utility of high-resolution genome-wide techniques in identifying neurodevelopmental and neurobehavioral genes, and it adds to the growing evidence, including a transgenic mouse model, that CELF4 is important for human brain development.
Keywords: CELF4, 18q12 deletion, developmental disorder, behavioral disorder, obesity, next-generation mate pair sequencing
Introduction
Only 20 patients with deletions of 18q12.2 have been reported in the literature.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 In 15 of these patients, the deletions were examined with chromosome analysis, and in the remaining 5 patients molecular investigations identified deletions ranging from 3.2 to 24 Mb in size.9, 10, 12, 13 Common clinical findings included intellectual disability, behavioral disorders, seizures, eye manifestations, obesity, and mild or absent dysmorphic facial features.
In recent years, the use of high-resolution cytogenetic techniques has led to the identification of numerous microdeletion syndromes, where patients with overlapping deletions share clinical features. However, the shared deletions often exceed hundreds of kb or even several Mb, encompassing many genes that hamper the identification of the exact gene(s) underlying the associated phenotype.14 In contrast, truncation of single genes – for example, by small intragenic deletions or by chromosomal translocation or inversion breakpoints – can directly identify a disease-causing gene.15
Here we describe a patient with a prenatally detected de novo translocation involving chromosomes 12 and 18, with febrile seizures in childhood, borderline intelligence, developmental and behavioral disorders, myopia, and obesity. We characterized the rearrangement with SNP array and next-generation sequencing (NGS) and found truncation of CELF4, which has previously been suggested as a neurodevelopmental candidate gene.13 This report illustrates the utility of high-resolution genome-wide techniques in identifying neurodevelopmental genes.
Patients and methods
The patient was identified though a nation-wide study of prenatally detected de novo balanced structural rearrangements. The study was approved by the Danish Scientific Ethics Committee and written informed consent was obtained.
Clinical report
The patient is a 27-year-old male. He is the second child to healthy unrelated parents. Amniocentesis was performed due to advanced maternal age, and a de novo reciprocal translocation involving chromosomes 12 and 18 was diagnosed. The child was born by uncomplicated vaginal delivery at 40 weeks of gestation with a birth weight of 4000 g and birth length 55 cm. The neonatal period was unremarkable. According to the mother, he walked independently at 12 months and there was no delay in language development. At 23 months of age, he was hospitalized due to a series of three febrile seizures. He did not experience recurrent seizures and an electroencephalography was not performed. Owing to minor neurological deficits and difficulties with complex motor tasks, he was diagnosed with minimal brain dysfunction at age 4. At the age of 6, he was diagnosed with hyperkinetic disorder and treated with methylphenidate. He was described as aggressive, with immature and oppositional behavior, having problems with impulsivity and attention shifting, and with a lack of emotional reciprocity. At the age of 13, he was tested with Weschler Intelligence Scale for Children-Revised and found to have an IQ of 71, and he was diagnosed with behavioral disorder, oppositional defiant disorder, and mixed specific developmental disorder (all according to ICD-10).
A physical examination at the age of 25 years revealed no dysmorphic features. His height was 180 cm, weight 110 kg, and head circumference 61 cm. He wore glasses for myopia (estimated −3 diopters). He was obese with a body mass index of 34; according to the mother, obesity has been present since the age of 6–7 years as a result of uninhibited eating behavior. At the age of 27 years, he was screened for major psychiatric disease using the Mini International Diagnostic Interview version 5.0.0 that refers to the DSM-IV. No single diagnosis could be pinpointed. Signs of low IQ and autistic behavior with indications of concrete thinking, impairment of social interaction skills, vagueness in description of others, lack of concentration over time, hyperactivity, and impulsive behavior were seen during the interview. There was significant impairment in occupational and social areas, and a general incapacity for labor market affiliation was manifest. In his youth, he was intensely preoccupied with trains and unable to develop peer relationships. He lives in a group home and his social interactions are limited.
Chromosome and copy number variation analyses
Standard G-banding chromosome analysis was performed on cultured peripheral lymphocytes. Genomic DNA was extracted by conventional methods from peripheral blood, and whole-genome copy number variation (CNV) analysis was performed with the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA). Raw intensity data files were analyzed with the Genotyping Console software (Affymetrix) according to the manufacturer's recommendations. CNVs larger than 1 kb and encompassing more than eight markers were evaluated.
Next-generation paired-end sequencing
Mate pair libraries were prepared using the Mate Pair Library v2 kit (Illumina, San Diego, CA, USA). Briefly, 10 μg DNA was sheared using a nebulizer. Two- to three-kilobase pair fragments were isolated, end-repaired using a mix of natural and biotinylated dNTPs, blunt-end ligated using circularization ligase, and fragmented to 200–400 bp. Biotinylated fragments were isolated and end-repaired and A-overhangs were added to the 3′-ends. Paired-end adapters were ligated to the fragments and the library was amplified by 18 cycles of PCR. Mate pair libraries were subjected to 2 × 36 bases paired-end sequencing on a Genome Analyzer IIx (Illumina), following the manufacturer's protocol. Reads were aligned to a reference genome using Bowtie16 allowing up to two mismatches in the seed region. Reads not aligning uniquely were discarded from further analysis. Paired reads aligning to different chromosomes or with unexpected strand orientation were extracted to identify potential translocation and inversion breakpoints, respectively. Breakpoints were only considered as candidates if they were confirmed by at least six independent paired reads with end-reads mapping within a 6-kb region. Predicted breakpoints were filtered against known in-house variants based on data from 30 individuals with known breakpoints. The translocation breakpoint was confirmed by PCR amplification and Sanger sequencing of the breakpoint-spanning fragments.
RESULTS
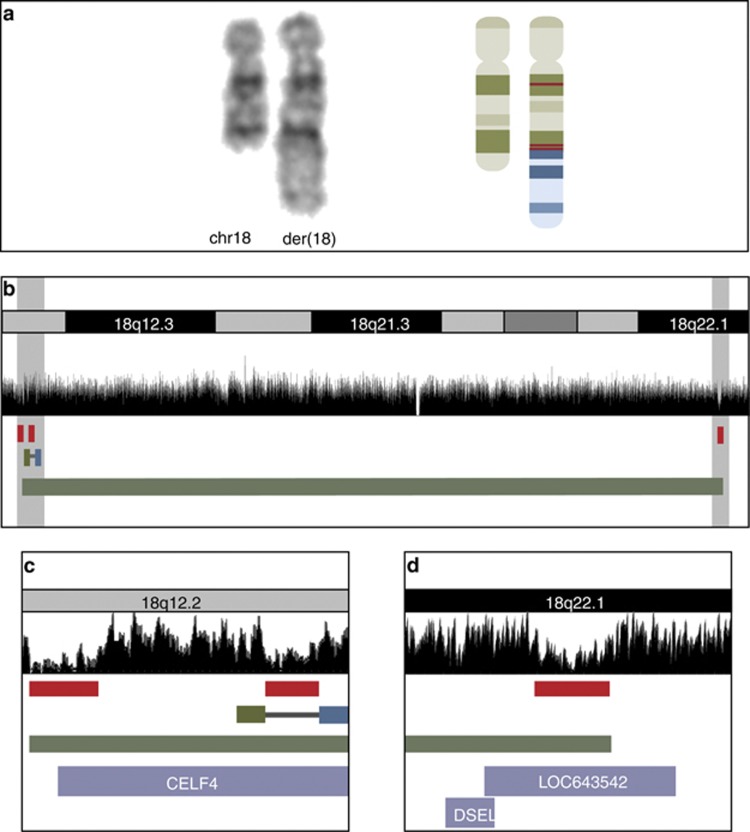
A total of 28 986 673 paired reads passed the chastity filter; 13 689 000 paired reads were aligned uniquely and 204 319 were chimeric pairs (end-reads mapping to different chromosomes). We removed non-clustering chimeric pairs leaving a total of 49 chimeric clusters genome-wide that were visually filtered against known variants. The translocation breakpoints were identified at 12q21.31 and 18q12.2, respectively. The breakpoint at chromosome 12 affected no annotated genes whereas the breakpoint at chromosome 18 truncated CELF4. The translocation breakpoint at chromosome 18 was confirmed by PCR amplification and subsequent Sanger sequencing of the breakpoint-spanning fragments that revealed a small sequence of micro homology (AGGA), likely to facilitate the translocation (Supplementary Figure 1). Additionally, NGS revealed an associated inversion at chromosome 18 with breakpoints at 18q12.2 and 18q22.1 that truncated CELF4 and LOC643542. Re-evaluation of the karyotype confirmed the cryptic complex rearrangement, and CNV analysis revealed small de novo deletions (<150 kb in size) corresponding to all four mapped breakpoints (Supplementary Table S1). The revised karyotype was t(12;18)(q21.31;q12.2),inv18(q12.2q22.1).arr12q21.31(83,761,458–83,857,594)x1,18q12.2(34,810,178–34,867,844)x1,18q12.2(35,007,705–35,049,456)x1,18q22.1(65,284,854–65,430,839)x1 dn (hg19). The molecular characterization of the rearrangements is depicted in Figures 1 and 2.
Figure 1.
Partial karyotype and molecular findings on the derivative chromosome 12. (a) A partial karyotype showing the normal and the derivative chromosome 12, and a schematic representation of the two chromosomes and the identified rearrangement; the red line represents the deletion associated with the translocation at 12q21.31. (b) The breakpoint region at 12q21.31. The mate pair coverage is shown in black. A deletion detected by SNP 6.0 array is represented by a red bar; note the corresponding low coverage of the mate pair reads. The translocation breakpoint, represented by blue reads mapping to 12q21.31 and green reads mapping to 18q12.2, flanks the deletion. (c) Zoom in on the translocation region (gray shaded area in b). No RefSeq genes were located at the breakpoint.
Figure 2.
Partial karyotype and molecular findings on the derivative chromosome 18. (a) A partial karyotype showing the normal and the derivative chromosome 18, and a schematic representation of the two chromosomes and the identified rearrangements; the red lines represent deletions associated with the translocation breakpoint at 18q12.2 and the inversion breakpoints at 18q12.2 and 18q22.1. (b) The breakpoint region at 18q. The mate pair coverage is shown in black. Deletions detected by SNP 6.0 array is represented by red bars; note the corresponding low coverage of the mate pair reads. The missing coverage of mate pair reads in 18q21.3 corresponds to a gap in the annotated reference genome (UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly). The translocation breakpoint is represented by green reads mapping to 18q12.2 and blue reads mapping to 12q21.31, and the inversion is represented by a green bar. (c, d) Zoom in on the breakpoint regions (gray shaded areas in b). The protein coding gene CELF4 is truncated by the translocation and two deletions at 18q12.2 (c), and the non-coding RNA LOC643542 is truncated by the inversion breakpoint and an associated deletion at 18q22.1 (d).
DISCUSSION
Here, we report a male patient with a translocation involving chromosomes 12q and 18q with borderline IQ, developmental and behavioral disorders, myopia, obesity, and a history of febrile seizures in childhood. Using SNP array and NGS, we identified a cryptic complex rearrangement that truncated 2 RefSeq genes: the protein coding gene CELF4 (also known as BRUNOL4) at 18q12.2 and the non-coding RNA LOC643542 at 18q22.1.
Few of the previously reported patients with deletions of 18q12.2 were characterized sufficiently both clinically and molecularly to allow direct phenotype–genotype comparisons. However, as illustrated in Table 1 there is a considerable phenotypic overlap with our patient. Gilling et al13 reported a patient with childhood autism, severe myopia, and a de novo translocation, t(5;18)(q34;q12). The patient had normal intelligence (IQ 88) and no dysmorphic facial features. The breakpoint region contained a 3.2-Mb deletion encompassing CELF4. McEntagart et al10 described a patient with a de novo del(18)(q11.2q12.2) and a phenotype remarkably like the one reported here consisting of mild intellectual disability (IQ 61), developmental delay, febrile seizures, clumsiness, behavioral difficulties including poor concentration and hyperactivity, distractibility and learning difficulties, and no facial dysmorphism. Tinkle et al11 described a 67-year-old female patient with del(18)(q12.2q21.1). This patient was only examined with conventional karyotyping but the reported phenotype included intellectual disability, a history of fine motor problems, and seizures in childhood. Her behavior was described as impulsive, aggressive, defiant, restless, and labile autistic-like. She was obese and had problems with food seeking obsession.
Table 1. Clinical features in patients with molecularly characterized and overlapping interstitial deletions involving the 18q12.2 region.
| Reference | Present report | Gilling et al13 | McEntagart et al10 | Published patientsa (not fine mapped and/or not phenotyped in detail) | Total |
|---|---|---|---|---|---|
| Number of patients | 1 | 1 | 1 | 18 | 21 |
| Examination method | SNP array, NGS | aCGH, FISH | aCGH | Karyotype/aCGH/FISH | — |
| Deletion | 18q12q12.2 | 18q12.1q12.2 | 18q11.2q12.2 | Unbalancedb | — |
| Age (years) | 27 | 38 | 4 | 2–67 | 2–67 |
| Gender | M | F | M | 6F, 12M | 7F, 14M |
| ID/DD | (+)c | − | + | 18/18 | 20/21 |
| Motor delay | + | + | + | 12/12 | 15/15 |
| Seizures | + | − | + | 11/16 | 13/19 |
| Hypotonia | + | − | NR | 8/11 | 9/13 |
| Behavioral difficulties | + | + | + | 11/13d | 14/16 |
| ASD/autistic traits | + | + | NR | 4/5 | 6/7 |
| Obesity | + | NR | NR | 9/13d | 10/14 |
| Strabismus | − | NR | NR | 8/11d | 8/12 |
| Myopia | + | + | NR | 1/4 | 3/6 |
| Hypermetropia | − | − | NR | 2/4 | 2/6 |
| Dysmorphic features | − | − | − | 15/16 | 15/19 |
Abbreviations: aCGH, array comparative genome hybridization; ASD, autism spectrum disorder; F, female; FISH, fluorescence in situ hybridization; ID/DD, intellectual disability or developmental delay; M, male; NR, not reported; SNP, single-nucleotide polymorphism.
Chudley et al1, 7; Engelen et al9; Feenstra et al12; Krasikov et al6; Poissonnier et al8; Schinzel et al4; Surh et al5; Wilson et al.2, 3
Cytogenetic unbalances (range 18q11–18q21.1), all include 18q12.2.
Intelligence quotient=71.
Feenstra et al12 note that common features in the studied group of 29 patients include obesity, strabismus, and behavioral problems. Since it is not specified if the 2 reported patients with deletions overlapping 18q12.2 have these features, they are not counted here.
Although CELF4 was previously found disrupted in patients with overlapping phenotypes,10, 13 no phenotype has been associated with the non-coding RNA LOC643542, thus we find it highly likely that haploinsufficiency of CELF4 causes the described phenotype. CELF4 belongs to a family of evolutionary highly conserved genes.17, 18 The encoded protein binds to RNA and act as a splicing regulator. CELF4 is expressed in various tissues with high expression in both fetal and adult human brain.19 In frog embryos (Xenopus laevis), Celf4 is solely expressed in the nervous system, including the optic veiscles,20 suggesting an important role in both brain and eye development. Furthermore, Celf4 is highly expressed in mouse brain13, 19, 21 and Celf4 insufficient transgenic mice develop convulsive seizures, hyperactivity, and late-onset body weight gain.21, 22 Not only is CELF4 conserved from Drosphila to mouse and human, but the genomic landscape next to CELF4 is also evolutionary conserved23 harboring ultra-conserved elements (UCE),24 and it has been suggested that UCEs may be directly involved in neurodevelopmental disorders.25, 26
We found CELF4 to be disrupted in a patient with borderline IQ, developmental and behavioral disorders, myopia, obesity, and a history of febrile seizures in childhood. The phenotype resembles that of patients with 18q12.2 deletions illustrating that CELF4 haploinsufficiency has a key role in explaining their phenotype. This report, and the finding of Celf4-deficient mice with overlapping traits, for example, seizures, hyperactivity, and obesity,21, 22 adds to the growing evidence that CELF4 is important for the brain development and in the disposition to obesity.
Acknowledgments
We thank the patient and his parents. We thank Elisabeth Larsen, Theresa Wass, Ingrid Kjær, and Hanne Mølgaard for technical assistance. This work was supported by the Danish National Research Foundation, the Lundbeck Foundation, the UNIK program ‘Food, Fitness and Pharma' supported by the Danish Ministry of Science, Technology, and Innovations, and by the Danish Strategic Research Council supporting the Center for Non-coding RNA in Technology and Health.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Chudley AE, Bauder F, Ray M, McAlpine PJ, Pena SD, Hamerton JL. Familial mental retardation in a family with an inherited chromosome rearrangement. J Med Genet. 1974;11:353–366. doi: 10.1136/jmg.11.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MG, Towner JW, Forsman I, Siris E. Syndromes associated with deletion of the long arm of chromosome 18[del(18q)] Am J Med Genet. 1979;3:155–174. doi: 10.1002/ajmg.1320030207. [DOI] [PubMed] [Google Scholar]
- Wilson GN, Al Saadi AA. Obesity and abnormal behaviour associated with interstitial deletion of chromosome 18 (q12.2q21.1) J Med Genet. 1989;26:62–63. doi: 10.1136/jmg.26.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel A, Binkert F, Lillington DM, et al. Interstitial deletion of the long arm of chromosome 18, del(18)(q12.2q21.1): a report of three cases of an autosomal deletion with a mild phenotype. J Med Genet. 1991;28:352–355. doi: 10.1136/jmg.28.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surh LC, Ledbetter DH, Greenberg F. Interstitial deletion of chromosome 18[del(18)(q11.2q12.2 or q12.2q21.1] Am J Med Genet. 1991;41:15–17. doi: 10.1002/ajmg.1320410105. [DOI] [PubMed] [Google Scholar]
- Krasikov N, Thompson K, Sekhon GS. Monosomy 18q12.1—21.1: a recognizable aneuploidy syndrome? Report of a patient and review of the literature. Am J Med Genet. 1992;43:531–534. doi: 10.1002/ajmg.1320430306. [DOI] [PubMed] [Google Scholar]
- Chudley AE, Kovnats S, Ray M. Recognizable behavioral and somatic phenotype in patients with proximal interstitial 18q deletion: report on a new affected child and follow-up on the original reported familial cases. Am J Med Genet. 1992;43:535–538. doi: 10.1002/ajmg.1320430307. [DOI] [PubMed] [Google Scholar]
- Poissonnier M, Turleau C, Olivier-Martin M, et al. Interstitial deletion of the proximal region of the long arm of chromosome 18, del(18q12) a distinct clinical entity? A report of two new cases. Ann Genet. 1992;35:146–151. [PubMed] [Google Scholar]
- Engelen JJ, Loots WJ, Albrechts JC, et al. Characterization of a de novo unbalanced translocation t(14q18q) using microdissection and fluorescence in situ hybridization. Am J Med Genet. 1998;75:409–413. [PubMed] [Google Scholar]
- McEntagart M, Carey A, Breen C, McQuaid S, Stallings RL, Green AJ, King MD. Molecular characterisation of a proximal chromosome 18q deletion. J Med Genet. 2001;38:128–129. doi: 10.1136/jmg.38.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinkle BT, Christianson CA, Schorry EK, Webb T, Hopkin RJ. Long-term survival in a patient with del(18)(q12.2q21.1) Am J Med Genet A. 2003;119A:66–70. doi: 10.1002/ajmg.a.10217. [DOI] [PubMed] [Google Scholar]
- Feenstra I, Vissers LE, Orsel M, et al. Genotype-phenotype mapping of chromosome 18q deletions by high-resolution array CGH: an update of the phenotypic map. Am J Med Genet A. 2007;143A:1858–1867. doi: 10.1002/ajmg.a.31850. [DOI] [PubMed] [Google Scholar]
- Gilling M, Lauritsen MB, Moller M, et al. A 3.2 Mb deletion on 18q12 in a patient with childhood autism and high-grade myopia. Eur J Hum Genet. 2008;16:312–319. doi: 10.1038/sj.ejhg.5201985. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Vries BB, Veltman JA. Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. J Med Genet. 2010;47:289–297. doi: 10.1136/jmg.2009.072942. [DOI] [PubMed] [Google Scholar]
- Mikhail FM, Lose EJ, Robin NH, et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A. 2011;155A:2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good PJ, Chen Q, Warner SJ, Herring DC. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J Biol Chem. 2000;275:28583–28592. doi: 10.1074/jbc.M003083200. [DOI] [PubMed] [Google Scholar]
- Meins M, Schlickum S, Wilhelm C, et al. Identification and characterization of murine Brunol4, a new member of the elav/bruno family. Cytogenet Genome Res. 2002;97:254–260. doi: 10.1159/000066619. [DOI] [PubMed] [Google Scholar]
- Wu J, Li C, Zhao S, Mao B. Differential expression of the Brunol/CELF family genes during Xenopus laevis early development. Int J Dev Biol. 2010;54:209–214. doi: 10.1387/ijdb.082685jw. [DOI] [PubMed] [Google Scholar]
- Wagnon JL, Mahaffey CL, Sun W, Yang Y, Chao HT, Frankel WN. Etiology of a genetically complex seizure disorder in Celf4 mutant mice. Genes Brain Behav. 2011;10:765–777. doi: 10.1111/j.1601-183X.2011.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Mahaffey CL, Berube N, Maddatu TP, Cox GA, Frankel WN. Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS Genet. 2007;3:e124. doi: 10.1371/journal.pgen.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Martinez F, Monfort S, Rosello M, et al. Enrichment of ultraconserved elements among genomic imbalances causing mental delay and congenital anomalies. BMC Med Genomics. 2010;3:54. doi: 10.1186/1755-8794-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitras L, Yu M, Lesage-Pelletier C, et al. An SNP in an ultraconserved regulatory element affects Dlx5/Dlx6 regulation in the forebrain. Development. 2010;137:3089–3097. doi: 10.1242/dev.051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.