Abstract
Recently, constitutional MLH1 epimutations have been identified in a subset of Lynch syndrome (LS) cases. The aim of this study was the identification of patients harboring constitutional MLH1 epimutations in a set of 34 patients with a clinical suspicion of LS, MLH1-methylated tumors and non-detected germline mutations in mismatch repair (MMR) genes. MLH1 promoter methylation was analyzed in lymphocyte DNA samples by MS-MLPA (Methylation-specific multiplex ligation-dependent probe amplification). Confirmation of MLH1 constitutional methylation was performed by MS-MCA (Methylation-specific melting curve analysis), bisulfite sequencing and pyrosequencing in different biological samples. Allelic expression was determined using heterozygous polymorphisms. Vertical transmission was evaluated by MS-MLPA and haplotype analyses. MS-MLPA analysis detected constitutional MLH1 methylation in 2 of the 34 individuals whose colorectal cancers showed MLH1 methylation (5.9%). These results were confirmed by bisulfite-based methods. Both epimutation carriers had developed metachronous early-onset LS tumors, with no family history of LS-associated cancers in their first-degree relatives. In one of the cases, the identified MLH1 constitutional methylation was monoallelic and results in MLH1 and EPM2AIP1 allele-specific transcriptional silencing. It was present in normal somatic tissues and absent in spermatozoa. The methylated MLH1 allele was maternally transmitted and methylation was reversed in a daughter who inherited the same allele. MLH1 methylation screening in lymphocyte DNA from patients with early-onset MLH1-methylated LS-associated tumors allows the identification of epimutation carriers. The present study adds further evidence to the emerging entity of soma-wide MLH1 epimutation and its heritability.
Keywords: Lynch syndrome, constitutional epimutation, MLH1, methylation, MS-MLPA, pyrosequencing
Introduction
Lynch syndrome (LS) is characterized by an autosomal dominant inheritance of early-onset colorectal cancer (CRC) and increased risk of other cancers.1, 2 It is caused by germline mutations in DNA mismatch repair (MMR) genes. MLH1 or MSH2 are the most commonly mutated MMR genes in LS, whereas mutations in MSH6 or PMS2 are significantly less common.3, 4 Occasionally, the presence of constitutional epimutations in MSH2 and MLH1 has been reported (reviewed in Hitchins and Ward5 and Kuiper et al6).
Constitutional epimutations are those stable changes in gene expression that do not affect DNA sequence and that are present in normal tissues of a given individual.7 An epimutation that occurs in the germline or early embryo can affect all or most of the soma, and phenocopy genetic disease. MSH2 epimutations, associated with a strong heritability, have been shown secondary to the presence of deletions in the neighboring EPCAM gene.6 The mutations lead to mosaic methylation of MSH2 in EPCAM-expressing cells.8
Approximately 40 index cases of constitutional MLH1 methylation have been reported.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23 However, the prevalence of MLH1 constitutional epimutations is still unknown. Most studies addressing this issue have enriched their sampling with patients affected with CRC showing loss of MLH1 protein expression.13, 17, 20, 22 In other cases, series were enriched for patients with CRC at an age of onset below 50 years.9, 14, 17, 23
In a very few cases genetic alterations in cis (gross rearrangements and variants in the promoter region) have been identified as responsible for the methylation.13, 16, 19 In these cases, an autosomal dominant pattern is readily observed. However, in most cases no genetic cause for the epimutation has been identified (reviewed in Hitchins and Ward5). In this context, the inheritance of the epimutation has only been experimentally confirmed in three cases.10, 17, 20 The functional impact of these epimutations seems clear. In the few cases analyzed, methylation has been linked to allele-specific silencing of MLH1 and EPM2AIP1.12, 13, 15, 17 This associates with an allele-specific methylation pattern.11, 17, 20, 21 In these cases, methylation seems to be widespread affecting all embryonic layers being mosaicism reported.10, 12, 20
The aim of our study was to investigate the prevalence of MLH1 epimutations in a series of 34 patients with MLH1-methylated CRC and no detected germline MLH1 mutations. We identified two bona fide MLH1 epimutation carriers and extensively characterized one of them. The epimutated allele is maternally transmitted, methylation is present in all embryonic layers, erased in spermatozoa and not transmitted to the next generation.
Materials and methods
Patients and samples
Patients were assessed through Cancer Genetic Counselling Units of the Institut Català d'Oncologia (ICO) and the University of Michigan (UM) from 1998 to 2010. A total of 34 individuals (30 ICO, 4 UM) presenting MLH1-methylated tumors (methylation levels above 20% in C or D regions) were included in this study (Table 1). The ICO patients were selected from a series of 56 individuals with MLH1-deficient CRC and no germline mutations identified in MLH1.24 In all, 29 patients met Bethesda criteria, 1 case met Amsterdam criteria and 4 cases showed other types of CRC familial aggregation. Clinico-pathological information was recorded. Informed consent was obtained from all individuals, and ethics committee approved this study. Sample processing is detailed in Supplementary Methods.
Table 1. Clinical and molecular features of patients with MLH1-methylated CRC.
| % somatic MLH1 methylation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Gender | Clinical criteria | CRC age of onset | CRC location | TNM | Grade | Mucinous component | Other tumors (age of onset) | BRAF | C region (−246) | D region (−13) | MLH1 rs1800734 (c.−93G/A) |
| 1 | M | BC | 32 | L | T3N0M0 | G1 | No | CRC (34) | wt | 57.6 | 59.7 | GA |
| 2 | F | BC | 49 | R | T2N2M0 | G3 | Yes | V600E | 24.9 | 36.9 | GA | |
| 3 | M | BC | 37 | L | T3N0M0 | G2 | No | wt | 29.3 | 31.7 | na | |
| 4 | F | BC | 73 | R | T4aN0M0 | na | Yes | wt | 73.5 | 70.5 | GA | |
| 5 | M | BC | 50 | R | T3N1M0 | G2 | No | wt | 28.6 | 33.6 | na | |
| 6 | F | FA | 62 | R | T3N0M0 | G3 | No | wt | 61.5 | 78.5 | AA | |
| 7 | M | BC | 42 | R | T4N2M0 | G2 | No | CRC (synch) | wt | 24.1 | 25.2 | GG |
| 8 | M | BC | 29 | R | T3N0M0 | G2 | No | wt | 25.1 | 27.6 | na | |
| 9 | F | BC | 47 | L | T3N1M0 | G2 | Yes | wt | 38.5 | 34.9 | na | |
| 10 | F | BC | 77 | R | T3N0M0 | na | Yes | wt | 38.2 | 24.1 | GG | |
| 11 | M | BC | 52 | R | T3N0M0 | G2 | No | V600E | 35.4 | 4.4 | AA | |
| 12 | F | BC | 62 | L | T3N0M0 | G2 | No | wt | 53.7 | 76.7 | GA | |
| 13 | F | BC | 59 | R | T3N0M0 | G2 | No | V600E | 39.4 | 45.8 | GG | |
| 14 | F | BC | 77 | R | T3N0M0 | G2 | No | V600E | 34.5 | 28.4 | GG | |
| 15 | F | BC | 52 | R | T4aN0M0 | G2 | Yes | V600E | 22.9 | 41.4 | GG | |
| 16 | F | BC | 24 | R | T3N0M0 | G3 | No | V600E | 57.5 | 75.1 | GG | |
| 17 | M | FA | 78 | R | T3N0M0 | G2 | No | wt | 12.5 | 24.0 | GG | |
| 18 | M | BC | 48 | R | na | na | No | wt | 32.8 | 34.8 | GA | |
| 19 | M | FA | 73 | R | T3N0M0 | G3 | Yes | V600E | 19.4 | 31.2 | GA | |
| 20 | F | BC | 50 | R | T3N0M0 | G2 | Yes | V600E | 35.8 | 27.0 | GG | |
| 21 | F | BC | 58 | R | T3N0M0 | G2 | No | 3 CRC (synch) | V600E | 40.6 | 66.6 | GG |
| 22 | M | FA | 85 | R | T4bN0M0 | G3 | No | V600E | 41.4 | 42.5 | GA | |
| 23 | F | BC | 47 | L | T3N0M0 | G3 | Yes | V600E | 20.3 | 39.3 | AA | |
| 24 | F | BC | 59 | R | T1N0M0 | G2 | No | CRC (29) | V600E | 11.4 | 20.6 | AA |
| 25 | M | BC | 69 | R | T4N0M0 | G3 | Yes | CRC (synch) | wt | 50.3 | 43.1 | GA |
| 26 | F | BC | 75 | R | T2N0M0 | G2 | No | CRC (64) | V600E | 27.1 | 30.3 | GA |
| 27 | M | BC | 47 | L | T3N0M0 | G1 | No | wt | 40.1 | 21.6 | AA | |
| 28 | M | BC | 31 | L | T4N0M0 | G2 | Yes | wt | 26.2 | 32.7 | GG | |
| 29 | F | BC | 23 | L | T4N1M0 | G2 | No | GC (26) | wt | 79.8 | 50.4 | GA |
| 30 | M | BC | 86 | R | T3N0M0 | na | na | BrC (69); RC (78) | wt | na | na | na |
| 31 | M | AMS | 68 | R | T3N0M0 | na | na | M (80) | wt | na | na | na |
| 32 | F | BC | 55 | R | T2N0M0 | na | na | wt | na | na | na | |
| 33 | F | BC | 52 | R | T3N1M0 | G3 | na | wt | na | na | na | |
| 34 | F | BC | 47 | R | T1N0M0 | na | No | CRC (29), EC (49) | wt | 26.1 | 37.3 | GG |
Abbreviations: AMS, Amsterdam criteria; BC, Bethesda criteria; FA, Familial aggregation; M, male; F, female; R, right; L, left; CRC, colorectal cancer; EC, endometrial cancer; GC, gastric cancer; BrC, breast cancer; RC, renal cancer; M, mesothelioma; synch, synchronous; wt, wild-type; na, not available.
MLH1 promoter methylation analyses
DNA from RKO colorectal tumor cell line (American Type Culture Collection, Manassas, VA, USA) was used as a biallelic MLH1 methylation control. To generate unmethylated DNA, peripheral blood lymphocyte (PBL) DNA was amplified using the REPLI-g kit (Qiagen, Valencia, CA, USA). A sample of CEPH DNA from the Coriell Institute was used as an unmethylated control in pyrosequencing analyses.25
Methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA)
SALSA MS-MLPA ME011 kit (MRC Holland, Amsterdam) is based on the use of probes that contain a digestion site specific for the methylation-sensitive HhaI enzyme. All reactions were carried out using 100 ng of DNA. The kit includes five probe pairs in MLH1 promoter (with the respective HhaI sites located at −659, −383, −246, −13 and +208 relative to the start codon; GenBank accession number U26559) that cover five independent regions: regions A to D of the promoter and intron 1.26
Methylation-specific melting curve analysis
Methylation-specific melting curve analysis method consists in a real-time PCR followed by temperature dissociation on DNA previously treated with sodium bisulfite,27 using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA). Experimental conditions and primers are detailed in Supplementary Methods, Figure S1 and Table S1.
Bisulfite sequencing
A total of 1 μl of bisulfite-converted DNA was used in a PCR reaction for the amplification and subsequently sequencing of MLH1 promoter regions C and D.26 Experimental conditions and primers are detailed in Supplementary Methods, Figure S1 and Table S1.
Clonal bisulfite sequencing
A total of 1 μl of bisulfite-modified DNA was amplified, cloned and sequenced. Experimental conditions and primers are detailed in Supplementary Methods, Figure S1 and Table S1.
Pyrosequencing
In all, 2 μl of bisulfite-converted DNA were used in a PCR reaction for the amplification of regions C and D of the MLH1 promoter26 using HotStarTaq master mix (Qiagen) and biotinylated primers (Supplementary Table S1 and Figure S1). Primers were designed using the Pyromark Assay Design Software 2.0 (Qiagen). Experimental conditions are detailed in Supplementary Methods.
MLH1 allelic expression analyses
For allelic expression analyses at the c.655A>G SNP (rs1799977) within MLH1 exon 8, the relative levels of the A/G alleles were determined in genomic DNA and cDNA by single-nucleotide primer extension (SNuPE) and pyrosequencing, as described in Supplementary Methods.
EPM2AIP1 allelic expression analysis
Amplification and sequencing of rs9311149 flanking region, within EPM2AIP1 gene, was performed as previously described.12 For allelic expression analysis at rs9311149, the relative levels of G/T alleles were determined in genomic DNA and cDNA by SNuPE as described in Supplementary Methods, using primers listed in Supplementary Table S1.
Direct sequencing of MLH1 promoter
Screening for mutations within the MLH1 promoter was performed by PCR amplification and sequencing as described.28 One reverse amplification primer has been modified (Supplementary Table S1).
Haplotype analysis
Haplotype analysis was performed using four intragenic MLH1 single-nucleotide polymorphisms (rs1800734, rs9876116, rs1799977 and rs4234259) and seven microsatellite markers (D3S1609, D3S1612, D3S2369, D3S1611, D3S3623, D3S1298, D3S3564) covering 12 Mb around MLH1, as previously described.29 To deduce the methylation-associated haplotype, intrafamilial segregation analysis was performed under the assumption that the number of crossovers between adjacent markers was minimal.
Second hit analysis
Loss-of-heterozygosity (LOH) analysis was performed on DNA extracted from paraffin-embedded tumor tissue and compared with PBL DNA at informative microsatellites (see haplotype analysis) and SNP rs1799977, either by genotyping or SNuPE (see Supplementary Methods), respectively. MLH1 somatic mutation status was assessed in tumor DNA by direct sequencing and multiplex ligation-dependent probe amplification (SALSA MLPA P003-B1; MRC Holland).
BRAF V600E screening
A 196-bp region of human BRAF gene spanning the hotspot mutation c.1799T>A (V600E) was amplified by PCR (Supplementary Table S1) as described.24 The PCR products were purified using Illustra GFX DNA and Gel Band Purification kit (GE Healthcare, Buckinghamshire, UK). BRAF V600E mutation detection was performed by SNuPE using the ABI PRISM SNaPshot Multiplex Kit (Applied Biosystems, Foster City, CA, USA) and a specific primer.
Results
Clinical and molecular features of patients with MLH1-methylated CRC
In all, 34 patients (15 males; 19 females) were analyzed (Table 1). Mean age at diagnosis was 55 (range 23–86 years). Twenty-six tumors (76%) were located in the right colon and ten (33%) were classified as mucinous. Only six patients (18%) had lymph node involvement and none of them had distal metastasis. BRAF mutations were detected in 13 tumors (38%). A common SNP rs1800734 (c.−93G>A) within the MLH1 promoter was found to be heterozygote in 10 cases (38%) and homozygote A in 5 (19%). In eight individuals (24%), additional LS-associated tumors were diagnosed, three synchronous and five metachronous (Table 1). Molecular characterization of these additional tumors (Table 2) allowed demonstrating the existence of two MLH1-methylated tumors in four individuals (cases 1, 7, 29 and 34).
Table 2. Molecular features of tumors from patients affected by multiple LS-associated tumors.
| IHC | % somatic MLH1 methylation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Tumor type | Age of onset | MSI analysis | MLH1 | MSH2 | MSH6 | PMS2 | BRAF | C region (−246) | D region (−13) |
| 1 | CRCa | 32 | + | − | + | + | − | wt | 57.6 | 59.7 |
| CRC | 34 | + | − | + | + | − | wt | 60.5 | 62.8 | |
| 7 | CRCa | 42 | + | − | + | + | ND | wt | 24.1 | 25.2 |
| CRC | 42 | + | − | + | + | − | wt | 28.9 | 24.2 | |
| 21 | CRCa | 58 | + | − | + | + | ND | V600E | 40.6 | 66.6 |
| CRC | 58 | − | + | + | + | + | ND | ND | ND | |
| CRC | 58 | − | + | + | + | + | ND | ND | ND | |
| CRC | 58 | − | ND | ND | ND | ND | ND | ND | ND | |
| 24 | CRC | 29 | NA | NA | NA | NA | NA | NA | NA | NA |
| CRCa | 59 | + | − | + | + | ND | V600E | 11.4 | 20.6 | |
| 25 | CRCa | 69 | + | − | + | + | ND | wt | 50.3 | 43.1 |
| CRC | 69 | NA | NA | NA | NA | NA | NA | NA | NA | |
| 26 | CRC | 64 | NA | NA | NA | NA | NA | NA | NA | NA |
| CRCa | 75 | + | − | + | + | − | V600E | 27.1 | 30.1 | |
| 29 | CRCa | 23 | + | − | + | + | ND | ND | 79.8 | 50.4 |
| GC | 26 | + | − | + | + | ND | ND | 63.0 | 73.0 | |
| 34 | CRC | 29 | NA | NA | NA | NA | NA | NA | NA | NA |
| CRCa | 47 | ND | − | ND | ND | − | wt | 55.5 | 48.8 | |
| EC | 49 | + | − | + | + | ND | wt | 26.1 | 37.3 | |
Abbreviations: NA, not available; ND, not done.
Tumors included in the initial series listed in Table 1.
Identification of new LS cases harboring a constitutional MLH1 epimutation
The methylation status of MLH1 promoter was analyzed by MS-MLPA in DNA extracted from PBLs. Constitutional methylation was only detected in 2 individuals (cases 1 and 34) of the 34 patients included (5.9%). It represented 2 out of 100 LS cases in ICO series (2%). In both cases, methylation in MLH1 promoter was detected in the five regions analyzed, including C and D promoter regions, which was correlated with transcriptional silencing26 (Table 3).
Table 3. Analysis of MLH1 methylation using MS-MLPA in samples from the proband and relatives.
| % MLH1 methylation | |||||||
|---|---|---|---|---|---|---|---|
| Family | Individual | Sample | A region (−659) | B region (−383) | C region (−246) | D region (−13) | Intron 1 (+208) |
| A | I.1 | PBL | 0 | 0 | 0 | 0 | 0 |
| II.1 (case 1) | CRC 1 | 61.2 | 83.7 | 57.6 | 59.7 | 60.9 | |
| CRC 2 | 62.3 | 86.9 | 60.5 | 62.8 | 63.5 | ||
| PBL | 60.5 | 76.7 | 56.0 | 56.2 | 60.2 | ||
| fibroblasts | 55.8 | 53.2 | 64.0 | 52.4 | 63.0 | ||
| colonic mucosa | 52.3 | 78.9 | 58.3 | 48.5 | 62.6 | ||
| sperm | 0 | 0 | 0 | 0 | 0 | ||
| II.2 | PBL | 0 | 0 | 0 | 0 | 0 | |
| III.1 | PBL | 0 | 0 | 0 | 0 | 0 | |
| III.2 | PBL | 0 | 0 | 0 | 0 | 0 | |
| B | case 34 | EC | 33.6 | 59.4 | 26.1 | 37.3 | 28.5 |
| CRC | 58.0 | 56.3 | 55.5 | 48.8 | 56.4 | ||
| PBL | 35.9 | 45.3 | 25.1 | 27.6 | 27.7 | ||
| RKO | 110.1 | 113.2 | 103.0 | 88.2 | 103.4 | ||
Peripheral blood lymphocytes (PBL), skin fibroblasts, colorectal tumors (CRC 1 and 2), normal adjacent mucosa and sperm from case 1 (II.1), PBL from his relatives, and PBL, CRC and endometrial cancer (EC) from case 34, were analyzed. DNA from RKO cell line (methylated in MLH1) is used as a positive control. Representative data from two independent experiments is shown. Methylation levels above 20% are shown in bold.
Sequencing analysis of the whole MLH1 promoter (from c.−1469 to intron 1) in PBL DNA from cases 1 and 34 did not detect any variant affecting the binding of MLPA probes nor HhaI restriction sites. Likewise, it revealed that case 1 was heterozygous for SNP rs1800734 (c.−93G>A) and case 34 was heterozygous for SNP rs34566456 (c.−607G>C). No other variants – including c.−27C>A and c.85G>T16 – were identified within the promoter region.
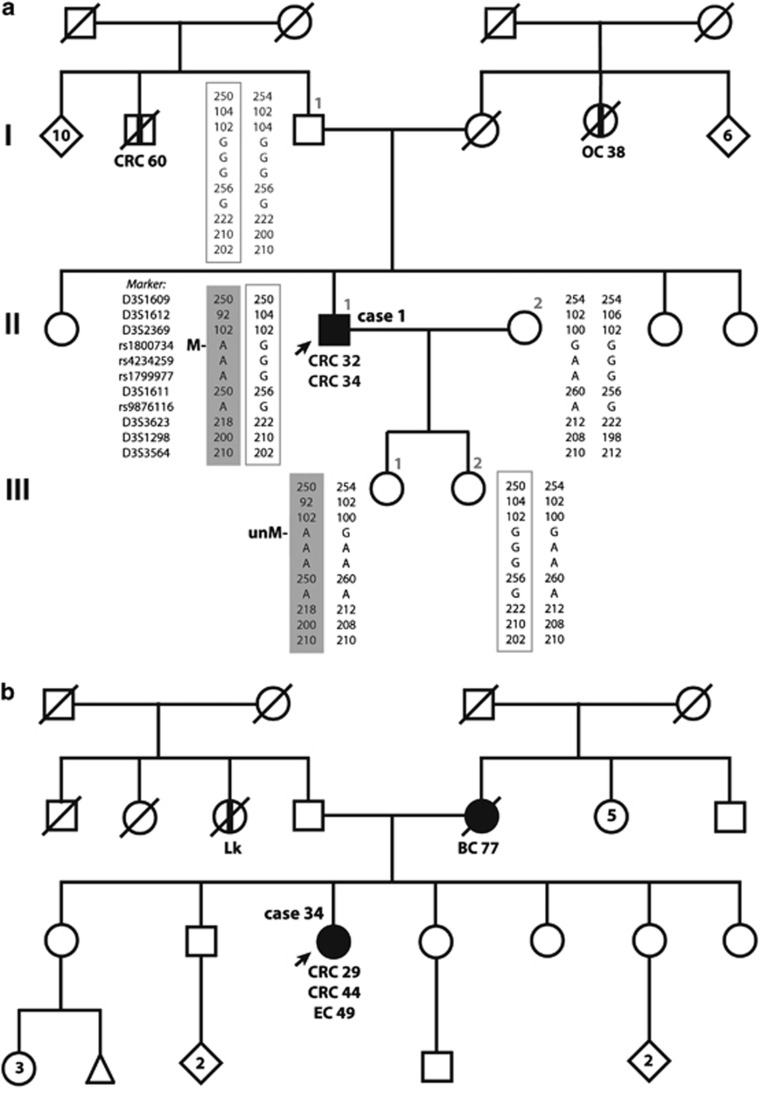
Case 1 is a 47-year-old male who underwent urgent sigmoidectomy due to intestinal occlusion secondary to a sigmoid adenocarcinoma (pT3N0M0, stage II) at the age of 32. After 2 years, the patient was diagnosed with an adenocarcinoma of the hepatic flexure (pT3N0M0, stage II) and a subtotal colectomy was carried out. Microsatellite analysis showed MSI, loss of MLH1 and PMS2 expression, absence of BRAF V600E mutation and somatic MLH1 methylation in both tumors (Table 2). The patient had no family history of cancer in his first-degree relatives as it is shown in his pedigree (Figure 1a).
Figure 1.
Family pedigree of the epimutation carriers. Circles, females; squares, males; filled, cancer affected; vertical line at center, non-confirmed cancer affected. Cancer localization (CRC, colorectal cancer; OC, ovarian cancer; EC, endometrial cancer; BC, breast cancer; Lk, leukemia) and age at diagnosis are indicated. (a) Pedigree and haplotypes of case 1. The epimutation carrier (II.1) is indicated by an arrow. Generations are indicated on the left margin in Roman numerals and analyzed relatives are identified by numbers. Haplotypes, generated by analyzing SNP and microsatellite markers flanking or within MLH1, are detailed according to the key indicated in individual II.1. The paternally inherited allele in II.1 is in a square and the maternally derived allele is highlighted in dark gray. The presence of methylation (M) or its absence (unM) is indicated. (b) Pedigree of case 34. The epimutation carrier is indicated by an arrow.
Case 34 is a 55-year-old female who was diagnosed of a sigmoid adenocarcinoma (pT3N1M0, stage III) at the age of 29 years and underwent a sigmoidectomy. After 15 years, the patient was diagnosed with an adenocarcinoma of the hepatic flexure (pT1N0M0, stage I). At the age of 49 years, she was diagnosed of an endometrial adenocarcinoma (pT1N0M0). Microsatellite analysis showed instability of the five analyzed markers in the second CRC, and instability of bat26 and MONO-27 in the endometrial cancer. Both second colorectal and endometrial tumors showed loss of MLH1 and PMS2 expression, absence of BRAF V600E mutation and somatic MLH1 methylation (Table 2). Patient's mother was affected by a breast cancer at the age of 77 years (Figure 1b).
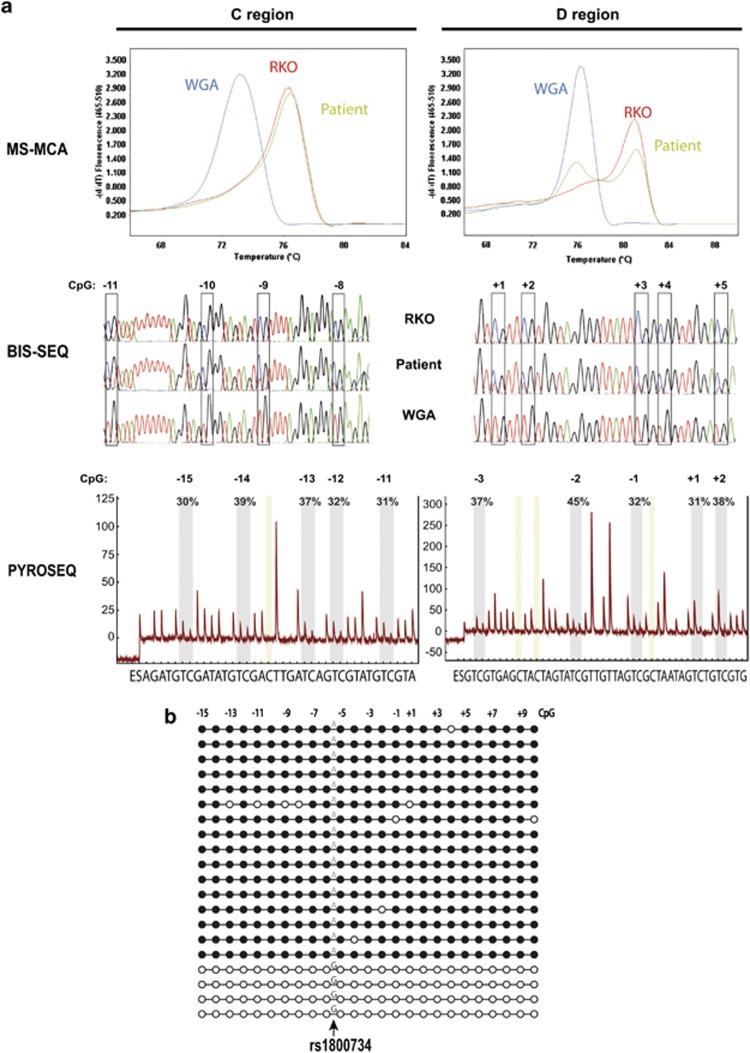
Methylation-specific melting curve analysis confirmed the presence of a methylated peak in C and D promoter regions in both cases (Figure 2a and Supplementary Figure S2). Likewise, bisulfite sequencing showed the presence of both methylated C as well as non-methylated T (bisulfite-converted non-methylated C) alleles at each CpG site in the samples of interest (Figure 2a and Supplementary Figure S2). Average methylation levels in PBL of the case 1 were 34% and 39% in C and D regions, respectively, as assesed by pyrosequencing (Figure 2a; Table 4). Clonal bisulfite sequence analysis confirmed hemiallelic methylation in PBL DNA confined to allele A of the rs1800734 (Figure 2b). In case 34, average methylation levels in PBL were 20% and 19% in C and D regions, respectively (Supplementary Figure S2; Table 4).
Figure 2.
Confirmation of the constitutional MLH1 epimutation of case 1. (a) Analysis of the MLH1 promoter C and D regions by methylation-specific melting curve analysis (MS-MCA), bisulfite sequencing (BIS-SEQ) and pyrosequencing (PYROSEQ). Top panel: MS-MCA of MLH1 promoter. In the analysis of C region, WGA DNA (unmethylated control) and RKO DNA (methylated control) show single melting peaks at 73 and 77° C, respectively. In D region, WGA and RKO melting peaks temperature are 76 and 82° C, respectively. Analysis by MS-MCA in PBL DNA from the patient 1 (green line) shows the presence of the methylated peak in both regions. Middle panel: sequence analysis of bisulfite-converted DNA. WGA DNA shows T at each CpG analyzed, consistent with complete modification of the DNA. RKO DNA shows C at each CpG. Patient DNA shows a mixture of T and C at CpG sites, attributable to partial methylation. Bottom panel: representative pyrograms obtained in the analysis of C and D MLH1 promoter regions in PBL DNA from the patient. The peaks within the shaded area of the pyrogram correspond to the CpG interrogated. Percentage methylation at each site is calculated as the C:T ratio of peak heights (representing methylated:unmethylated cytosine). x axis represents the nucleotide dispensation order. y axis units are arbitrary representing light intensity. (b) Clonal bisulfite sequencing of the MLH1 promoter in PBL DNA from the epimutation carrier 1. Each horizontal line represents a single allele. CpG dinucleotides are depicted by circles. Black and white circles indicate methylated and unmethylated CpG, respectively. The allele at rs1800734 (c.−93G>A) is indicated as A or G. Methylation is confined to the A allele. Each CpG analyzed is numbered according to its position relative to the translation initiation codon.
Table 4. Quantification of MLH1 promoter methylation by pyrosequencing.
| MLH1 promoter C region | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG position | |||||||||||
| Family | Individual | Sample | −15 | −14 | −13 | −12 | −11 | Mean | SD | Min | Max |
| A | II.1 (case 1) | PBL | 32.0 | 38.1 | 36.1 | 31.7 | 33.6 | 34.3 | 2.8 | 31.7 | 38.1 |
| sperm | 2.1 | 0.0 | 3.8 | 2.1 | 1.4 | 1.9 | 1.4 | 0.0 | 3.8 | ||
| B | case 34 | PBL | 22.1 | 21.6 | 20.1 | 17.1 | 17.7 | 19.7 | 2.3 | 17.1 | 22.1 |
| RKO | 95.5 | 96.5 | 94.2 | 92.6 | 95.9 | 94.9 | 1.6 | 92.6 | 96.5 | ||
| CEPH | 2.2 | 2.15 | 3.6 | 2.55 | 2.3 | 2.6 | 0.6 | 2.2 | 3.6 | ||
| MLH1 promoter D region | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG position | ||||||||||||||
| Family | Individual | Sample | −6 | −5 | −4 | −3 | −2 | −1 | 1 | 2 | Mean | SD | Min | Max |
| A | II.1 (case 1) | PBL | 39.0 | 50.0 | 38.9 | 36.4 | 43.8 | 33.3 | 32.1 | 39.4 | 39.1 | 5.8 | 32.1 | 50.0 |
| sperm | 0.0 | 5.3 | 0.0 | 1.6 | 6.5 | 2.9 | 0.0 | 1.7 | 2.3 | 2.5 | 0.0 | 6.5 | ||
| B | case 34 | PBL | 19.7 | 20.5 | 19.1 | 19.2 | 17.2 | 18.3 | 19.8 | 19.2 | 19.1 | 1.0 | 17.2 | 20.6 |
| RKO | 95.5 | 92.6 | 84.0 | 90.2 | 76.1 | 72.7 | 81.0 | 93.7 | 85.7 | 8.6 | 72.7 | 95.5 | ||
| CEPH | 3.4 | 5.7 | 0.0 | 0.0 | 9.4 | 3.8 | 2.7 | 2.9 | 3.5 | 3.0 | 0.0 | 9.4 | ||
Each sample was run in triplicates. Methylation at each specific CpG was calculated as the mean of the triplicates. Values for each specific CpG within the region are given in percentage. Average percentage of methylation of the whole region was calculated as the mean for the five CpGs analyzed in C region and the eight CpGs in the D region. Both peripheral blood lymphocytes (PBL) and sperm from the proband (II.1) were analyzed. DNA from the colorectal cancer cell line RKO was used as positive control. CEPH DNA was used as negative control. Each CpG analyzed is numbered according to its position relative to the translation initiation codon.
Functional impact of the MLH1 epimutations
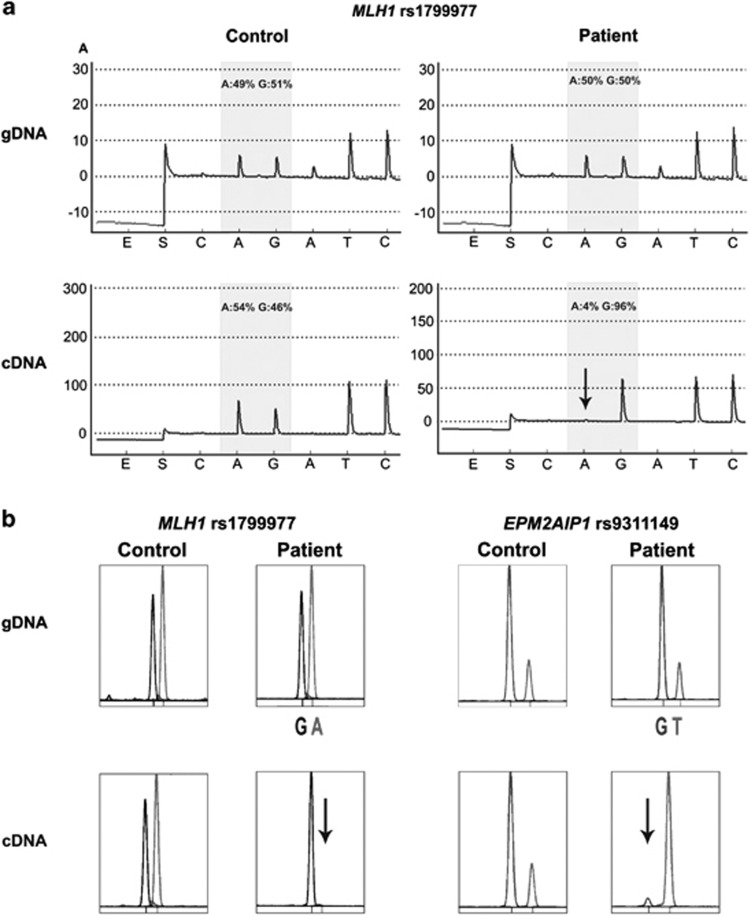
The MLH1 promoter is bi-directional for transcription of MLH1 and EPM2AIP1 genes. In case 1, the neutral heterozygous polymorphism c.655G>A (rs1799977) within MLH1 exon 8 was used to determine the effect of the epimutation on MLH1 transcriptional activity. Monoallelic expression of MLH1 transcript, associated to G allele, was demonstrated by pyrosequencing and SNuPE (Figure 3). ASE (allele-specific expression) values obtained in patient and control sample were 0.05 and 1.17 when analyzed by pyrosequencing, and 0.02 and 0.98 by SNuPE, respectively. In case 34, the absence of coding heterozygous polymorphisms in MLH1 prevented its transcriptional analysis.
Figure 3.
Transcriptional inactivation of MLH1 and EPM2AIP1 alleles. (a) Illustrative example of the pyrogram across the expressible MLH1 rs1799977 (c.655A>G) in genomic DNA (gDNA) (top panels) and cDNA (bottom panels) derived from a heterozygous healthy control (left panels) and the epimutation carrier (right panels). The peaks within the shaded area of the pyrogram are the nucleotides at the SNP site, quantified with respect to neighboring nucleotides. Their relative values are given as percentage values above the pyrogram trace. There was a transcriptional inactivation of the ‘A' allele (indicated with a downward arrow) in the cDNA of the patient with the MLH1 epimutation. x axis represents the nucleotide dispensation order. y axis units are arbitrary representing light intensity. (b) Representative results of the SNuPE analysis at MLH1 rs1799977 (c.655A>G) (left panel) and EPM2AIP1 rs9311149 (right panel) in gDNA and cDNA derived from a heterozygous control and the epimutation carrier. Transcriptional silencing of the A allele at MLH1 rs1799977 and T allele at EPM2AIP1 rs9311149 in the cDNA of the patient was observed.
SNuPE analysis at rs9311149 of EPM2AIP1 evidenced complete silencing of EPM2AIP1 G allele in case 1 (Figure 3b, right panel) and partial silencing of the same allele in case 34 (Supplementary Figure S2b), further reinforcing the functional impact of the constitutional methylation. The obtained ASE values were 0.02 in case 1, 0.48 in case 34 and 1.00 in control sample.
Characterization of the MLH1 epimutation
MLH1 methylation pattern
Follow-up of case 34 and her family has proved difficult. Thus, for the purpose of detailed characterization, we have focused in the characterization of case 1. First, we wanted to explore whether methylation was present in all embryonic layers and in the germline of case 1. MS-MLPA analysis in skin fibroblasts (ectoderm) and colorectal mucosa (endoderm) revealed similar levels of MLH1 methylation than in PBL (Table 3), indicating hemiallelic methylation in all embryonic layers. In contrast, no methylation was detected in patient sperm as evidenced by MS-MLPA and pyrosequencing analyses (Tables 3 and 4). Direct sequencing of the PBL and sperm for MLH1 promoter C region evidenced the presence of both alleles at rs1800734 in both samples (data not shown). These results indicate the reversion of the epimutation in patient spermatozoa.
Inheritance pattern of the epimutant allele
To further investigate the inheritance pattern of the allele harboring the epimutation, we analyzed the MLH1 promoter methylation status as well as a haplotype of 12 Mb around MLH1 in available PBL DNA from patient's first-degree relatives. MS-MLPA analysis showed no evidence of MLH1 methylation in relatives (Table 3). Haplotype analysis revealed that the epimutated allele is only shared by the patient and one of his daughters (Figure 1a). The lack of availability of biological material from the mother has precluded us from analyzing the presence of the epimutation in her. These results confirmed that the epimutated allele is maternally inherited in the patient, and that methylation is erased in the patient's daughter who inherited the same allele.
Inactivation of the non-methylated allele in tumor tissue
We explored the nature of the putative second hit in the patient's sigmoid colon cancer. Full exonic sequencing of the MLH1-coding region did not identify any additional mutation. LOH was evidenced at MLH1 rs1799977 and D3S1611 (data not shown). Retention of heterozygosity was observed at the distal marker D3S3564, whereas LOH was not evaluable at markers D3S1612, D3S3623 and D3S1298 due to their instability. These results point to the loss of the wild-type MLH1 allele in tumor DNA. MLPA analysis in tumor DNA was not conclusive, probably owing to the poor quality of tumor FFPE-DNA.
Discussion
We identified two bona fide MLH1 epimutations and one of them has been extensively characterized. In previous reports, MLH1 epimutations were detected in 8–13% of patients with tumors showing MLH1 loss of expression.13, 17, 20, 22 We have detected this alteration in 2 out of 30 patients with MLH1-methylated CRC meeting Bethesda or Amsterdam criteria (6.7%) and in 2 of 14 patients with an age of onset below 50 years (14.2%), in whom no germline MLH1 mutation was identified. This is in line with the prevalence reported by van Roon et al23 in patients with MLH1-methylated tumors enriched for cases with an early age of onset. If we take into consideration only the ICO series, MLH1 epimutations represent so far 2% of all LS cases.
In accordance with previous reports (reviewed in Hitchins and Ward5), the cases identified in this study had developed multiple LS tumors at an early age. This may not only reflect the phenotype associated with the epimutation but also the selection criteria used so far in most studies. Of note, methylation was not only detected in metachronous colon tumors but also in endometrial carcinomas as well. BRAF mutation was absent in four analyzed tumors from the identified epimutation carriers. However, the presence of somatic BRAF V600E mutation has been previously reported in tumors from three epimutation carriers,10, 12, 23 representing 15.8% (3/22) of the reported cases. In our set of cases, the degree of MLH1 methylation is highly variable among tumors from both epimutation carriers and the remaining patients. Epimutations have been detected in two of four cases where multiple tumors showed somatic MLH1 hypermethylation.
PBL methylation levels correlated with the observed transcriptional silencing, suggesting the presence of mosaicism in case 34. Dosage of the methylated allele is important. In line with previous observations, approximately 50% of the alleles were methylated in case 1.10, 11, 14, 17, 20, 21 As reported, the functional impact of the epimutation seems clear, as it associates with monoallelic expression of MLH1 and EPM2AIP1 transcripts12, 13, 15, 17 and an allele-specific methylation pattern.14, 17, 20, 21 LOH in an intragenic MLH1 microsatellite marker was detected, consistent with somatic loss of the unmethylated allele. In fact, LOH has been found to be the most frequent mechanism of inactivation of wild-type allele in tumors from epimutation carriers.12
So far, in all cases identified but one, the methylated allele was of maternal origin.10, 12, 15, 17, 20 The epimutation was found in the maternally inherited allele. Although we were unable to definitively demonstrate whether the epimutation was inherited or de novo, this may further support the notion that this type of aberration is more likely to accumulate during the oogenesis. We were able to perform a more detailed study of the index case and descendants. While MLH1 methylation was present in every embryonic layer of the index case, a complete erasure was observed in the spermatozoa, as reported by Hitchins et al.17, 30 The lack of methylation in spermatozoa does not necessarily mean that inheritance cannot occur. In fact, this was clearly demonstrated in one descendant who inherited the epimutation out of three harboring the same allele.17 In our case, the epimutated allele was transmitted unmethylated to one of his daughters.
In spite of an extensive search, we have not been able to identify a genetic alteration underlying the epimutated allele. Genetic aberrations in cis (gross rearrangements in two cases (one deletion of MLH1 exons 1 and 2, and one duplication involving the whole gene) and in a third one the variant c.−27C>A within the promoter region) have been identified as responsible for MLH1 methylation.13, 16, 19 Dominant transmission pattern is observed in these cases. Dominant inheritance has been also observed in cases where no genetic alterations are detected.10, 12, 15, 17, 20 In these cases, methylation was mosaic and associated to a shared haplotype.
Although we cannot completely rule out that aberrations have been missed, the lack of family history and the lack of vertical transmission are compatible with a de novo methylation occurred in the early embryo, where there is no apparent predisposing genetic mechanism that would allow for the restoration of methylation after the gametogenesis. However, this is an unsettled issue. The epimutation carrier identified in this study showed methylation confined to the A allele at rs1800734, although allele-specific methylation is not restricted to either A or G allele in other reported cases.14, 17, 20, 21 It is intriguing that the A allele at rs1800734 associates with somatic MLH1 promoter methylation and increased risk of MSI CRC.23, 31, 32, 33, 34, 35 In addition, it has been shown that this polymorphism modifies the efficiency of MLH1/EPM2AIP1 transcription.36
It is difficult to translate these findings into specific recommendations for these patients and their relatives. At this time caution is mandatory. In the presence of a detected constitutional epimutation, genetic screening of descendants is important. However, in the presence of an inherited non-methylated allele in lymphocyte DNA, two options are available. On the one hand, descendants can be counseled as relatives of a LS case where direct genetic testing has been non-informative. In this setting, it is assumed that lack of methylation in the inherited allele does not rule out that a mosaic status is present in the patient or that a non-detected genetic alteration predisposing to a late acquisition of methylation is present in this family. Alternatively, recommendation can be made based on the degree of personal and familial history of cancer. Further knowledge is needed to translate these research findings into useful information for management of patients and families.
The increasing detection of epimutations has lead to the suggestion that the diagnostic algorithm of LS might be improved. So far, the detection of somatic MLH1 hypermethylation is often used to exclude patients from further MMR mutation analysis, based on cost effectiveness considerations.24, 37 The patients with somatic MLH1 hypermethylation could now be considered as candidates to screen for constitutional MLH1 epimutations. Based on the clinical presentation of the reported cases5 and our experience, this screening could be restricted to those diagnosed earlier than 50 years or with multiple tumors the first one before the age of 60. If this was the case, MS-MLPA could be a good methodological approach. The robustness and informativeness already shown for paraffin-embedded tissues24 has been confirmed when being used in the germline. In any case, confirmation with at least another technique (ie, pyrosequencing) would be mandatory.
In summary, MLH1 methylation screening in PBL from patients with early-onset MLH1-methylated CRC allows the identification of epimutation carriers. Using this strategy we have identified two bona fide MLH1 epimutations. In one of them, the methylated allele is from maternal origin, is present in all embryonic layers and is absent in spermatozoa. The characterization of these cases provides further evidence of the emerging entity of soma-wide MLH1 epimutation and its heritability.
Acknowledgments
We thank the patients who participated in this study, Gemma Aiza for technical assistance, Javier Carmona for his assistance with pyrosequencing and Dr Juana Fernández for her assistance in skin fibroblast isolation and culture. This work was supported by grants from Ministerio de Ciencia e Innovación (SAF 06-06084; 09-07319), Fundació Gastroenterologia Dr Francisco Vilardell (F05-01), Ministerio de Educación y Ciencia Spanish Networks RTICCC (RD06/0020/1050, 1051), Acción en Cáncer (Instituto de Salud Carlos III), Fundación Científica AECC and NCI U19 CA 148107-02.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Lynch HT, Lynch JF, Lynch PM, Attard T. Hereditary colorectal cancer syndromes: molecular genetics, genetic counseling, diagnosis and management. Fam Cancer. 2008;7:27–39. doi: 10.1007/s10689-007-9165-5. [DOI] [PubMed] [Google Scholar]
- Vasen HF, Moslein G, Alonso A, et al. Guidelines for the clinical management of Lynch syndrome (hereditary non-polyposis cancer) J Med Genet. 2007;44:353–362. doi: 10.1136/jmg.2007.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer: the syndrome, the genes, and historical perspectives. J Natl Cancer Inst. 1995;87:1114–1125. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition -- update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP, Ward RL. Constitutional (germline) MLH1 epimutation as an aetiological mechanism for hereditary non-polyposis colorectal cancer. J Med Genet. 2009;46:793–802. doi: 10.1136/jmg.2009.068122. [DOI] [PubMed] [Google Scholar]
- Kuiper RP, Vissers LE, Venkatachalam R, et al. Recurrence and variability of germline EPCAM deletions in Lynch syndrome. Hum Mutat. 2011;32:407–414. doi: 10.1002/humu.21446. [DOI] [PubMed] [Google Scholar]
- Hesson LB, Hitchins MP, Ward RL. Epimutations and cancer predisposition: importance and mechanisms. Curr Opin Genet Dev. 2010;20:290–298. doi: 10.1016/j.gde.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Ligtenberg MJ, Kuiper RP, Chan TL, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- Auclair J, Vaissiere T, Desseigne F, et al. Intensity-dependent constitutional MLH1 promoter methylation leads to early onset of colorectal cancer by affecting both alleles. Genes Chromosomes Cancer. 2011;50:178–185. doi: 10.1002/gcc.20842. [DOI] [PubMed] [Google Scholar]
- Crepin M, Dieu MC, Lejeune S, et al. Evidence of constitutional MLH1 epimutation associated to transgenerational inheritance of cancer susceptibility. Hum Mutat. 2012;33:180–188. doi: 10.1002/humu.21617. [DOI] [PubMed] [Google Scholar]
- Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925–3928. [PubMed] [Google Scholar]
- Goel A, Nguyen TP, Leung HC, et al. De novo constitutional MLH1 epimutations confer early-onset colorectal cancer in two new sporadic Lynch syndrome cases, with derivation of the epimutation on the paternal allele in one. Int J Cancer. 2011;128:869–878. doi: 10.1002/ijc.25422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gylling A, Ridanpaa M, Vierimaa O, et al. Large genomic rearrangements and germline epimutations in Lynch syndrome. Int J Cancer. 2009;124:2333–2340. doi: 10.1002/ijc.24230. [DOI] [PubMed] [Google Scholar]
- Hitchins M, Owens S, Kwok CT, Godsmark G, Algar U, Ramesar R. Identification of new cases of early-onset colorectal cancer with an MLH1 epimutation in an ethnically diverse South African cohort. Clin Genet. 2011;80:428–434. doi: 10.1111/j.1399-0004.2011.01660.x. [DOI] [PubMed] [Google Scholar]
- Hitchins M, Williams R, Cheong K, et al. MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;129:1392–1399. doi: 10.1053/j.gastro.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Hitchins MP, Rapkins RW, Kwok CT, et al. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5′UTR. Cancer Cell. 2011;20:200–213. doi: 10.1016/j.ccr.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Hitchins MP, Wong JJ, Suthers G, et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- Miyakura Y, Sugano K, Akasu T, et al. Extensive but hemiallelic methylation of the hMLH1 promoter region in early-onset sporadic colon cancers with microsatellite instability. Clin Gastroenterol Hepatol. 2004;2:147–156. doi: 10.1016/s1542-3565(03)00314-8. [DOI] [PubMed] [Google Scholar]
- Morak M, Koehler U, Schackert HK, et al. Biallelic MLH1 SNP cDNA expression or constitutional promoter methylation can hide genomic rearrangements causing Lynch syndrome. J Med Genet. 2011;48:513–519. doi: 10.1136/jmedgenet-2011-100050. [DOI] [PubMed] [Google Scholar]
- Morak M, Schackert HK, Rahner N, et al. Further evidence for heritability of an epimutation in one of 12 cases with MLH1 promoter methylation in blood cells clinically displaying HNPCC. Eur J Hum Genet. 2008;16:804–811. doi: 10.1038/ejhg.2008.25. [DOI] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- Valle L, Carbonell P, Fernandez V, et al. MLH1 germline epimutations in selected patients with early-onset non-polyposis colorectal cancer. Clin Genet. 2007;71:232–237. doi: 10.1111/j.1399-0004.2007.00751.x. [DOI] [PubMed] [Google Scholar]
- van Roon EH, van Puijenbroek M, Middeldorp A, et al. Early onset MSI-H colon cancer with MLH1 promoter methylation, is there a genetic predisposition. BMC Cancer. 2010;10:180. doi: 10.1186/1471-2407-10-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gausachs M, Mur P, Corral J, et al. MLH1 promoter hypermethylation in the analytical algorithm of Lynch syndrome: a cost-effectiveness study Eur J Hum Genet 2012(in press). [DOI] [PMC free article] [PubMed]
- Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- Deng G, Peng E, Gum J, Terdiman J, Sleisenger M, Kim YS. Methylation of hMLH1 promoter correlates with the gene silencing with a region-specific manner in colorectal cancer. Br J Cancer. 2002;86:574–579. doi: 10.1038/sj.bjc.6600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara D, Rodriguez-Moranta F, de Oca J, et al. Novel methylation panel for the early detection of colorectal tumors in stool DNA. Clin Colorectal Cancer. 2010;9:168–176. doi: 10.3816/CCC.2010.n.023. [DOI] [PubMed] [Google Scholar]
- Mueller J, Gazzoli I, Bandipalliam P, Garber JE, Syngal S, Kolodner RD. Comprehensive molecular analysis of mismatch repair gene defects in suspected Lynch syndrome (hereditary nonpolyposis colorectal cancer) cases. Cancer Res. 2009;69:7053–7061. doi: 10.1158/0008-5472.CAN-09-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borras E, Pineda M, Blanco I, et al. MLH1 founder mutations with moderate penetrance in Spanish Lynch syndrome families. Cancer Res. 2010;70:7379–7391. doi: 10.1158/0008-5472.CAN-10-0570. [DOI] [PubMed] [Google Scholar]
- Hitchins MP, Ward RL. Erasure of MLH1 methylation in spermatozoa-implications for epigenetic inheritance. Nat Genet. 2007;39:1289. doi: 10.1038/ng1107-1289. [DOI] [PubMed] [Google Scholar]
- Whiffin N, Broderick P, Lubbe SJ, et al. MLH1-93G>A is a risk factor for MSI colorectal cancer. Carcinogenesis. 2011;32:1157–1161. doi: 10.1093/carcin/bgr089. [DOI] [PubMed] [Google Scholar]
- Allan JM, Shorto J, Adlard J, et al. MLH1 -93G>A promoter polymorphism and risk of mismatch repair deficient colorectal cancer. Int J Cancer. 2008;123:2456–2459. doi: 10.1002/ijc.23770. [DOI] [PubMed] [Google Scholar]
- Chen H, Taylor NP, Sotamaa KM, et al. Evidence for heritable predisposition to epigenetic silencing of MLH1. Int J Cancer. 2007;120:1684–1688. doi: 10.1002/ijc.22406. [DOI] [PubMed] [Google Scholar]
- Mrkonjic M, Roslin NM, Greenwood CM, et al. Specific variants in the MLH1 gene region may drive DNA methylation, loss of protein expression, and MSI-H colorectal cancer. PLoS One. 2010;5:e13314. doi: 10.1371/journal.pone.0013314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PT, Curtin K, Ulrich CM, et al. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut. 2009;58:661–667. doi: 10.1136/gut.2007.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera S, Mrkonjic M, Rawson JB, Bapat B. Functional effects of the MLH1-93G>A polymorphism on MLH1/EPM2AIP1 promoter activity. Oncol Rep. 2011;25:809–815. doi: 10.3892/or.2010.1129. [DOI] [PubMed] [Google Scholar]
- Perez-Carbonell L, Alenda C, Paya A, et al. Methylation analysis of MLH1 improves the selection of patients for genetic testing in Lynch syndrome. J Mol Diagn. 2010;12:498–504. doi: 10.2353/jmoldx.2010.090212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.