Abstract
While there has been remarkable progress in understanding the biology of HIV-1 and its recognition by the human immune system, we have not yet developed an efficacious HIV-1 vaccine. Vaccine challenges include the genetic diversity and mutability of HIV-1 which create a plethora of constantly changing antigens, the structural features of the viral envelope glycoprotein that disguise conserved receptor-binding sites from the immune system, and the presence of carbohydrate moieties that shield potential epitopes from antibodies. Despite these challenges, there has been significant scientific progress in recent years. In 2009, a large-scale clinical trial known as RV144 demonstrated that a HIV-1 vaccine could modestly reduce the incidence of HIV-1 infection. Further, the identification of broadly neutralizing monoclonal antibodies (such as VRC01, a human monoclonal antibody capable of neutralizing over 90% of natural HIV-1 isolates, as well as PG and PGT antibodies that recognize conserved glycopeptide epitopes) has revealed new opportunities for vaccine design. Our ability to understand HIV-1 structure and antibody epitopes at the atomic level, the rapid advance of computational and bioinformatics approaches to immunogen design, and our newly acquired knowledge that it is possible for a vaccine to reduce the risk of HIV-1 infection, have all opened up new and promising pathways towards the development of an urgently needed effective HIV-1 vaccine. This article summarizes challenges to the development of an HIV-1 vaccine, lessons learned from scientific investigation and completed vaccine trials, and promising developments in HIV-1 vaccine design.
Keywords: HIV-1, HIV-1 clinical trials, vaccine design, structural biology, antibody response, somatic maturation
Introduction
Although insight into HIV-1 pathogenesis has been gained since the identification of HIV-1, the successful development of an effective vaccine has been elusive. HIV-1 has a high degree of antigenic and genetic diversity. In addition, the virus has evolved multiple mechanisms to inhibit elicitation of and neutralization by antibodies. New molecular and structural technologies have been applied to gain a better understanding of HIV-1 as an immune target and to provide new insights into the development of improved immunogens capable of eliciting immune responses that prevent infection by circulating strains of HIV-1.
Challenges in developing an effective HIV-1 vaccine
Unlike currently licensed vaccines, which are typically designed to elicit neutralizing antibodies against a limited number of viral surface proteins, HIV-1 vaccines must counteract a swarm of viruses. The genetic diversity and mutability of HIV-1 creates a plethora of antigens that are constantly changing. Within infected individuals, the struggle between the virus and the immune system is persistent, such that the virus continually escapes host immunity and replicates.
In addition to the genetic diversity and mutability of the HIV-1 Envelope (Env), structural features of Env create inherent difficulties in the ability of the immune system to develop an effective neutralizing antibody. HIV-1 is an enveloped virus with a lipid bilayer surrounding and protecting its core structural proteins. The virus spikes protrude through this protective lipid, and every spike is composed of three gp120 proteins, each of which is non-covalently associated with a gp41 transmembrane glycoprotein molecule. HIV-1 entry into host cells is mediated by binding of gp120 to its primary receptor, the CD4 glycoprotein on the cell surface. Binding to CD4 induces conformational changes in gp120, leading to the exposure and/or formation of a binding site for specific chemokine receptors, mainly CCR5 and CXCR4, which serve as secondary receptors for virus entry [1]. Structurally, the gp120 glycoprotein is divided into three parts, an inner domain, an outer domain, and a bridging sheet. The bridging sheet is the part of the molecule that is responsible for binding to both chemokine receptor and CD4. The CD4 binding site is highly conserved, since the virus needs a conserved region to recognize CD4.
HIV-1 gp120 contains a number of features that help the virus evade the host's humoral immunity, including variable loops [2], N-linked glycosylation [3,4], and conformational flexibility [5,6]. The conformational flexibility of gp120 disguises the conserved receptor-binding sites from the humoral immune system. The presence of carbohydrate moieties on gp120 physically shields potential epitopes from eliciting or binding to antibodies, an obstacle that is further complicated by the extensive diversity of N-linked glycans.
Lessons from completed clinical trials
Early efforts at developing an HIV-1 vaccine attempted to elicit protective antibodies against the viral envelope and used forms of recombinant glycoprotein 120 (rgp120) as the immunogen. VAX004, the first efficacy trial for an HIV-1 vaccine, began recruitment in 1998 and used two rgp120 HIV-1 envelope antigens, derived from two subtype B strains. Results from this double-blind, placebo-controlled trial showed no efficacy in 5403 volunteers. The vaccine did not prevent disease acquisition or impact the level of viremia in those infected [7]. The first HIV-1 vaccine efficacy trial in Asia, VAX003, also contained two rgp120 HIV-1 envelope antigens, one from subtype B and one from subtype E. Again, the vaccine did not prevent HIV-1 infection or delay HIV-1 disease progression [8].
Following the initial failure of attempts to elicit protective antibodies, the next set of antigens used in clinical efficacy studies were designed to test whether T cells, or the cellular arm of the immune system, could protect against HIV-1 infection. The Step trial was designed as a proof-of-concept study for the efficacy of a cell-mediated immunity vaccine to protect against HIV-1 infection, or to reduce early plasma HIV-1 levels. Step was a multicenter, double-blind, randomized, placebo-controlled, phase II trial of the Merck replication-defective adeno 5 viral vector MRKAd5 HIV-1 clade B Gag/Pol/Nef vaccine 3000 HIV-seronegative participants were randomized (1:1) to receive 3 injections of vaccine or placebo. Randomization was pre-stratified by gender, baseline Ad5 titer, and study site. Participants were tested approximately every 6 months for HIV acquisition; early plasma HIV RNA was measured ~3 months post-HIV diagnosis. The study was halted following an interim analysis that showed the vaccine to be ineffective. Moreover, post-hoc analysis suggested a trend towards an increased HIV-1 infection rate in certain subgroups of vaccine recipients (men who were uncircumcised and had antibodies to adenovirus type 5 [Ad5] at enrolment). The results were further confounded because a companion study (Phambili) of the same vaccine in a different population did not show increased susceptibility to HIV-1 infection. The reasons for these unexpected results remain unclear [9,10].
The next large scale clinical study, known as RV144, was an efficacy trial that showed for the first time that an HIV-1 vaccine could significantly reduce the incidence of HIV-1 infection. In this study, priming with the Aventis Pasteur live recombinant viral vector ALVAC-HIV-1 (vCP1521) and boosting with protein-based vaccine VaxGen gp120 B/E (AIDSVAX B/E) yielded a 31% reduction in HIV-1 acquisition in a modified intent to treat analysis. Although the duration of protection was limited and the effect modest, this study provided proof of concept that it is possible for a vaccine to elicit protective immunity that blocks infection [11]. A number of investigative teams are now working to identify correlates of immune protection from this study, with an emphasis on antibody-mediated mechanisms, such as binding antibodies, and antibody-dependent cell-mediated cytotoxicity [12–15].
The latest large scale clinical study, HVTN 505, is a proof-of-concept study of a Vaccine Research Center (VRC) product involving a multiclade HIV-1 DNA plasmid (EnvA, EnvB, EnvC, gagB, polB, nefB) boosted by a recombinant adenovirus vector (Ad5 EnvA, EnvB, EnvC, gag/polB). The study is being conducted in HIV-1-uninfected, adenovirus type 5 seronegative, circumcised men who have sex with men (MSM). Study enrolment began in 2009 and is estimated to be complete in 2012. This trial was initially designed to assess whether the VRC's multiclade vaccine regimen can reduce viral load in subjects who become infected despite vaccination; its scope has been expanded to detect whether the experimental vaccine regimen is at least 50% effective at preventing HIV-1 acquisition during the 18 months following immunization. This vaccine protocol is anticipated to elicit both cellular and humoral immune responses [16].
Identification and characterization of VRC01, a broadly-reactive neutralizing antibody
Among the large number of antibodies generated during natural infection by HIV-1, most are highly strain specific or non-neutralizing. The non-neutralizing antibodies are often directed against the gp120 or gp41 regions that are occluded on the assembled trimeric spike and exposed only upon disassembly of the spike. The accessible surface of gp120 in the trimer is largely composed of variable, heavily glycosylated core and loop structures that surround the receptor-binding regions. Despite this barrier, broadly reactive, potently neutralizing sera have been found in select individuals living with HIV, demonstrating that humans are capable of making broadly neutralizing antibodies [17–19]. Although a number of monoclonal antibodies against HIV-1 have now been identified, only a few display the combination of potent neutralization and breadth of reactivity that is desired for an efficacious vaccine response [20–22].
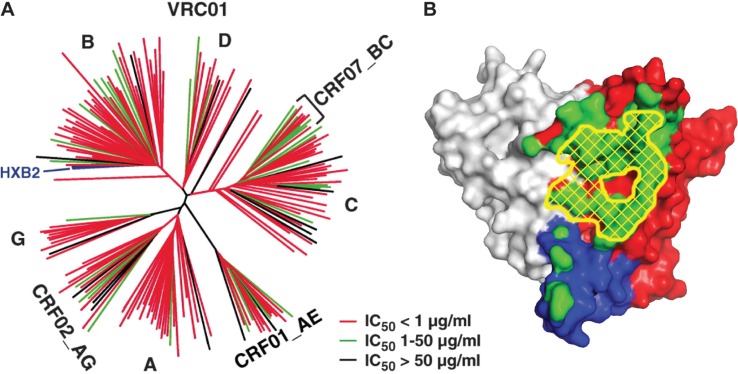
AIDS vaccine research investigators have developed strategies for identifying broadly reactive sera, isolating individual B cells from these sera, and cloning potent and broadly neutralizing antibodies from the B cells. High throughput assays to measure HIV-1 neutralization in large panels of sera revealed that approximately 25% of individuals living with HIV make relatively broadly reactive neutralizing antibodies during the course of HIV-1 infection [18]. Molecular probes used to isolate these antibodies were made by altering amino acids of the gp120 inner domain and exposing neutralization. These probes were designed to interact with antibodies specific for the highly conserved CD4 binding site of gp120 and are termed resurfaced stabilized cores, as the amino acid changes stabilize the desired gp120 structure and prevent the conformational flexibility that is normally present on the virus surface. Using peripheral blood B cells from a donor with potent serum neutralizing antibodies, three monoclonal antibodies were found to bind specifically to the CD4 binding site. One of these antibodies, designated VRC01, when cloned and expressed, neutralizes more than 90% of naturally circulating strains [23]. The broad neutralization capability of VRC01 can be seen (Figure 1a). Before VRC01 was identified, b12 was considered to be the best of the CD4 binding site neutralizing antibodies, but it was only able to neutralize 40% of clade B viruses [24].
Figure 1.
Broadly neutralizing antibodies against HIV-1. The VRC01 antibody is capable of near pan-neutralization of HIV-1, which it achieves by recognition of the initial contact site of the CD4 receptor on HIV-1 gp120. (a) Neutralization dendrogram of VRC01. Strains of HIV-1 are displayed according to their genetic distance and coloured red, green and black according to their sensitivity to neutralization by VRC01 (Reproduced with permission from AAAS; Wu et al. [23]). (b) Recognition by VRC01. The surface of the gp120 glycoprotein is coloured grey for inner domain, red for outer domain and blue for bridging sheet region. The initial contact surface for CD4 is highlighted by yellow cross-hatching, and the recognition surface of VRC01 is shown in green. Note the concordance between VRC01 recognition and CD4 contact (Reproduced with permission from AAAS; Zhou et al. [26]).
Purified VRC01, produced in the laboratory and passively administered to nonhuman primates, has shown protection against SHIV in rectal and vaginal challenge models [25]. The crystal structure of VRC01 in complex with an HIV-1 gp120 core shows that VRC01 binds directly over the site of initial attachment of CD4 to the virus, and this specificity may account for the potency and broad neutralization attributes of VRC01 (Figure 1b) [26].
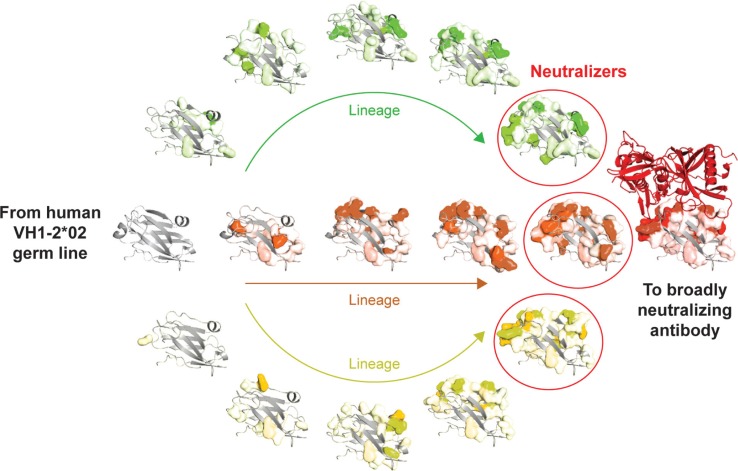
If VRC01-like antibodies are to be elicited by a vaccine, it is important to understand how they are generated in infected individuals. Although there are large quantities of gp120 antigen in persons living with HIV-1, the human immune system takes several years to generate VRC01-like antibodies. While the delay in the evolution of broadly reactive antibodies is not yet well understood, we know that antibodies arise through a process that involves three steps: recombination of germline antibody genes to form unique B-cell receptors on naïve B cells, deletion of autoreactive B-cell clones, and antigen-driven affinity maturation. The third step of this process, somatic mutations leading to affinity maturation, is illustrated (Figure 2) [17,19]. By using deep sequencing technology, it has been possible to study the precursor and maturation process of HIV-1 specific antibodies in an individual living with HIV. The VRC01-like antibodies identified to date have arisen most commonly from specific heavy chain genes, generally IGHV1-2*02. While this is a common precursor gene allele, extensive somatic maturation involving 70 to 90 changes in IGHV1-2*02 gives rise to mature broadly neutralizing VRC01-like antibodies. This maturation process focuses the developing antibodies onto a conserved site of HIV-1, a focusing that may be a common process in the generation of broadly neutralizing antibodies [27]. More recently, additional CD4 binding site antibodies have been defined through deep sequencing and structure-based probes, both from the IGHV1-2*02 germline as well as closely related germlines [27,28]. These additional antibodies will help to define more completely the alternative mechanisms used to generate this response and provide additional probes to define how alternative antibodies recognize the highly conserved CD4 binding site.
Figure 2.
Maturation of broadly neutralizing HIV-1 immunity. VRC01-like antibodies mature from the human VH1-2*02 germline (grey ribbon at left) and acquire over 50 somatic mutations while evolving to effectively recognize HIV-1 gp120 (red ribbon at right). The gp120-interactive surface of antibody VRC01 corresponds to the “top” of the domain in this orientation, with the binding surface for the light chain, on the surface facing the reader. Shown are variable heavy chain domains from three lineages of maturing antibodies (green, orange and yellow), with somatic mutations in each lineage highlighted in amino acid-surface representation coloured according to lineage, with fully saturated colours corresponding to charged changes and less saturated colours corresponding to hydrophobic changes.
How can all this information be used to develop a vaccine? The lengthy time required to generate VRC01-like antibodies suggests that the human immune system can generate such antibodies, but the process is not efficient. Whether this feature is due to the recessed nature of the immunogen, underlying immunodeficiency, or other factors is unclear. Analysis of germline and intermediate antibodies suggests at least one potential block exists at an early stage of B-cell receptor recognition. One approach then is to design modified gp120s that have high affinity for B cells expressing early, or germ line precursor, forms of VRC01. Such modified gp120s may help to stimulate the induction and appropriate maturation of VRC01-like antibodies, to guide the vaccine-induced elicitation of VRC01-like antibodies. The focus of current efforts to engage germline immunoglobulins is to remove structures that introduce potential clashes with the germline antibody. Based on knowledge of the mature VRC01-Env structure, attention has been focused on Loop D and the V5 region. These domains are responsible for VRC01 resistance in insensitive strains [25], and the initial recognition of these sites by germline precursors may affect the ability to drive proteins down this pathway. To effect this change, modified forms of the resurfaced cores, the core gp120, trimers, and outer domain proteins are being evaluated with and without different adjuvants to engage the relevant B-cell receptors.
Other approaches to generating broadly neutralizing antibodies
Other investigative teams have also been interested in generating and studying broadly neutralizing antibodies. The IAVI Neutralizing Antibody Center at the Scripps Research Institute has reported that antibodies directed to the V1 and V2 region of HIV-1 gp120 are quite broadly neutralizing [29]. Two such antibodies, PG9 and PG16, are able to neutralize 70 to 80% of circulating HIV-1 isolates. Additionally, scientists at the Center for HIV/AIDS Vaccine Immunology have identified a clonal lineage of four V1V2-directed broadly neutralizing antibodies (CH01-CH04) from a Tanzanian elite neutralizer. These antibodies were screened for their binding to a series of gp120 envelope monomers. Unlike some V1V2-directed antibodies that do not bind to Env monomers, these antibodies can bind select Env monomers. These gp120 monomers are being considered as potential immunogen candidates for induction of V1V2 epitope-specific antibodies [30]. More recently, additional broadly neutralizing antibodies to this region and an alternative site, glycans at the base of the V3 region, have been identified [31] and may serve as guides for additional immunogen designs.
Vaccine Research Center investigators have focused on a structure-guided approach to design immunogens to elicit potent antibodies against other HIV-1 antigens. Efforts have largely centered on strategies to stimulate an immune response to the highly conserved CD4 binding site on HIV-1 Env similar to the VRC01 antibody. Alternative forms of the protein, including trimers, monomers, resurfaced stabilized cores, or outer domain subregions of the protein have been designed to maximize exposure of the CD4 binding site and to mask irrelevant regions of the proteins that may detract from a focused immune response to this site. A discussion of these efforts has been detailed elsewhere [32]. With these approaches, it has been possible to elicit antibody responses directed to the CD4 binding site, which can neutralize tier 1 viruses. It has not yet proven possible to elicit such antibodies to tier 2 viruses (moderate sensitivity to antibody-mediated neutralization) but knowledge gleaned from the atomic level structure of Env is facilitating these efforts.
This knowledge is also being used to target other sites on HIV-1 Env. For example, antibodies against the HIV-1 gp41 epitope that partially mimic the specificity of the broadly neutralizing antibody 2F5 have been elicited. This epitope is one of the few conserved sites of vulnerability that allows broad neutralization of diverse strains of HIV-1. To accomplish this goal, the 2F5 epitope was transplanted onto several non-HIV-1 acceptor scaffold proteins that were termed epitope scaffolds (ES), and an ES prime:boost immunization regimen was shown to elicit specific serum antibody titers by ELISA and B cell responses by ELISpot analysis. Similar strategies could be used to target conserved but poorly immunogenic sites, or transient sites present not only on the HIV-1 Env, but also on other structurally defined pathogen targets [33,34].
Even with optimally designed immunogens, a separate goal of HIV vaccine research is to identify the best strategy for immunization. This effort requires a fundamental understanding of B-cell activation, proliferation and cell death to ensure that immunogens engage the appropriate B-cell receptors and induce antibodies of the desired specificity. Autoreactive B cells are eliminated by clonal deletion, and immunization likely must drive somatic mutations necessary for affinity maturation and development of high affinity antibodies. The use of relevant adjuvants and/or delivery matrices can facilitate the generation of antibody diversity and production, though such delivery agents must have the necessary safety and immunogenicity profiles. Potential adjuvants include alum, saponin-based emulsions, ASO1A and B, ASO2, and MF59. In addition, nanoparticles and multimeric viral carriers such as Qβ may help to stimulate appropriate B-cell activation. Finally, relevant animal models are needed for preclinical testing; not all species show similar degrees of somatic mutation or the ability to make long CDR3 regions found in humans. Candidates would be optimally tested in relevant humanized mouse models and NHP before progression into phase 1 human clinical trials.
Conclusions and the path forward
Exciting new technologies and the recent identification of broadly neutralizing HIV-1 antibodies enable us to approach HIV-1 vaccine development in ways that were previously unanticipated. It is clear, for example, that broadly neutralizing antibodies are not as rare as previously thought, and that the immune system can effectively target highly conserved regions on diverse strains of HIV-1. Not only can these antibodies guide rational vaccine design but they can also be used directly to confer protection, either by passive immunity or by gene-based antibody delivery. The lessons learned from such efforts will pay dividends not only in protecting against HIV-1 but also for vaccination against other infectious diseases that show broad genetic diversity and immune evasion, such as influenza, hepatitis C, and dengue viruses.
Over the last 10 years, there has been great progress in advancing AIDS preventive strategies, including male circumcision, use of antiretroviral chemoprophylaxis, and topical microbicides. Thus, one may ask whether a vaccine is still needed. The unequivocal answer remains, yes. A safe and efficacious vaccine could be administered simply and would provide long-term protection. Such a vaccine could have a dramatic effect on HIV-1 infection rates and is viewed widely as the most pragmatic, efficient, and cost-effective way to control the global HIV-1 pandemic.
Recent research identifying potent and broadly neutralizing antibodies has revealed new vistas for immunogen design. While significant hurdles remain for the development of an efficacious HIV-1 vaccine product, the scientific opportunities have never been more promising, and the need for a highly effective AIDS vaccine remains as urgent as ever.
Acknowledgements
The authors thank Jonathan Stuckey for his contribution to Figure 2 in the paper. The authors acknowledge Mythreyi Shastri and Susan Spring, the science writers who assisted in the process. The authors also thank Abraham Mittelman for his editing and comments.
Competing interests
The authors are named on intellectual property applications on aspects of this work filed by the National Institutes of Health.
Authors' contributions
Dr Gary Nabel presented a plenary session of “The Changing Face of HIV Vaccine Research” at the Sixth IAS Conference on HIV Pathogenesis, Treatment, and Prevention in Rome, Italy. With this presentation as the starting point, Dr Nabel wrote the initial draft of the paper, which then underwent revision and final approval by all authors.
Abbreviations
ES, epitope scaffolds; MSM, men who have sex with men; VRC, Vaccine Research Center.
Funding sources
This article was written with funding provided by the intramural program of the NIH.
References
- 1.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starcich BR, Hahn BH, Shaw GM, McNeely PD, Modrow S, Wolf H, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-WLAV, the retrovirus of AIDS. Cell. 1986;45:637–46. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 3.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–11. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 4.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 5.Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, et al. Energetics of the HIV gp120-CD4 binding reaction. PNAS. 2000;97(16):9026–31. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV gp120. Science. 2009;326:1123–7. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The rgp120 HIV Vaccine Study Group. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV infection. J Infect Dis. 2005;191:654–65. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 8.Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 9.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, et al. Efficacy assessment of a cell-mediated immunity HIV vaccine (the Step Study): a double-blind, randomized, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–93. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray G, Buchbinder S, Duerr A. Overview of STEP and Phambili trial results: two phase IIb test-of-concept studies investigating the efficacy of MRK adenovirus type 5 gag/pol/nef subtype B HIV vaccine. Curr Opin HIV AIDS. 2010;5:357–61. doi: 10.1097/COH.0b013e32833d2d2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209–20. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 12.Robb M. The Thai trial (RV 144); Presented at XVIII International AIDS Conference; Vienna, Austria. 2010. Jul 18–23, [abstract TUSY0603] [Google Scholar]
- 13.Karnasuta C, Paris RM, Cox JH, Nitayaphan S, Pitisuttithum P, Thongcharoen P, et al. Antibody-dependent cell-mediated cytotoxic responses in participants enrolled in a phase I/II ALVAC-HIV/AIDSVAX B/E prime-boost HIV vaccine trial in Thailand. Vaccine. 2005;23:2522–9. doi: 10.1016/j.vaccine.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs JD, Sobieszczyk ME, Hammer SM, Buchbinder SP. Lessons drawn from recent HIV vaccine efficacy trials. J Acquir Immune Defic Syndr. 2010;55(Suppl 2):S128–31. doi: 10.1097/QAI.0b013e3181fbca02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JH, Rerks-Ngarm S, Excler JL, Michael NL. HIV vaccines: lessons learned and the way forward. Curr Opin HIV AIDS. 2010;5:428–34. doi: 10.1097/COH.0b013e32833d17ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Institute for Allergy and Infectious Diseases [Internet] Bethesda, MD: National Institutes of Health; The HVTN 505 HIV Vaccine Regimen Study. [last updated 2009 Aug 24; accessed 2012 Jun 8]. Available from: http://www.niaid.nih.gov/news/qa/pages/hvtn505qa.aspx. [Google Scholar]
- 17.Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, et al. Broad HIV neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–4. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV infection: good news for an HIV vaccine? Nat Med. 2009;15(8):866–70. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 19.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV infected individuals. PLoS Pathog. 2010;6(8):1–14. doi: 10.1371/journal.ppat.1001028. Epub 1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–44. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 21.Haynes BF, Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV vaccine candidates. Expert Rev Vaccines. 2006;5(4):579–95. doi: 10.1586/14760584.5.4.579. [DOI] [PubMed] [Google Scholar]
- 22.Pantophlet R, Burton DR. GP120: target for neutralizing HIV antibodies. Annu Rev Immunol. 2006;24:739–69. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 23.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV. Science. 2010;329:856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, et al. Structural definition of a conserved neutralization epitope on HIV gp120. Nature. 2007;445:732–37. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pegu A, Yang ZY, Chen X, Todd JP, McKee K, Rao S, et al. VRC01 provides sterilizing protection to non human primates from mucosal SHIV challenges [abstract] J Immunol. 2011;186:155.11. [Google Scholar]
- 26.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, et al. Structural basis for broad and potent neutralization of HIV by antibody VRC01. Science. 2010;329:811–7. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–7. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV vaccine target. Science. 2009;326(5950):285–9. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, et al. Analysis of a clonal lineage of HIV envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85(19):9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–70. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwong PD, Mascola JR, Nabel GJ. Rational design of vaccines to elicit broadly neutralizing antibodies to HIV-1. Cold Spring Harb Perspect Med. 2011;1(1):007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guenaga J, Dosenovic P, Ofek G, Baker D, Schief WR, Kwong PD. Heterologous epitope-scaffold prime: boosting immuno-focuses B cell responses to the HIV gp41 2F5 neutralization determinant. PLoS One. 2011;6(1):1–12. doi: 10.1371/journal.pone.0016074. Epub 16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ofek G, Guenaga FJ, Schief WR, Skinner J, Baker D, Wyatt R, et al. Elicitation of structure-specific antibodies by epitope scaffolds. PNAS. 2010;107(42):17880–7. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]