Abstract
Drylands are extensive across sub-Saharan Africa, socio-economically and ecologically important yet highly sensitive to environmental changes. Evolutionary history, as revealed by contemporary intraspecific genetic variation, can provide valuable insight into how species have responded to past environmental and population changes and guide strategies to promote resilience to future changes. The gum arabic tree (Acacia senegal) is an arid-adapted, morphologically diverse species native to the sub-Saharan drylands. We used variation in nuclear sequences (internal transcribed spacer (ITS)) and two types of chloroplast DNA (cpDNA) markers (PCR-RFLP, cpSSR) to study the phylogeography of the species with 293 individuals from 66 populations sampled across its natural range. cpDNA data showed high regional and rangewide haplotypic diversity (hT(cpSSR)=0.903–0.948) and population differentiation (GST(RFLP)=0.700–0.782) with a phylogeographic pattern that indicated extensive historical gene flow via seed dispersal. Haplotypes were not restricted to any of the four varieties, but showed significant geographic structure (GST(cpSSR)=0.392; RST=0.673; RST>RST (permuted)), with the major division separating East and Southern Africa populations from those in West and Central Africa. Phylogenetic analysis of ITS data indicated a more recent origin for the clade including West and Central African haplotypes, suggesting range expansion in this region, possibly during the Holocene humid period. In conjunction with paleobotanical evidence, our data suggest dispersal to West Africa, and across to the Arabian Peninsula and Indian subcontinent, from source populations located in the East African region during climate oscillations of the Plio-Pleistocene.
Keywords: aridity, gum arabic, hybridisation, long-distance dispersal, phylogeny, refugia
Introduction
Modern dry woodland and savannah biomes, dominated by grasses and woody species, are key ecosystems in sub-Saharan tropical Africa (Plana, 2004). With a species composition and diversity that is driven by low rainfall and pronounced dry seasons (Jacobs, 2004), they evolved during the Miocene (23-5 Mya), a period associated with drastic lowering of global temperatures and increasing aridity that saw the replacement of once expansive lowland rainforests with savannah woodland (Plana, 2004). From the Plio-Pleistocene onwards (from ∼5 Mya), the species that occupy these biomes have experienced cyclical range expansions and contractions due to climatic oscillations (Maley, 2001; Plana, 2004), restricted by aridification on one hand and moist forest expansion on the other. Thus, the genetic structure of widely distributed dryland tree species is likely to reflect the influence of these processes.
The interplay of major geomorphological features with climate oscillations has also played a key role in shaping the genetic patterns and phylogeography of flora and fauna since the end of the Pliocene (∼3.5 Mya; deMenocal, 1995). For example, contraction and expansion of the Sahara desert in Central and West Africa, and interaction of climate with the Great Rift Valley and the elevated topographies of East and Southern Africa produced wide altitudinal ranges forming a complex mosaic of landscapes and localised climate regimes that functioned as refugia during extreme climate conditions (Plana, 2004; Lorenzen et al., 2010).
Previous phylogeographic studies of tropical African tree species have largely focussed on moist forests (montane, lowland or rainforests) and have typically interpreted results with respect to Pleistocene refugia (for example, Hagenia abyssinica, Ayele et al., 2009; Milicia excelsa, Daïnou et al., 2010; Irvingia gabonensis, Lowe et al., 2010). It has been suggested that previously continuous rainforests fragmented into refugia within mountainous areas and became separated by the expansion of savannah vegetation during glacial maxima associated with cooler and drier conditions (Plana, 2004). Dry woodlands and savannahs are likely to have experienced the same processes in reverse, and fragmented dryland habitats may have acted as refugia for arid-adapted species during warmer and wetter periods (Plana, 2004). Relics of xeric vegetation or savannah enclaves persisting today in the Central and West African rainforest since the last glacial maximum may represent such refuges (Maley, 2001).
A few savannah-type or dry woodland tree species have been studied, mainly in the Brazilian cerrado (for example, Caryocar brasilensis, Collevatti et al., 2003; Hymenaea stigonocarpa, Ramos et al., 2007; Astronium urundeuva, Caetano et al., 2008) and others in the seasonally dry tropical forests (Pennington et al., 2004; 2009). Species in the latter show patterns of diversity, endemism and phylogeny that indicate historically stable and dispersal-limited systems, caused partly by the widespread patchy distribution of this biome and its persistence over evolutionary timescales regarded as Pleistocene refugia (Collevatti et al., 2003; Pennington et al., 2004; Caetano et al., 2008). Although seasonally dry tropical forests share some of the ecological characteristics of dry woodlands and savannahs of sub-Saharan Africa (Lock, 2006; Pennington et al., 2009), direct comparisons are limited by phytogeographical differences. In Africa, there have been three major phylogeographic studies of African tree species found in these biomes. A study of Acacia nilotica populations showed genetic differences that broadly matched subspecific designations and a phylogeographic separation of North and West Africa from East and Southern Africa (Wardill et al., 2005). The other two studies, of the baobab tree (Adansonia digitata L., Malvaceae; Pock Tsy et al., 2009) and the shea tree (Vitellaria paradoxa C. F. Gaertn, Sapotaceae; Allal et al., 2011) showed strong phylogeographic structure, distinguishing Eastern and Western populations within the Sudano-Sahelian region. However, the ecological distribution of the baobab tree only partially matches that of the dry woodlands and savannahs, and the shea tree has a limited distribution in the Sudano-Sahelian zone. In contrast to plants, there are numerous studies of vertebrates that show development of intraspecific differentiation that coincides with formation of the woodland and savannah biomes in Africa (Jacobs, 2004), for example, plains zebra, Equus quagga (Lorenzen et al., 2008) and common eland antelope, Taurotragus oryx (Lorenzen et al., 2010).
Here, we study the phylogenetics and phylogeography of Acacia senegal (L.) Willd. (Leguminosae, Mimosoideae) commonly known as the ‘gum arabic' tree. Although renowned for its commercial gum, which has been traded for centuries (Fagg and Allison, 2004), A. senegal also provides other key ecosystem services such as sand stabilisation, shade, fodder and forage for livestock and wildlife as well as N2-fixation, which enhances soil fertility and sustains food crop production in the gum arabic agroforestry systems. A. senegal is not only drought tolerant and can occur in extremely dry habitats, but also has a wide climatic (temperature and rainfall) and altitudinal range (Supplementary Table S1). It has an inter-continental distribution extending from Africa eastwards to the Arabian Peninsula, Pakistan and India (Figure 2; Fagg and Allison, 2004). Reports from common garden experiments have shown wide variations in survival, physiology, growth and gum production traits among provenances, reflecting the adaptive variation that exists across its natural distribution range (Sprent et al., 2010).
According to a recent phylogeny of African acacias, the genus Acacia (Syn. Senegalia) is estimated to have inhabited the open habitats (woodlands and savannahs) of Africa since the Miocene epoch (∼21 Mya, Bouchenak-Khelladi et al., 2010). This also marks the period of rapid diversification within the Leguminosae family (for example, see Richardson et al., 2001; Lavin et al., 2005). As important drivers of the evolutionary processes of diversification, hybridisation and introgression were likely to have contributed to the adaptation of the African acacias to the new, sometimes extreme environments experienced in the sub-Saharan regions during the Plio-Pleistocene (Maley, 2001; Jacobs, 2004; Plana, 2004; Bouchenak-Khelladi et al., 2010). A. senegal belongs to a group of more than 20 closely related species referred to as the A. senegal complex characterised by spicate inflorescences and prickles that are mostly in threes, the central one hooked downwards and the laterals hooked upwards (Ross 1979; Fagg and Allison, 2004). It forms a natural hybrid (A. laeta R.Br. ex Benth.) with A. mellifera (Vahl) Benth. (El Amin, 1976). We focus on A. senegal documented to have four putative varieties (A. senegal var. senegal, A. senegal var. kerensis Schweinf, A. senegal var. rostrata Brenan and A. senegal var. leiorhachis Brenan) whose delimitations rely largely on growth and morphological characters (Ross, 1979; Fagg and Allison, 2004). In Eastern Africa, regarded as the centre of diversity for the species, the intraspecific delimitation can be less resolved, especially among varieties senegal, kerensis and rostrata due to variation and overlaps in morphological characters (Ross, 1979; Ross, 1981; Fagg and Allison, 2004; Table 1). However, var. leiorhachis has some unique growth, pod and phenological features that separate it from the other varieties. The key growth and morphological characters often used to distinguish the varieties include growth form, type of stem or trunk texture, inflorescence axis pubescence and shape of pods (Table 1).
Table 1. Growth and morphological characters for distinguishing Acacia senegal varieties (source: Ross, 1979; Fagg and Allison, 2004).
| Key characters | Variety senegal | Variety kerensis | Variety rostrata | Variety leiorhachis |
|---|---|---|---|---|
| Inflorescence axis | Sparsely to densely pubescent | Sparsely to densely pubescent | Sparsely to densely pubescent | Glabrous or subglabrous, except sometimes for some basal hairs |
| Tree, shrub or bush | Tree with a distinct trunk | Shrub or bush without a distinct trunk | Tree with a distinct trunk | Slender spindly tree, whippy |
| Shape of pod | Rounded to acute (sharply pointed) at apex | Rounded to acute, seldom acuminate | Markedly acuminate or rostrate (beaked) at apex | Rounded to acute |
| Phenology | Flowers usually produced after the leaves | Not known | Not known | Flowers often produced before or with the young leaves |
A. senegal is mainly insect pollinated, and predominantly outcrossing. Flowering occurs annually (or biannually in some regions) followed by a good seed crop, which is dispersed by wind and to some extent animals, especially ungulates (Fagg and Allison, 2004). Cytological studies on A. senegal var. senegal indicated that it is a diploid, 2n=26 (Bukhari, 1997). The species is reproductively active within 2–4 years of establishment. Previous genetic studies of Kenyan populations using both chloroplast and nuclear microsatellites found high genetic diversity within populations and low genetic differentiation among populations, indicating extensive gene flow (Omondi et al., 2010). These attributes demonstrate effective pollen and seed dispersal mechanisms with potential for long-distance dispersal.
Given the historical range changes that A. senegal has probably experienced, the complex landscape it occupies and its effective dispersal ecology, we predict the following:
The divergence events in A. senegal will reflect those of major climatic shifts because sub-Saharan woodlands and savannahs are ancient and have been subject to fluctuations in size and distribution out of phase with those of the moist forests during climate oscillations of the Plio-Pleistocene.
As regional differentiation has been seen in previous studies of dryland species (for example, baobab, shea), we predict similar differentiation for A. senegal. Within regions we expect low genetic structure due to life history characteristics that predispose the species to effective dispersal.
There will be high genetic diversity and presence of basal groups in the regions of Eastern and Southern Africa due to extant taxonomic diversity and a complex mosaic of landscapes (lowlands to highlands covered by forests, woodlands and savannahs) and climate regimes predating the Pleistocene.
To address these hypotheses, we use variation in the internal transcribed spacer (ITS) of the nuclear ribosomal DNA (nrDNA) and the cpDNA (trnH-psbA and cpSSR ) to (i) assess phylogenetic relationships among the A. senegal varieties, (ii) to test for phylogeographic structuring within A. senegal and (iii) to examine evolutionary history of A. senegal in terms of colonisation of the dryland habitats from its origins and hypothesised refugia in the light of environmental and habitat distributional changes since the original diversification that produced the species in the Plio-Pleistocene.
Materials and methods
Collection and DNA extraction
Samples were collected from a total of 293 A. senegal individuals from wild populations, experimental trials and herbarium specimens representing 66 sites across its natural range in sub-Saharan Africa, the Arabian Peninsula, Pakistan and India (Table 2). Field samples were collected from well-known and documented sources; samples of doubtful or unverified intraspecific affiliations are indicated (Table 2). Fresh leaf or stem tissue samples were dried on silica gel in ziplock bags. For each sample, roughly 1 cm2 of dried tissue was ground to a fine powder using a Retsch Tissuelyser (Haan, Germany). DNA extraction was carried out using QIAGEN DNeasy 96 Plant kits (Hilden, Germany) following the manufacturer's protocol. Extracted DNA was assessed for quality and concentration on a 1% agarose gel before storage at −20 °C.
Table 2. Locations of all sampled populations of Acacia senegal.
| Region | Country | Population | Variety | n | Latitude | Longitude |
|---|---|---|---|---|---|---|
| East Africa | Ethiopia | Sodera | senegal | 3 | 08°24′N | 39°23′E |
| Kenya | Archer's Post | kerensis | 2 | 00°39′N | 37°39′E | |
| Daaba | kerensis | 2 | 00°32′N | 37°46′E | ||
| Kajiado | senegal | 7 | 01°54′S | 36°45′E | ||
| Kakuma | kerensis | 5 | 03°45′N | 34°40′E | ||
| Kaleing' | kerensis | 6 | 04°24′N | 35°31′E | ||
| Kargi | kerensis | 1 | 02°39′N | 37°28′E | ||
| Kibwezi | kerensis | 5 | 02°13′S | 38°04′E | ||
| Kulamawe | leiorhachis | 6 | 00°32′N | 37°60′E | ||
| Koriema | senegal | 7 | 00°27′N | 35°52′E | ||
| Lokichar | kerensis | 3 | 02°22′N | 35°38′E | ||
| aMachakos | ckerensis | 1 | 01°31′S | 37°16′E | ||
| Magadi | leiorhachis | 7 | 01°32′S | 36°35′E | ||
| Marigat | kerensis | 6 | 00°28′N | 35°54′E | ||
| Merille | kerensis | 3 | 01°32′N | 37°73′E | ||
| Ngarendare | kerensis | 8 | 00°28′N | 37°25′E | ||
| Ngurunit | leiorhachis | 6 | 01°43′N | 37°17′E | ||
| Ntumburi | senegal | 6 | 00°12′N | 37°31′E | ||
| Rimoi | senegal | 7 | 00°40′N | 35°34′E | ||
| Serolipi | kerensis | 5 | 01°15′N | 37°59′E | ||
| Taita-Taveta | senegal | 2 | 03°27′S | 38°29′E | ||
| Somalia | aHargesia | kerensis | 1 | 09°30′N | 44°03′E | |
| aAfgoi | ckerensis | 1 | 02°06′N | 45°08′E | ||
| Tanzania | Kigwe | leiorhachis | 15 | 06°06′S | 35°29′E | |
| Wangingombe | senegal | 8 | 08°51′S | 34°38′E | ||
| Sudan | Fallatu Forest | senegal | 12 | 13°06′N | 30°08′E | |
| Kordofan | senegal | 6 | 12°44′N | 29°35′E | ||
| aKhartoum | senegal | 1 | 15°38′N | 32°32′E | ||
| aKundoura, Nyala | senegal | 1 | 12°52′N | 30°13′E | ||
| aBora | senegal | 1 | 08°49′N | 26°11′E | ||
| aWad Medani | senegal | 1 | 14°24′N | 33°31′E | ||
| aEl Haraza | senegal | 1 | 11°18′N | 24°11′E | ||
| aSobar | senegal | 1 | 08°48′N | 32°54′E | ||
| aRodom | senegal | 1 | 12°05′N | 23°03′E | ||
| aKassala | senegal | 1 | 15°27′N | 36°24′E | ||
| aEl Felaya | senegal | 1 | 09°36′N | 28°26′E | ||
| Central Africa | Cameroon | Maroua | senegal | 10 | 10°15′N | 14°14′E |
| Chad | Tourba | senegal | 2 | 12°49′N | 15°18′E | |
| West Africa | Burkina Faso | Bissiga Fc | senegal | 6 | 12°26′N | 00°32′W |
| Di (Sousou) | senegal | 7 | 13°10′N | 03°25′W | ||
| Mali | Aïte | senegal | 4 | 15°05′N | 11°39′W | |
| Kirane | senegal | 50 | 15°23′N | 10°15′W | ||
| Somo | senegal | 5 | 13°17′N | 04°54′W | ||
| Mauritania | Djiguéni | senegal | 6 | 15°44′N | 08°40′W | |
| Kankossa | senegal | 1 | 15°56′N | 11°27′W | ||
| Niger | Karofane | senegal | 2 | 14°18′N | 06°11′E | |
| Senegal | Daïba | senegal | 6 | 15°22′N | 13°08′W | |
| Diaménar | senegal | 6 | 16°00′N | 15°54′W | ||
| Kidira | senegal | 4 | 14°28′N | 12°13′W | ||
| Ngane | senegal | 3 | 14°08′N | 16°12′W | ||
| Southern Africa | Botswana | aTsau | rostrata | 1 | 21°47′S | 24°04′E |
| aMbeleapudi | rostrata | 1 | 20°09′S | 22°19′E | ||
| South Africa | aJozinidam | rostrata | 3 | 27°32′S | 31°58′E | |
| aKamlushwa | rostrata | 1 | 25°42′S | 31°45′E | ||
| aSokwe | rostrata | 1 | 26°52′S | 32°13′E | ||
| aZululand | rostrata | 1 | 27°08′S | 31°59′E | ||
| aPongolo | rostrata | 1 | 27°25′S | 32°04′E | ||
| aHlabisa | rostrata | 1 | 28°08′S | 31°52′E | ||
| Arabian Peninsula, Pakistan and India | India | bJodhpur | csenegal | 15 | 26°43′N | 73°09′E |
| India | Jodhpur (Inde50) | senegal | 4 | 26°19′N | 79°31′E | |
| Jodhpur (Inde60) | senegal | 3 | 26°19′N | 79°31′E | ||
| aOld Delhi Ridge | senegal | 1 | 28°42′N | 77°13′E | ||
| Oman | aDhofar | senegal | 3 | 16°52′N | 53°47′E | |
| Pakistan | aSind | senegal | 2 | 25°19′N | 68°04′E |
n, Number of samples.
Herbarium specimens held by Royal Botanic Garden, Edinburgh; further details are presented in Supplementary Table S1.
Collections from World Agroforestry Centre, formerly ICRAF.
Samples with uncertain intraspecific affiliations.
PCR and sequencing
We used data from both nuclear and organelle genomes to test for phylogeographic patterns. The ITS of the nrDNA was used for phylogenetic reconstruction. Besides its utility in phylogenetic studies in angiosperms, the high sequence variation found among conspecifics and allopatric populations also makes it suitable for studying intraspecific relationships (Baldwin et al., 1995). The ITS region, alone or concatenated with other spacer regions, has been successfully used in previous studies to resolve phylogenetic and taxonomic issues among other Acacia taxa (for example, Ariati et al., 2006; Brown et al., 2008). Chloroplast (cp) DNA was also used to analyse geographical patterns of diversity and population genetic structure across the distribution range of the species. The cpDNA is maternally inherited in angiosperms and has a slow rate of evolution (compared with nrDNA), making it ideal for studying historical patterns of gene flow, colonisation and migration events (Cavers et al., 2003).
For ITS sequence analysis, a subsample of 65 individuals representing the four different recognised varieties and the full extent of the geographic range were selected (Table 1). The 5.8S subunit and flanking spacers ITS 1 and ITS 2 were amplified together with part of the 18S and 26S gene regions as a single fragment using primers 17SE and 26SE and PCR conditions as detailed by Sun et al. (1994). Direct sequencing was carried out at the NERC Biomolecular analysis facility at the University of Edinburgh, UK.
Three chloroplast regions were selected for screening of the whole collection after preliminary screening in a subset of 24 geographically and taxonomically representative samples. These were: the trnH-psbA spacer (Shaw et al., 2005), restricted with DraI (selected after testing ApaI, AluI, BamHI, DraI, MseI and RsaI), and universal microsatellite primer pairs ccmp5 and 10 (Weising and Gardner, 1999, after screening primer pairs ccmp 1–10). The fragment trnH-psbA was amplified in 25 μl reactions containing: 2 μl DNA (∼20 ng of genomic template DNA), 2.5 μl of 10 × buffer (New England Biolabs, Hitchin, Herts, UK), 0.5 μl dNTPs (Promega, Madison, WI, USA), 0.5 μl each primer (MWG Biotech, Ebersberg, Germany), 0.2 μl (0.5 U) Taq (New England Biolabs), 0.4 μl BSA (Sigma, St Louis, MO, USA) and 13.4 μl water. PCR was carried out on a Thermo MBS thermal cycler (Asheville, NC, USA) with the following protocol: 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 45 °C for 30 s, 72 °C for 1 min and finally 72 °C for 10 min. Amplicons were digested with DraI and restriction fragment patterns were visualised as described by Cavers et al. (2003).
The two cpSSR loci were amplified in 25 μl reactions containing: 0.5 μl DNA (∼20 ng of genomic template DNA), 2 μl of 10X PCR buffer (Promega), 1.6 μl dNTPs (Promega), 0.4 μl each primer (MWG Biotech), 0.2 μl (0.5 U) Taq (New England Biolabs), 0.4 μl BSA and 19.5 μl water. The PCR products were electrophoresed, band sizes determined and characterised for cpDNA haplotype following the procedure described by Omondi et al. (2010).
Statistical analysis
ITS sequences were edited and assembled in CodonCode Aligner 3.5.7 (CodonCode Corporation, Dedham, MA, USA). Sequences were lodged with GenBank under the accession numbers HQ605042-HQ605077 (Supplementary Table S2, Supporting information). Insertions/deletions (indels) were coded for presence or absence. At ITS loci, multiple copies, paralogues or pseudogenes can confound phylogenetic inference (Álvarez and Wendel, 2003). Therefore, consistency index, retention index and GC content were assessed and sequences were checked for large indels, which can indicate the presence of these variant sequences; none of these parameters suggested the occurrence of paralogues or pseudogenes.
Phylogenetic analysis of ITS sequence data were conducted using parsimony approaches in the computer package PAUP* v4.0b10 (Swofford, 2003). The heuristic search option was employed, using tree bisection-reconnection, with 1000 random stepwise addition replicates and two trees held at each cycle. Branch support values were calculated using a faststep heuristic search with 10 000 bootstrap replicates. A bootstrap majority-rule consensus tree was calculated in PAUP. We included an ITS sequence from GenBank; EF638213.1 (Acacia (Senegalia) senegal) from Zimbabwe. Sequences of Vachellia (Syn. Acacia) farnesiana (EF638219.1) from Australia and Vachellia (Syn. Acacia) collinsii (EF638216.1) from Mexico were included as outgroups. Bayesian inference was performed in MrBayes version 3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). Four Markov Chain Monte Carlo simulations (three heated, one cold) were run with sampling every 100 generations for 5 000 000 generations with the first 10% discarded as burn in. Trees remaining after burn in were used to calculate posterior probabilities for nodes in the majority-rule consensus tree.
The geographical structure of genetic variation at the cpSSR loci was explored using the program SAMOVA (spatial analysis of molecular variation, Dupanloup et al., 2002). The method uses a simulated annealing procedure to define K groups of populations that are geographically homogenous and maximally differentiated from each other. The method requires the a priori definition of the number of groups (K) and generates F statistics (FSC, FST and FCT) based on analysis of molecular variance (AMOVA). One hundred simulated annealing processes were used for values of K from 2 to 10. Herbarium specimens were also included in SAMOVA analysis where we had at least three geographically proximate individuals to constitute a quasi population. The most likely number K of groups was identified from FCT, the proportion of total genetic variance due to differences among groups of populations. Population structure was further investigated by a Bayesian-based assignment algorithm using the STRUCTURE program version 2.3.3 (Pritchard et al., 2000). We used the admixture model with correlated allele frequencies and run analysis with and without prior sample location information. We performed 10 independent repetitions for each K (ranging from 1 to 20), with 100 000 MCMC repetitions and a 100 000 burn-in period. The minimum number of K was evaluated using the ΔK procedure (Evanno et al., 2005). The geographical distribution of ITS, trnH-psbA haplotypes and SAMOVA (cpSSR) delineated groups were mapped using the ESRI software ArcMap 10 (ESRI, Redlands, CA, USA).
Although the cpDNA molecule is non-recombining and therefore functions as a single locus, data derived from RFLP and SSR loci were treated separately due to their different modes of evolution. The program PERMUT (Pons and Petit, 1996) (www.pierroton.inra.fr/genetics/labo/Software) was used to calculate the mean within-population genetic diversity (hS), the total gene diversity (hT) and the proportion of diversity resulting from genetic differentiation among populations (GST), as well as the corresponding ordered parameters (taking into account similarities between haplotypes, NST and RST for RFLP and SSR, respectively), to test for phylogeographic structure at rangewide and regional geographic scales. Differentiation parameters were tested and compared with means from 1000 permutations. For all population-based analysis, samples were pooled where possible to form populations of n ⩾3; highly isolated individual samples were excluded. As ITS phylogeny, SAMOVA and STRUCTURE analyses indicated an East and West African phylogeographic pattern, PERMUT analysis was also carried out at the African geographic scale. In addition to the PERMUT analysis (Pons and Petit, 1996), a hierarchical AMOVA and pairwise FST between geographic groups was computed for both RFLP and cpSSR data using ARLEQUIN version 3.5 (Excoffier et al., 2005).
Results
ITS sequences and phylogeny
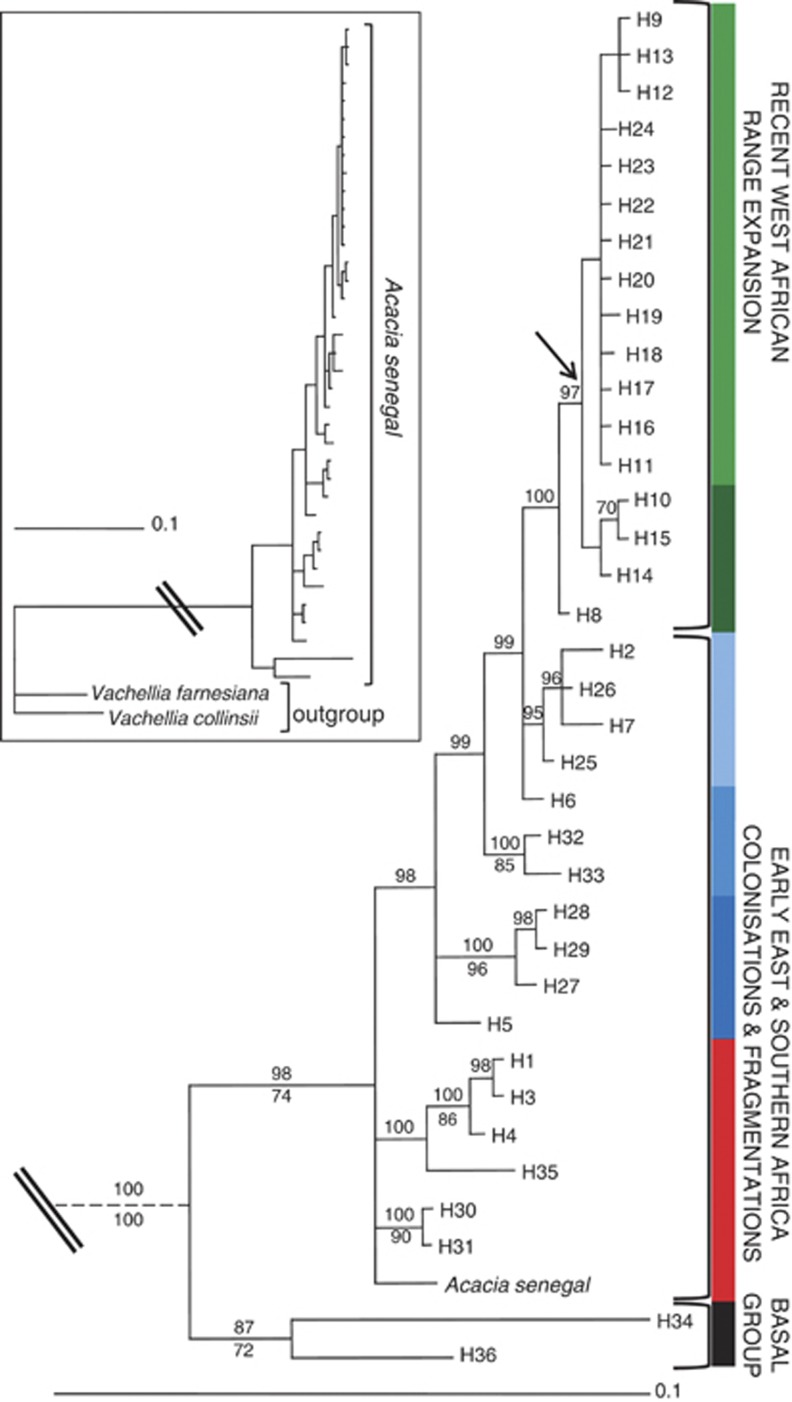
A total of 65 sequences were obtained averaging 570 bp aligned length in all individuals. The ITS region provided 109 parsimony informative characters (with outgroup), including four indels ranging from 2 to 4 bp. Parsimony analysis of these data found 1 111 625 trees of length 251–576; consistency index=0.831, retention index=0.843. The strict consensus tree (Figure 1) of 141 604 trees resolved 17 nodes, of which 10 were supported by bootstrap values >50%. The A. senegal specimens formed a monophyletic group with respect to the outgroup taxa, V. collinsii and V. farnesiana. The basal group comprised two geographically disparate individuals (one from Karofane in Niger, West Africa and the other a herbarium specimen from Botswana, Southern Africa), which was highly divergent from the main branch (72% and 87% bootstrap and posterior probability support, respectively). The main branch exhibited high genetic variation across the geographic range of the species with a topology displaying sequential Eastern and Southern Africa–West Africa organisation for the African sampled populations (Figure 1). The basal clades comprise mainly East and Southern Africa populations. They consist of two groups affiliated to var. leiorhachis (including Genbank sequence EF638213.1 from Zimbabwe), strongly separated from the third group (98% posterior probability) that contains samples affiliated to vars. senegal, kerensis and rostrata. The terminal clades comprise mainly West African samples affiliated to var. senegal, majority of which are in the least resolved clade (see Figure 1); it is notable that the few samples of East African origin are also mostly affiliated to var. senegal.
Figure 1.
Bayesian 50% majority-rule consensus phylogram of ITS sequences from Acacia senegal individuals sampled across its distribution range. Branches are labelled with ⩾70% bootstrap support (below) and posterior probability (above) values. Arrow indicates presence of a single clade in the maximum parsimony strict consensus tree. The tree was rooted with Acacia (Vachellia) farnesiana and Acacia (Vachellia) collinsii (see inset tree, double line denotes the point at which the break is in the main tree) obtained via GenBank (accession numbers EF638219 and EF638216, respectively). Acacia (Senegalia) senegal sequence is accession number EF638213 from Zimbabwe. Each haplotype is code-labelled HX where X refers to a number 1–36 as described in Supplementary Table S2, Supporting information. Hypothesised colonisation or range expansion events are indicated. Clades are identified by different colours on the vertical bar (black, basal group; red, var. leiorhachis; dark blue, var. rostrata; blue–light blue, vars. kerensis-senegal; dark green–light green, var. senegal); clades that are phylogenetically related are shown with different shades of one colour. The same colour coding is used for Figure 2a. Scale bar signifies 0.1 substitutions per nucleotide site.
Geographic distribution of trnH-psbA haplotypes
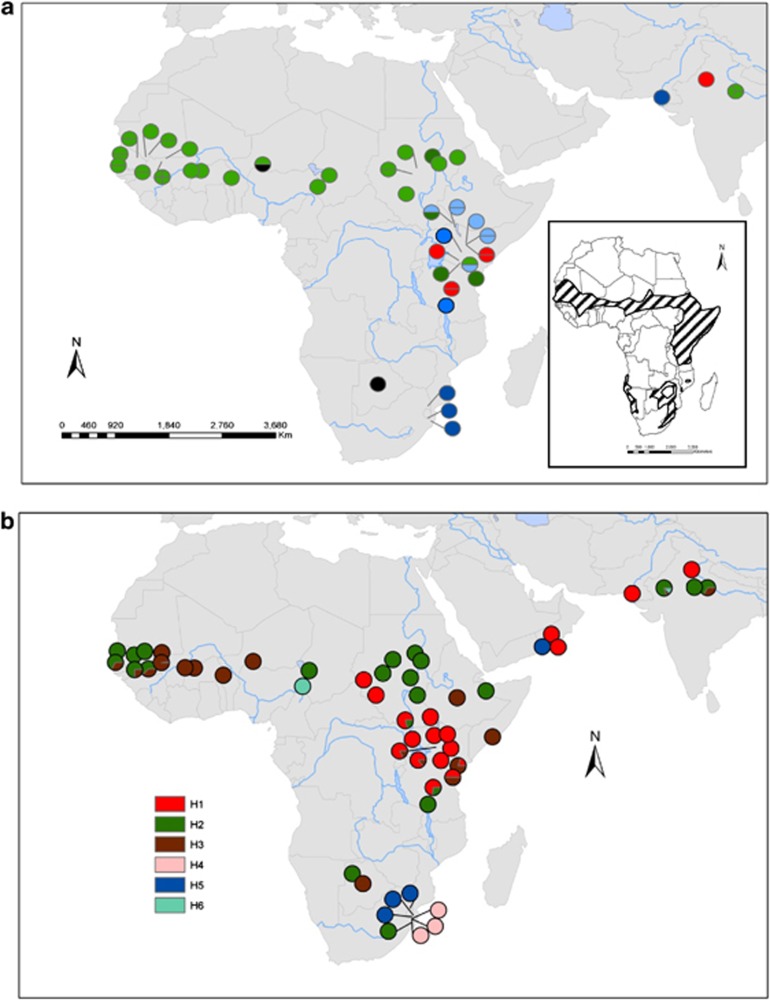
The trnH-psbA amplicons were ∼490 bp in size. Six RFLP trnH-psbA haplotypes were detected (Figure 2b; Supplementary Table S3, Supporting information). Most populations were fixed for a single haplotype. Haplotype 1 was the most frequent, highly dominant in East Africa (mainly Kenya, Tanzania and Sudan), and present in Oman and Pakistan. Haplotypes 2 and 3 were the most widespread, occurring throughout Africa and the Indian subcontinent. Haplotypes 4, 5 and 6 were rare: haplotype 4 was restricted to Southern Africa; haplotype 5 was present in Southern Africa and Oman and haplotype 6 occurred in two geographically distant populations—in Maroua (Cameroon) and Jodhpur (ICRAF general collection, India). The Maroua population was fixed for haplotype 6 while in Jodhpur it was mixed with another haplotype, constituting only a small proportion (13%). All of the haplotypes found in India, Pakistan and Oman were shared with continental African populations.
Figure 2.
Rangewide distribution of nuclear (a) and chloroplast (b) haplotypes in Acacia senegal. Data are: (a) haplotypes from ITS of the nuclear ribosomal DNA and (b) PCR-RFLP haplotypes of chloroplast trnH-psbA intergenic spacer of A. senegal; each circle represents a single population with colour denoting haplotype variation. The number of samples per population is presented in Supplementary Tables S2 and S3, respectively. Inset: approximate distribution of A. senegal in Africa (hatch-shaded area).
Geographic distribution of chloroplast microsatellite haplotypes
Thirty-six haplotypes were identified (Supplementary Table S5, Supporting information). Unlike the trnH-psbA haplotypes, most populations with at least two analysed individuals/population were polymorphic (mixed). The distribution of haplotypes across the range showed a pattern similar to that seen for trnH-psbA haplotypes, in that the majority were regionally fixed in either East or West Africa. Haplotypes 1–21 (21 haplotypes) occurred in East Africa, of which haplotypes 3, 4, 6 and 7 were shared with Southern Africa. Haplotypes 19–36 (18 haplotypes) mainly occurred in West Africa. Populations in Sudan were the most diverse. Populations from Oman, Pakistan and India predominantly shared haplotypes with West and East Africa. Haplotypes 10 and 14 were the most prevalent, occurring mainly in the East African region and accounting for 11.4% and 11.1% of samples, respectively. Haplotypes 24 and 26 were the most prevalent in the West African region, accounting for 7.6% and 8.7% samples, respectively. Several private haplotypes were found, most notably in the Fallatu population from Sudan (5), but also in the quasi population from Oman (3) and one each in Koriema (Kenya), Kigwe (Tanzania), Maroua (Cameroon), Somo (Mauritania) and Jodhpur (India, World Agroforestry Centre general collection). Populations Di (Burkina Faso), Daaba and Rimoi (Kenya), Jodhpur Inde50 and Inde60 (India) were all fixed for a single haplotype.
Population genetic structure and phylogeographic patterns
The SAMOVA analysis showed little change in differentiation among groups from the lowest to the highest FCT values of 0.77 and 0.81 for K=2 and 7, respectively. The biggest change occurred between K=2 and 3. We retained K=3 (FCT 0.79, P<0.001) because single population group membership appeared from K=4. It also detected a key substructure that distinguished the Southern Africa, some East African and Arabian Peninsula populations from the core members of the East African group 1(also supported by strong pairwise FST values, Supplementary Table S7). The three SAMOVA groups primarily separated into East (group 1) and Southern (group 3) from West (group 2) African regions (Table 3 and Figure S1, Supporting information). There were a few exceptions: two West African populations (Diamenar, Senegal; and Tourba, Chad) were assigned to the East African group while populations from Oman and India were grouped with either East or West African groups; two Kenyan populations (Kulamawe and Ntumburi) were grouped with the quasi population from South Africa. The STRUCTURE and the ΔK analysis identified K=2 as the most likely minimum number of clusters but only with prior sample location information (Supplementary Figures S2a and b). As with SAMOVA groups, populations mainly grouped into East and West African clusters, with Southern Africa populations assigned to the East African cluster, whereas those from Arabian Peninsula and Indian subcontinent assigned either to East or West African clusters. Most populations had >74% ancestry, but with evidence of strong admixture, particularly Ngane, Tourba and some Sudanese populations. Cluster 1 (=SAMOVA groups 1 and 3) had lower differentiation (FST=0.188) than Cluster 2 (=SAMOVA group 2, FST=0.283, Figure S1, Supporting information).
Table 3. Haplotype diversity (within-population diversity h S, v S and total diversity h T, v T) and differentiation (G ST, N ST and R ST) for Acacia senegal populations across its range and within SAMOVA delineated groups 1 (mainly East Africa), 2 (mainly West Africa) and 3 (East and Southern Africa).
| Marker | Region | n | hS | hT | GST | vS | vT | NST |
|---|---|---|---|---|---|---|---|---|
| (a) trnH-psbA | Mainly East Africa | 21 | 0.162 (0.0487) | 0.545 (0.0838) | 0.703 (0.0924) | 0.159 (0.0530) | 0.545 (0.0904) | 0.709 (0.0987), <0.736 (permuted) |
| Mainly West Africa | 16 | 0.138 (0.0610) | 0.584 (0.0534) | 0.764 (0.1100) | 0.146 (0.0646) | 0.584 (0.0534) | 0.750 (0.1121), <0.806 (permuted) | |
| East and Southern Africa | 3 | 0.200 (0.2000) | 0.667 (0.3289) | 0.700 (NC)a | 0.200 (0.2000) | 0.667 (0.3289) | 0.850 (NC)a, <0.850 (permuted) | |
| Rangewide | 40 | 0.155 (0.0370) | 0.711 (0.0263) | 0.782 (0.0514) | 0.162 (0.0413) | 0.711 (0.0421) | 0.772 (0.0555), <0.799 (permuted) | |
| East Africa | 22 | 0.138 (0.0467) | 0.523 (0.0972) | 0.737 (0.0835) | 0.120 (0.0433) | 0.524 (0.1077) | 0.771 (0.0782), =0.771 (permuted) | |
| West Africa | 11 | 0.155 (0.0787) | 0.613 (0.0782) | 0.747 (0.1358) | 0.169 (0.0860) | 0.612 (0.0608) | 0.723 (0.1393), <0.805 (permuted) | |
| Africa-wide | 33 | 0.143 (0.0400) | 0.693 (0.0406) | 0.793 (0.0562) | 0.150 (0.0443) | 0.693 (0.0606) | 0.784 (0.0589), <0.807 (permuted) | |
| RST | ||||||||
| (b) cpSSR | Mainly East Africa | 21 | 0.641 (0.0646) | 0.903 (0.0255) | 0.290 (0.0726) | 0.779 (0.5359) | 0.901 (0.3768) | 0.136 (0.2649), <0.437 (permuted) |
| Mainly West Africa | 15 | 0.501 (0.0869) | 0.925 (0.0203) | 0.458 (0.0927) | 0.287 (0.1039) | 0.937 (0.2443) | 0.694 (0.0383), >0.641 (permuted), P=0.05 | |
| East and Southern Africa | 3 | 0.500 (0.2646) | 0.833 (0.0912) | 0.400 (0.4243) | 0.452 (0.2609) | 0.853 (0.0639) | 0.471 (0.4404), <0.759 (permuted) | |
| Rangewide | 39 | 0.576 (0.0516) | 0.948 (0.0092) | 0.392 (0.0537) | 0.312 (0.1842) | 0.954 (0.1773) | 0.673 (0.1555), >0.576 (permuted), P=0.01 |
n, Number of populations; s.e. in parentheses.
NC, not computed due to small sample size.
Structuring of the two cpDNA markers was fairly similar: populations in SAMOVA group 1 predominantly had trnH-psbA haplotypes 1 and 2 and were located in East Africa; populations in SAMOVA group 2 predominantly had trnH-psbA haplotypes 2 and 3 and were located in West Africa (Supplementary Table S4; Figures 2a and b). Overall, cpDNA variation did not segregate with variety, particularly in the East and Southern Africa species range, where the four varieties co-occur; regional location was more important than taxonomy in determining haplotype. Hierarchical AMOVA showed contrasting marker resolution at the rangewide scale (FCT, trnH-psbA=0.156, cpSSR=0.789, Supplementary Table S7; Supporting information). Differentiation was greatest between groups 2 vs 3 (FST, trnH-psbA=0.850, cpSSR=0.964) compared with groups 1 vs 2 or 1 vs 3.
Levels and structure of genetic diversity
At the trnH-psbA locus, most diversity indices had generally similar levels of magnitude among regions, except East and Southern Africa due to small sample size. Differentiation among populations was greater in West than East Africa (GST, 0.764 vs 0.703, Table 3). The contribution of phylogenetic relationships between haplotypes to among population differentiation was not significant at the various geographic scales (NST>NST (permuted), P>0.05). Similar trends were also observed when only Africa populations were analysed (Supplementary Table S4; Supporting information). At the rangewide scale, the cpSSR data showed high within-population diversity (hS, 0.576–0.641) and high total diversity (hT, 0.903–0.948). Population differentiation neglecting haplotype order was GST=0.392, but taking microsatellite evolution into account was RST=0.673 (RST>RST (permuted), P<0.01; Table 3), indicating clear phylogeographic structuring (Pons and Petit, 1996). However, this varied considerably among the regions—with strong phylogeographic structure present in West Africa (SAMOVA group 2: RST=0.694, RST>RST (permuted), P<0.05), and no significant structure in East Africa (SAMOVA group 1: RST=0.136, RST<RST (permuted), P>0.05) and East and Southern Africa (SAMOVA group 3: RST=0.471, RST<RST (permuted), P>0.05). In contrast, strong phylogeographic structure was obtained in both East and West African populations when only African dataset was analysed (Supplementary Table S4; Supporting information).
Discussion
Taking A. senegal as a whole, our data show a significant geographic structuring of genetic variation, with the major division separating East and Southern African populations from those in West and Central Africa. Patterns in the nuclear ITS and chloroplast data were largely concordant. Genetic data poorly reflected the taxonomic subdivision of A. senegal vars. kerensis and senegal, suggesting few barriers to hybridisation among these varieties where they co-occur. The occurrence of highly divergent ITS haplotypes suggests hybridisation among Acacia species may be more frequent than has been observed to date. The early and multiple evolutionary divergence events within the species support the hypothesis that East Africa was the centre of diversification, and that the current wide distribution has arisen largely following past colonisation, migration and range expansion events from Eastern Africa since the late Pleistocene.
Phylogenetic relationships: intraspecific taxonomy, migration and hybridisation
In our analysis of ITS data, var. leiorhachis was distinct and contained a significant amount of variation, suggesting its divergence from the other varieties (senegal, kerensis and rostrata) was ancient. Throughout its recorded distribution var. leiorhachis is found in association with or in close proximity to other varieties, therefore genetic distinctiveness at the nuclear ITS locus appears to be maintained even in the face of potential gene flow. The lack of differentiation among the varieties at chloroplast loci may therefore be due to retained ancestral variation or to chloroplast capture if hybridisation among varieties occurs. However, it is reported that var. leiorhachis is phenologically asynchronous with other varieties, even where they co-occur as proximate populations (Fagg and Allison, 2004). Given the geographically close proximity of the varieties in East Africa, it seems likely that the divergence is ecologically driven and further detailed studies on the distribution should be undertaken. In contrast, variety rostrata is largely confined to Southern Africa. Rather than being ecologically driven, as seems to be the case for var. leiorhachis, it seems more likely that the origins of var. rostrata lie in dispersal to Southern Africa and subsequent independent evolution due to drift.
Of the other varieties, var. kerensis is restricted to East Africa, whereas var. senegal has the widest geographic distribution, occurring throughout the range from East to West Africa and to India. The terminal clade mainly comprised West African var. senegal, suggesting a relatively recent expansion of the range, probably from an origin in Eastern Africa via Sudan and Central Africa to the West and via the Horn of Africa and the Arabian Peninsula to the East. According to the recent phylogenetic chronogram of African acacias by Bouchenak-Khelladi et al. (2010), var. leiorhachis diverged from var. rostrata approximately 1 Mya. Although such estimates are often associated with wide error margins, it suggests that the A. senegal species complex predates the Pleistocene and that the early splits within var. leiorhachis and diversifications within A. senegal coincide with a period marked by prolonged glacial cycles (100 ky periodicity) and extreme aridity (Maley, 2001; Plana, 2004). On the other hand, the unresolved terminal clade (Figure 1) depicts a rapid expansion in the Late Pleistocene by var. senegal, which could be as recent as the last glacial maximum. This period encompasses major glacial maxima with prolonged aridity phases (deMenocal, 1995; Plana, 2004), which offers the necessary driver of restriction and expansion of vegetation that could explain the relatively close relationships among populations across such a wide geographic range.
In a number of cases, hybridisation appears to have been important. In the basal group that comprised an individual from Karofane in Niger (West Africa) and one from Botswana (Southern Africa), similar, highly divergent, sequences were found despite wide geographic separation. It seems at least possible that this is the outcome of interspecific hybridisation—of the 20 other related species that form the A. senegal species complex, most co-occur or share its ecological range (Ross, 1981). Alternatively, the phylogenetic association between the West African and Southern African individuals could also suggest chance retention of ancestral variation. In addition, trnH-psbA haplotypes were variably distributed among ITS clades, which could indicate introgressive hybridisation (Figure 1; Supplementary Table S6). Byrne et al. (2002) also reported haplotype sharing among clades/subspecific taxa within A. acuminata in the mesic and arid zones of Western Australia, which they largely attributed to retention of ancestral polymorphism or incomplete lineage sorting. Both introgression or hybridisations and incomplete lineage sorting may account for the observations in A. senegal. Besides the reported natural hybrid (A. laeta) with A. mellifera, allotetraploids have recently been found in some members of the A. senegal complex indicating that hybridisation may not be uncommon (Assoumane et al., 2012). Although sharing of haplotypes between the regions may also suggest homoplasy, the unexpected shared ITS and chloroplast haplotypes between individuals from East Africa's var. leiorhachis populations and one of the Jodhpur Indian accessions (see H4, Figure 1 and Table 2) is likely to be due to human-mediated dispersal. These regions have had a long history of human migrations and trade links, which may have included gum arabic as a commodity and possible germplasm exchanges or introductions such as was the case for the widely domesticated drumstick tree (Moringa oleifera, Moringaceae; Muluvi et al., 1999).
Genetic diversity, population structure and phylogeography of A. senegal (refugia and gene flow barriers)
Recognising the utility and limitations of different markers on interpretations of diversity and differentiation indices within and among species (for example, see Petit et al., 2005; Meirmans and Hedrick, 2010), the two chloroplast markers used in this study showed considerable complementarity. Most populations were fixed for single chloroplast RFLP trnH-psbA haplotypes but contained multiple cpSSR haplotypes. Although RFLP trnH-psbA haplotypes had less resolution than cpSSR haplotypes at the rangewide scale, both showed similar regional genetic structuring and phylogeographic patterns (Table 3, Figure 2, Supplementary Figure S1). The levels of within-population and total diversity estimated from cpSSR data (hS=0.576, hT=0.948) were comparable to those reported by Byrne et al. (2002) within A. acuminata populations (cf. hS=0.442; hT=0.920) in Western Australia. Of particular interest is comparisons with the boabab tree that also showed a similar East-West African phylogeographic pattern; they had contrasting within-population, total diversity and population differentiation with RFLP data (for example, A. senegal: hS, 0.155; hT, 0.711; GST, 0.793 vs boabab: hS, 0.017; hT, 0.58; GST, 0.970; Pock Tsy et al., 2009). These differences are likely to be due to the contrasting life histories and modes of seed or fruit dispersal between the two species. A. senegal is pollinated mainly by bees and seeds dispersed by wind (Fagg and Allison, 2004). Baobab is pollinated by bats and seed dispersed by large mammals and humans (Pock Tsy et al., 2009). Baobab's long domestication history, significant human-mediated dispersal and reported longevity of ∼1300 years are particularly important distinguishing factors.
The distribution patterns of cpDNA haplotypes suggest that colonisation, migration or expansion events may have happened more than once originating from an East African source population. The Horn of Africa is an important centre of speciation in the A. senegal complex and also has the highest concentration of African Acacia species (Ross, 1981; Fagg and Allison, 2004). In A. senegal, this is the only region where all four varieties are reportedly distributed, in some cases as co-occurring populations (Fagg and Allison, 2004). Diversification of the species may have been driven by an interaction between the climate changes of the late Pleistocene and the complex topography of Eastern Africa, which provides potential refugia in areas that have historically been sheltered against the impacts of extreme climate oscillations (Plana, 2004). During enhanced and prolonged aridity phases the Acacia range may have shifted or migrated to higher altitudes, tracking moist forest (Hamilton, 1982) and causing fragmentation. These sites could have acted as source populations for (re-) colonisations or migrations into other regions during favourable climatic conditions. The genetic and morphological diversity of A. senegal in this region, as well as fossil pollen data, supports this hypothesis. Acacia-type pollen, predating the Pleistocene, has been recorded from the Rift Valley floor—at the Lokichar Basin (near one of our sampling sites, see Table 1) in the southwestern part of Lake Turkana Basin, Kenya (Vincens et al., 2006). The Lake Turkana Basin still harbours several Acacia spp., including the predominant A. senegal. There is also pollen evidence of montane Acacia woodlands (up to 4040 masl) in the East African highlands predating the arid period at ∼10 000 years before present (BP; Hamilton, 1982). These pollen records suggest that East Africa may have harboured refugia for Acacia taxa predating the Pleistocene. The haplotype-rich populations of Sudan are found in this region. Of these populations, Fallatu forest had an exceptionally high number of private cpSSR haplotypes (5), possibly suggesting an ancient refugium. The Sudanese populations occur in the so-called gum belt from which the bulk of the internationally traded gum is produced in the traditional ‘gum orchard' agroforestry systems. Although it is possible that domestication has resulted in incorporation of extra diversity, importation of seed would be uncharacteristic because the traditional gum production systems are normally established with local collections or through natural regenerations (Fagg and Allison, 2004). Alternatively, if the region has long acted as a refugium, the potential for hybridisation and chloroplast capture from congeners is higher and may offer an explanation. In Southern Africa, the presence of a private trnH-psbA haplotype suggests long-term persistence and isolation of the population in this region. The expansion and contraction of moist rainforest across East Africa is likely to have been the driving force behind isolation of Southern African populations (Cowling et al., 2008). Similar patterns have been found in phylogeographic studies of other savannah-adapted species, such as the plains zebra (Lorenzen et al., 2008). Further sampling of A. senegal in Southern Tanzania and Mozambique would help to test this hypothesis.
In West Africa, population differentiation was higher and had a stronger phylogeographic structure than that of East Africa (cpSSR data, GST=0.458 vs 0.290; RFLP, 0.764 vs 0.703, Table 3; also see Supplementary Figure S2, STRUCTURE analysis, FST=0.283 vs 0.188), which also indicates high admixture of A. senegal organelle lineages. This suggests that dispersal from refugia occurred over relatively short distances, probably due to the influence of geographic barriers, as invoked to explain the genetic structuring of other dry woodland and savannah species in the Sudano-Sahelian region (for example, shea tree, Allal et al., 2011). For example, in the Chad basin, an inundation during the Quaternary pluvial, followed by the formation of a Mega-Lake was suggested to have isolated baobab populations of West Africa from those of East Africa (Pock Tsy et al., 2009). The historical Mega-Lake zone also coincides with the haplotype disjunctions observed in this study between Central-West and West African A. senegal populations. The two populations that mark this contact zone are Tourba, Chad (east of Lake Chad) and Maroua, Cameroon (south of Lake Chad). However, the Maroua population (fixed trnH-psbA haplotype 6) would have also been isolated or restricted either on the fringes of tropical rainforest or riparian conditions. Further West, dry riverine woodlands and gallery forests along the extensive West African river systems (Senegal, Niger, Volta and Gambia) may have also acted as refugia during periods of extreme aridity that also promoted southward expansion of the Sahara and regression of moist tropical forest. The possible refugial zones for A. senegal in this region would therefore include the westernmost part of West Africa (Western Senegal) and Central Africa (north of Cameroon). Although poorly represented in pollen cores, Acacia-type pollen records have also been reported for the Sudano-Sahelian region during the Holocene humid period (for example, Jikariye Lake, Nigeria, ∼11 000 years BP, Waller et al., 2007; Lake Yoa, Chad, ∼6000 years BP, Lézine, 2009). However, lack of pollen data predating the Holocene and last glacial maximum may not preclude the likely occurrence of ancient Acacia refugia in West Africa. In other parts of the range beyond Africa, Acacia-type pollen have also been recorded from the semi-desert around Kwar al Jaramah (Oman, Arabian Peninsula) and Makran on the Pakistani coast and the Indian subcontinent (∼6000 years BP, Lézine, 2009).
Conclusions
The combined nrDNA and cpDNA analysis of A. senegal have shown high haplotypic variation with both regional and rangewide phylogeographic patterns, suggesting a long history of colonisation and expansion events characterised by extensive, recurrent gene flow among populations and regions. The phylogenetic analysis revealed multiple evolutionary divergence events that also separated var. leiorhachis from the other three (senegal, kerensis and rostrata). The phylogeographic structure separating East and Southern Africa from West African populations reflects a pattern reported for other drylands species straddling the Sudano-Sahelian region (for example, baobab, Pock Tsy et al., 2009; shea, Allal et al., 2011). However, contrary to our prediction and in contrast to the other species, within-region differentiation of A. senegal was greater in West Africa than East and Southern Africa, clearly showing species differences at the regional scale. Spatial studies at the population and landscape scales will be necessary to elucidate factors that have influenced the observed regional differences. Cytological studies within the species complex would provide evidence for introgression or hybridisation events and show whether certain morphological variants (intraspecies) have been more successful than others in colonisation and range expansion in the more arid conditions. In general, new studies of a range of species, in particular those that take a comparative approach, will be required to understand how plant species have dispersed and adapted to dryland habitats and coped with the effects of repeated historical range shifts. Studying the evolutionary history of organisms can provide valuable insight into how species have responded to past environmental and population changes. In dryland biomes, likely to be highly sensitive to the effects of climate change but socio-economically and ecologically vital across sub-Saharan Africa, such knowledge is essential for guiding strategies to promote resilience to future changes.
Data archiving
Sequence data have been deposite at GenBank: accession numbers HQ605042-HQ605077. Chloroplast marker data are publicly available via the CEH Information Gateway (http://dx.doi.org/10.5285/de1f9a43-dd0d-428c-af7b-2dfdfc8c127a).
Acknowledgments
This work was supported by a Marie Curie Fellowship Contract MIIF-CT-2006-39216 ASGEN and the ACACIAGUM Project EC FP6 Contract No. 032233. We thank Alan Gray (CEH Edinburgh) and the Royal Botanic Garden Edinburgh for herbarium specimens and the World Agroforestry Centre (ICRAF) for supplying seeds. We also thank Stephen Omondi and John Gicheru of KEFRI, and Aliou Faye, Mayecor Diouf and Mame Sokhna of ISRA for field assistance during sample collection. We thank Stephen Harris (University of Oxford), the three anonymous reviewers and the subject editor for their constructive criticisms and suggestions.
Author contributions: DWO led the draft of the manuscript. DWO, SC, AT and JW designed the work, acquired and analysed data; AG led field design, sample and data collection in Senegal. All authors contributed ideas, comments and revised the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Allal F, Sanou H, Millet L, Vaillant A, Camus-Kulandaivelu L, Logossa ZA, et al. Past climate changes explain the phylogeography of Vitellaria paradoxa over Africa. Heredity. 2011;107:174–186. doi: 10.1038/hdy.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Ariati SR, Murphy DJ, Udovicic F, Ladiges PY. Molecular phylogeny of three groups of acacias (Acacia subgenus Phyllodineae) in arid Australia based on the internal and external transcribed spacer regions of nrDNA. Syst Biodivers. 2006;4:417–426. [Google Scholar]
- Assoumane A, Zoubeirou AM, Rodier-Goud R, Favreau B, Bezançon G, Verhaegen D.2012Highlighting the occurrence of tetraploidy in Acacia senegal (L.) Willd. and genetic variation patterns in its natural range revealed by DNA microsatellite markers Tree Genet Genomes(e-pub ahead of print 26 June 2012; doi: 10.1007/s11295-012-0537-0) [DOI]
- Ayele TB, Gailing O, Umer M, Finkeldey R. Chloroplast DNA haplotype diversity and postglacial recolonization of Hagenia abyssinica (Bruce) J.F. Gmel. in Ethiopia. Plant Syst Evol. 2009;280:175–185. [Google Scholar]
- Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. The its region of nuclear ribosomal region: a valuable source of evidence on angiosperm phylogeny. Ann Mo Bot Gard. 1995;82:242–277. [Google Scholar]
- Bouchenak-Khelladi Y, Maurin O, Hurter J, et al. The evolutionary history and biogeography of Mimosoideae (Leguminosae): an emphasis on African acacias. Mol Phylogenet Evol. 2010;57:495–508. doi: 10.1016/j.ympev.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Brown GK, Murphy DJ, Miller JT, Ladiges PY. Acacia s.s. and its relationship among tropical legumes, Tribe Ingeae (Leguminosae: Mimosoideae) Syst Bot. 2008;33:739–751. [Google Scholar]
- Bukhari YM. Cytoevolution of taxa in Acacia and Prosopis (Mimosaceae) Aust J Bot. 1997;45:879–891. [Google Scholar]
- Byrne M, Macdonald B, Coates D. Phylogeographical patterns in chloroplast DNA variation within the Acacia acuminata (Leguminosae: Mimosoideae) complex in Western Australia. J Evol Biol. 2002;15:576–587. [Google Scholar]
- Caetano C, Prado D, Pennington RT, Beck S, Olivera-Filho A, Spichiger R, et al. The history of seasonally dry forests in eastern South America: inferences from the genetic structure of the tree Astronium urundeuva (Anacardiaceae) Mol Ecol. 2008;17:3147–3159. doi: 10.1111/j.1365-294X.2008.03817.x. [DOI] [PubMed] [Google Scholar]
- Cavers S, Navarro C, Lowe AJ. Chloroplast DNA phylogeography reveals colonization history of Cedrela odorata L., in Mesoamerica. Mol Ecol. 2003;12:1451–1460. doi: 10.1046/j.1365-294x.2003.01810.x. [DOI] [PubMed] [Google Scholar]
- Collevatti RG, Grattapaglia D, Hay JD. Evidences for multiple maternal lineages of Caryocar brasiliense populations in the Brazilian Cerrado based on the analysis of chloroplast DNA sequences and microsatellite haplotype variation. Mol Ecol. 2003;12:105–115. doi: 10.1046/j.1365-294x.2003.01701.x. [DOI] [PubMed] [Google Scholar]
- Cowling SA, Cox PM, Jones CD, Maslin MA, Peros M, Spall SA. Simulated glacial and interglacial vegetation across Africa: implications for species phylogenies and trans-African migration of plants and animals. Glob Change Biol. 2008;14:827–840. [Google Scholar]
- Daïnou K, Bizoux J-P, Doucet J-L, Mahy G, Hardy OJ, Heuertz M. Forest refugia revisited: nSSRs and cpDNA sequences support historical isolation in a wide-spread African tree with high colonization capacity, Milicia excelsa (Moraceae) Mol Ecol. 2010;19:4462–4477. doi: 10.1111/j.1365-294X.2010.04831.x. [DOI] [PubMed] [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Mol Ecol. 2002;11:2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- El Amin HM. Acacia laeta (R. Br.) ex. Benth., considered as a species of hybrid origin. Sudan Silva. 1976;3:14–23. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fagg CW, Allison GE.2004Acacia Senegal and the Gum Arabic Trade. Monograph and Annotated BibliographyTropical forestry papers no. 42Oxford Forestry Institute: Oxford [Google Scholar]
- Hamilton AC. Environmental History of East Africa. A Study of the Quaternary. Academic Press: London; 1982. [Google Scholar]
- Huelsenbeck JP, Ronquist. F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jacobs BF. Palaeobotanical studies from tropical Africa: relevance to evolution of forest, woodland and savannah biomes. Philos T Roy Soc B. 2004;359:1573–1583. doi: 10.1098/rstb.2004.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Bot. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Lézine A-M. Timing of vegetation changes at the end of the Holocene humid period in desert areas at the northern edge of the Atlantic and Indian monsoon systems. CR Geoscience. 2009;341:750–759. [Google Scholar]
- Lock JM.2006The seasonally dry vegetation of Africa: parallels and comparisons with the neotropicsIn: Pennington RT, Lewis GP, Ratter JA, (eds)Neotropical Savannas and Dry Forests: Diversity, Biogeography, and Conservation Taylor and Francis Ltd: London; 449–467. [Google Scholar]
- Lorenzen ED, Arctander P, Siegismund HR. High variation and very low differentiation in wide ranging plains zebra (Equus quagga): insights from mtDNA and microsatellites. Mol Ecol. 2008;17:2812–2824. doi: 10.1111/j.1365-294X.2008.03781.x. [DOI] [PubMed] [Google Scholar]
- Lorenzen ED, Masembe C, Arctander P, Siegismund HR. A long-standing Pleistocene refugium in Southern Africa and a mosaic of refugia in East Africa: insights from mtDNA and the common eland antelope. J Biogeogr. 2010;37:571–581. [Google Scholar]
- Lowe AJ, Harris D, Dormont E, Dawson IK. Testing putative tropical forest refugia using chloroplast and nuclear phylogeography. Trop Plant Biol. 2010;3:50–58. [Google Scholar]
- Maley J.2001The impact of arid phases on the African rain forest through geological historyIn: Weber W, White L, Vedder A, Naughton-Treves L, (eds)African Rain Forest Ecology and Conservation Yale University Press: New Haven, Connecticut; 68–87. [Google Scholar]
- Meirmans PG, Hedrick PW. Assessing population structure: FST and related measures. Mol Ecol Res. 2010;11:5–18. doi: 10.1111/j.1755-0998.2010.02927.x. [DOI] [PubMed] [Google Scholar]
- deMenocal PB. Plio-Pleistocene African climate. Science. 1995;270:53–59. doi: 10.1126/science.270.5233.53. [DOI] [PubMed] [Google Scholar]
- Muluvi GM, Sprent JI, Soranzo N, Provan J, Odee D, Folkard G, et al. Amplified fragment length polymorphism (AFLP) analysis of genetic variation in Moringa oleifera Lam. Mol Ecol. 1999;8:463–470. doi: 10.1046/j.1365-294x.1999.00589.x. [DOI] [PubMed] [Google Scholar]
- Omondi SF, Kireger E, Dangasuk OG, Chikamai B, Odee DW, Cavers S, et al. Genetic diversity and population structure of Acacia senegal (L.) Willd. in Kenya. Trop Plant Biol. 2010;3:59–70. [Google Scholar]
- Pennington RT, Lavin M, Oliveira-Filho A. Woody plant diversity, evolution and ecology in the tropics: perspectives from seasonally dry tropical forests. Annu Rev Ecol Evol Syst. 2009;40:437–457. [Google Scholar]
- Pennington RT, Lavin M, Prado DE, Pendry CA, Pell SK, Butterworth CA. Historical climate change and speciation: neotropical seasonally dry forest plants show patterns of both tertiary and quaternary diversification. Philos T Roy Soc B. 2004;359:515–537. doi: 10.1098/rstb.2003.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol. 2005;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- Plana V. Mechanisms and tempo of evolution in the African Guineo-Congolian rainforest. Philos T Roy Soc B. 2004;359:1585–1594. doi: 10.1098/rstb.2004.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pock Tsy J-ML, Lumaret R, Mayne D, Vall AOM, Abutaba YIM, Sagna M, et al. Chloroplast DNA phylogeography suggests a West African centre of origin for the baobab, Adansonia digitata L. (Bombacoideae, Malvaceae) Mol Ecol. 2009;18:1707–1715. doi: 10.1111/j.1365-294X.2009.04144.x. [DOI] [PubMed] [Google Scholar]
- Pons O, Petit RJ. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996;144:1237–1245. doi: 10.1093/genetics/144.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos ACM, Lemos-Filho JP, Ribeiro RC, Santos FC, Lovato MD. Phylogeography of the tree Hymenaea stigonocarpa (Fabaceae: Caesalpinioideae) and influence of quaternary climate changes in the Brazilian cerrado. Ann Bot. 2007;100:1219–1228. doi: 10.1093/aob/mcm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM. Rapid diversification of a species-rich genus of neotropical rain forest trees. Science. 2001;293:2242–2245. doi: 10.1126/science.1061421. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ross JH. A conspectus of the African Acacia species. Mem Bot Surv S Afr. 1979;44:1–155. [Google Scholar]
- Ross JH. An analysis of the African Acacia species: their distribution, possible origins and relationships. Bothalia. 1981;13:398–413. [Google Scholar]
- Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, et al. The tortoise and the hare: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Sprent JI, Odee DW, Dakora FD. African legumes: a vital but under-utilized resource. J Exp Bot. 2010;61:1257–1265. doi: 10.1093/jxb/erp342. [DOI] [PubMed] [Google Scholar]
- Sun Y, Skinner DZ, Liang GH, Hulbert SH. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theor Appl Genet. 1994;89:26–32. doi: 10.1007/BF00226978. [DOI] [PubMed] [Google Scholar]
- Swofford DL.2003PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods)Version 4.0 b10, version 4.0 beta 10Sinauer Associates: Sunderland [Google Scholar]
- Vincens A, Tiercelin J-J, Buchet G. New Oligocene-early Miocene microflora from the southwestern Turkana Basin. Palaeoenvironmental implications in the northern Kenya Rift. Palaeogeogr Palaeocl. 2006;239:470–486. [Google Scholar]
- Waller MP, Street-Perrot FA, Wang H. Holocene vegetation history of the Sahel: pollen, sedimentological and geochemical data from Jikariya Lake, north-eastern Nigeria. J Biogeogr. 2007;34:1575–1590. [Google Scholar]
- Wardill TJ, Graham GC, Zalucki N, Palmer WA, Playford J, Scott KD. The importance of species identity in the biocontrol process: identifying the subspecies of Acacia nilotica (Leguminosae: Mimosoideae) by genetic distance and the implications for biological control. J Biogeogr. 2005;32:2145–2159. [Google Scholar]
- Weising K, Gardner RC. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome. 1999;42:9–19. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.