Abstract
Local adaptation is important in evolutionary processes and speciation. We used multiple tests to identify several candidate genes that may be involved in local adaptation from 1026 loci in 14 natural populations of Cryptomeria japonica, the most economically important forestry tree in Japan. We also studied the relationships between genotypes and environmental variables to obtain information on the selective pressures acting on individual populations. Outlier loci were mapped onto a linkage map, and the positions of loci associated with specific environmental variables are considered. The outlier loci were not randomly distributed on the linkage map; linkage group 11 was identified as a genomic island of divergence. Three loci in this region were also associated with environmental variables such as mean annual temperature, daily maximum temperature, maximum snow depth, and so on. Outlier loci identified with high significance levels will be essential for conservation purposes and for future work on molecular breeding.
Keywords: adaptation, conifer, environment, linkage map, selection
Introduction
Local adaptation is important in evolutionary processes and speciation (Hoffmann and Willi, 2008; Stapley et al., 2010). It occurs over relatively long periods of time in plant populations with long generation times, such as forest tree species. The process of local adaptation involves the genetic divergence of specific loci in populations inhabiting different environments. Once identified, these loci can be used for conservational purposes, for future forestation, and to study the mechanisms by which trees adapt to specific environments. Forest tree species have not been as extensively domesticated as crop species, and natural populations are still abundant. By identifying and studying adaptive genes in these natural populations, it is possible to gain new insights into the adaptive mechanisms that allow them to flourish in the wild (Neale and Savolainen, 2004; González-Martínez et al., 2006; Neale, 2007; Savolainen and Pyhäjärvi, 2007; Savolainen et al., 2007; Neale and Ingvarsson, 2008; Grattapaglia et al., 2009). This is facilitated by certain properties of forest tree species: they typically have relatively low genetic differentiation between populations on average, are widely distributed in different environments and have relatively large population sizes. Consequently, the selective pressures acting on a population in a given environment tend to result in adaptation by selecting for a relatively small number of genotype changes at a few specific loci (Howe et al., 2003; Holliday et al., 2010; Pelgas et al., 2011).
Genome scanning is an effective and useful method for detecting genetic differentiation and candidate genes associated with specific phenotypes or environmental variables in forest tree species (Eveno et al., 2008; Namroud et al., 2008; Eckert et al., 2010; Holliday et al., 2010; Neale and Kremer, 2011; Prunier et al., 2011). Recently developed multiple genotyping methods have greatly reduced the cost of single-nucleotide polymorphism (SNP) genotyping. Thus, multiple-marker-based neutrality tests are frequently used for detecting functional DNA variations. The most widely used ‘outlier tests' are multiple-population tests such as the FST-based test (Beaumont and Nichols, 1996), the coalescent-based simulation approach (Vitalis et al., 2001) and the lnRv test based on the Gaussian null distribution (Kauer et al., 2003). The Bayesian method implemented in BayeScan has the advantage that it estimates population-specific FST coefficients, thereby accounting for different demographic histories and different amounts of genetic drift between populations (Foll and Gaggiotti, 2008). When identifying outlier loci, it is generally necessary to use multiple tests in order to minimize the false-positive rate; however, false positives remain common even when using multiple methods and multitest correction (Pérez-Figueroa et al., 2010). It is therefore important to validate the detected outlier loci in multiple ways to determine whether or not they genuinely are adaptive. Luikart et al. (2003) suggested several ways to confirm outlier behavior and selection, including considering their genomic location, quantitative trait locus mapping and testing for genotyping errors.
Cryptomeria japonica is an allogamous coniferous species that relies on wind-mediated pollen and seed dispersal. Modern natural forests of the species are distributed across various different environments in the Japanese Archipelago, from Aomori Prefecture (40° 42′ N) to Yakushima Island (30° 15′ N) (Hayashi, 1960). However, its distribution is discontinuous and scattered across small, restricted areas as a result of its extensive exploitation over the last thousand years (Ohba, 1993). The geographical variation between natural forests of C. japonica has been investigated, focusing on both morphological traits (needle length, needle curvature and other features; Murai, 1947) and diterpene components (Yasue et al., 1987). The results of these studies suggest that there are two main lines: ura-sugi (C. japonica var. radicans, found near the Sea of Japan) and omote-sugi (C. japonica, located near the Pacific Ocean). The ura-sugi variety has slender branchlets with soft leaves, while the omote-sugi variety has rough branchlets with hard leaves (Yamazaki, 1995). A previous study focusing on 148 cleaved amplified polymorphic sequence loci revealed genetic differentiation between the two varieties, and four outlier loci were identified as potential local adaptation genes (Tsumura et al., 2007) that may be associated with the genetic differentiation between the varieties.
In the study reported herein, we identified potential local adaptation genes at 1026 loci using two tests and also studied the relationships between specific SNPs and environmental variables such as climate data to investigate the selective pressures acting on the studied populations. The linkage map positions of outlier loci that were associated with specific environmental variables are also discussed.
Materials and methods
Investigated populations
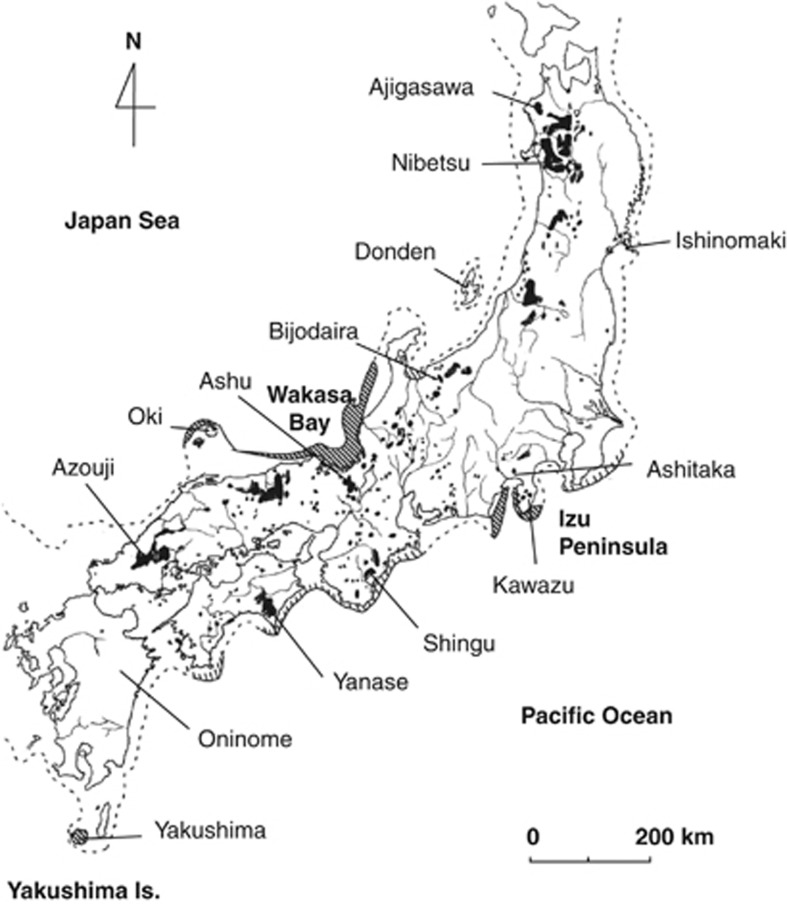
We examined 14 populations in this study with 186 individuals in total (the average number of individuals per population was 13.29): seven from the Japan Sea side of Japan and seven from the Pacific Ocean side (Table 1). All trees sampled were growing in national forests that were candidates for in situ gene-conservation programs. The locations of the sampled populations covered most of the natural distribution of C. japonica (Figure 1, Tsumura et al., 2007).
Table 1. Locations and environment variables for 14 investigated natural populations in C. japonica.
| Population | Abbreviation | Latitude | Longitude | Altitude (m) | Number of individuals | Mean annual temperature (°C) | Mean daily maximum temperature (°C) | Mean daily minimum temperature (°C) | Annual precipitation (mm) | Annual sunshine hours (h) | Amount of global solar radiation (MJ m−2) | Deepest snow (cm) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ajigasawa | AJG | 40.44.53 | 140.12.54 | 110 | 14 | 9.7 | 13.7 | 5.9 | 1312.3 | 1711.5 | 12.3 | 70 |
| Nibetsu | NBT | 39.48.22 | 140.15.36 | 320 | 15 | 11.3 | 15.2 | 7.6 | 1701.2 | 1593.7 | 11.8 | 37 |
| Ishinomaki | ISN | 38.19.43 | 141.29.31 | 190 | 13 | 11.1 | 14.3 | 8.5 | 1276.3 | 1972.4 | 13.1 | 7 |
| Donden | DND | 38.08.23 | 138.23.00 | 730 | 14 | 9.9 | 13.0 | 6.8 | 2184.0 | 1658.7 | 12.7 | 99 |
| Bijodaira | BJD | 36.34.55 | 137.27.55 | 1000 | 16 | 7.8 | 11.6 | 3.7 | 2884.4 | 1552.2 | 12.9 | 171 |
| Ashitaka | AST | 35.12.30 | 138.49.90 | 1000 | 17 | 8.2 | 11.4 | 4.3 | 2886.1 | 1857.7 | 14.0 | 37 |
| Kawazu | KWZ | 34.49.53 | 139.00.00 | 650 | 8 | 15.9 | 19.1 | 12.9 | 2921.6 | 1830.1 | 12.5 | 1 |
| Ashu | ASH | 35.18.28 | 135.46.26 | 700 | 18 | 9.3 | 13.8 | 5.4 | 2379.4 | 1523.4 | 12.4 | 55 |
| Oki | OKI | 36.16.60 | 133.19.45 | 360 | 9 | 11.9 | 15.7 | 8.3 | 1873.9 | 1782.5 | 13.2 | 29 |
| Azouji | AZJ | 34.46.70 | 132.06.12 | 360 | 16 | 12.0 | 17.3 | 7.1 | 2220.4 | 1635.9 | 12.8 | 43 |
| Shingu | SNG | 33.49.24 | 135.45.36 | 170 | 16 | 14.2 | 20.0 | 9.3 | 2870.5 | 1834.0 | 12.8 | 2 |
| Yanase | YNS | 33.35.40 | 134.05.80 | 500 | 8 | 12.9 | 18.1 | 8.8 | 3739.7 | 2004.6 | 14.2 | 2 |
| Oninome | ONN | 32.42.06 | 131.31.43 | 960 | 6 | 10.2 | 15.1 | 5.9 | 2762.2 | 1884.7 | 14.0 | 2 |
| Yakushima | YKU | 30.18.17 | 130.34.17 | 1100 | 16 | 13.3 | 15.6 | 11.1 | 4119.7 | 1559.8 | 13.6 | 0 |
Figure 1.
Natural distribution of C. japonica in Japan (shaded areas; from Hayashi, 1960) and the locations of the 14 natural populations surveyed in this study. The dotted line indicates the coastline ca. 18 000 years ago. Areas shaded in bold or within thin diagonal lines show established refugia (Izu Peninsula, Wakasa Bay, Oki Island and Yakushima Island) and probable refugia, respectively, at that time (Tsukada, 1986).
SNP genotyping
We selected one SNP each from 1536 unique expressed sequence tag contigs in C. japonica. This assumes implicitly that SNPs within different expressed sequence tag contigs are independent (that is, unlinked). The identification of SNPs was carried out previously and was accomplished through the resequencing of 5170 unique expressed sequence tag contigs in a discovery panel of four C. japonica individuals collected from a range-wide sample of trees (Uchiyama et al., 2012). Multiplexed genotyping of SNP markers for natural populations was carried out using Illumina's 1536-plex GoldenGate array (Illumina Inc., San Diego, CA, USA) according to the protocol recommended by the manufacturer. A total of 0.5–1.0 μg of genomic DNA per sample (at a rate of 100–200 ng μl−1) was used in each GoldenGate assay. Only SNPs with Illumina design scores above 0.6 were used. When multiple SNPs were available within the same sequence, a single highly polymorphic SNP was selected as the target for that sequence. The GoldenGate assay uses highly multiplexed allele-specific extension methods and universal PCR amplification reactions. The PCR products, which were fluorescently labeled by using the 5′-labeled primers P1 (Cy3) and P2 (Cy5), were hybridized to capture probes on the beads in the array. The ratio of the fluorescent signals from two allele-specific ligation products was used to determine the genotype. Signal intensities were quantified and matched to specific alleles, using the Genome Studio v2010.2 software package in Illumina BeadArray Reader (Illumina Inc.). The quality of the GoldenGate genotype scores for individual SNPs was assessed based on their GenTrain cluster and GenCall genotype scores in GenomeStudio (Illumina Inc.). A minimum GenCall50 (GC50) score of 0.25 was chosen as the threshold for inclusion of SNP loci in the final data set, and genotypic clusters were edited manually when necessary. In the present study, this threshold corresponded to SNPs with accurate scoring for at least 95% of the individuals, with most successful SNPs scored for over 99% of the individuals analyzed.
Genetic structure
To evaluate the within-population variation, we used the proportion of polymorphic loci (Pl) at the 95% probability level, the unbiased heterozygosity (He), (Nei, 1977) and allelic richness (Rs) calculated from the allele frequencies of all loci analyzed. Allelic richness was measured by the method of El Mousadik and Petit (1996) with a reference sample size of 6. The fixation indices, FIS=1−He/Ho, for polymorphic loci and their averages over all loci were determined to compare the observed genotype frequencies with expectations based on the Hardy–Weinberg equilibrium (Wright, 1922; Nei, 1977; Nei and Chesser, 1983). Deviations from such expectations were analyzed using Fisher's exact test. Coefficients of gene differentiation, FST, among populations, were calculated to determine how gene diversity was partitioned at each level (Wright, 1978). These analyses were done using the FSTAT software package (Goudet, 2000) and GenAlEx (Peakall and Smouse, 2006). To detect population structure and infer the most appropriate number of subpopulations (K) for interpreting the data without prior information on the number of locations at which the populations were sampled, we used the F-model of Bayesian clustering approach proposed by Pritchard et al. (2000). For this analysis, we excluded 184 loci with high linkage disequilibrium (LD) values (χ2 test, significant at 99% level after Bonferroni correction), 141 outlier loci that were detected by Fdist2 analysis and a locus that was distorted from the Hardy–Weinberg equilibrium. Ten independent runs with K values ranging from 1 to 10 were performed using 2 × 106 MCMC (Marcov chain Monte Carlo) sampling after a burn-in period of 50 000 iterations. The posterior probability was then calculated for each value of K using the estimated log likelihood of K to identify the optimal K value. The optimal number of K was defined as the one at which the log likelihood of the data, ln P(X|K) (Pritchard et al., 2000) or ΔK, the rate of change of ln P(X|K) between successive K values (Evanno et al., 2005), was maximal. To examine the genetic differentiation between two groups representing the two varieties, C. japonica and C. japonica var. radicance, we performed a hierarchical analysis of molecular variance (Excoffier et al., 1992) in which the significance levels for variance components were tested using permutations.
Methods for detecting candidate loci for divergence
The following methods were used to detect signs of natural selection and to tentatively identify the environmental parameters associated with the corresponding selective pressures acting on C. japonica populations. We also compared the distributions of the FST values over all loci to their expected distributions under the assumption of neutrality. Beaumont and Nichols (1996) have shown that the distribution of FST as a function of heterozygosity in the context of an island model is quite robust, that is, insensitive to variations in factors such as population structure, demographic structure and mutation level. We therefore used this method to identify markers deviating from the null hypothesis of neutral evolution. All calculations for identifying potential non-neutral genes in the studied populations were performed using the Fdist2 (Beaumont and Nichols, 1996) and Arlequin programs (Excoffier and Lischer, 2010). Arlequin program allows us the hierarchical analysis using two genetic lines such as ura-sugi and omote-sugi varieties. In the analysis using Fdist2, we identified the outlier loci using not only all 14 populations but also each variety populations such as seven ura-sugi populations and seven omote-sugi populations.
An alternative method for identifying candidate loci is BayeScan, which uses a Bayesian method to directly estimate a posterior probability for each locus and a reversible-jump MCMC approach to selection (Foll and Gaggiotti, 2008). The main advantage of BayeScan is that it estimates population-specific FST coefficients and therefore accommodates differences in demographic history and the extent of genetic drift between populations. This method is an extension of that proposed by Beaumont and Balding (2004), and it is based on a logistic regression model that decomposes genetic variation into population- and locus-specific effects. Preliminary tests were conducted using a burn-in of 10 000 iterations, a thinning interval of 50 and a sample size of 10 000 (Foll and Gaggiotti, 2008). Four independent runs were performed for each of the two data sets to account for the consistency of the detected outliers. The loci were ranked according to their estimated posterior probability and all loci with a value over 0.990 were retained as outliers. This corresponds to a log10 Bayes factor of more than 2, making it possible to be confident in the validity of the model.
The gathering of environmental data for each population and the identification of correlations with the genetic data
Environmental information on the area of origin of each population, including longitude, latitude, altitude, mean annual temperature, mean annual precipitation, annual sunshine hours, amount of global solar radiation and maximum snow depth over 30 years (1971–2000) was obtained from the Japan Meteorological Agency (2002), in the form of detailed 1-km mesh data. We used information from the 1-km mesh data to estimate the mean annual temperature, precipitation and snowfall, and so on, for each site using the program in Mesh climatic data of Japan of the Japan Meteorological Agency (2002).
To calculate correlations between SNP allele frequencies and climate variables, we used the Bayesian linear model method of Coop et al. (2010), which controls for population history by incorporating a covariance matrix of populations and that accounts for differences in sample size among populations. Using the full set of SNPs, we estimated a covariance matrix of allele frequencies across populations. This covariance matrix was used as the basis of the null model for the transformed allele frequencies at each SNP to be tested. For each tested SNP, the method generates a Bayes factor as a measure of the support for the alternative model relative to the null model, in which the transformed population allele frequency distribution is dependent on population structure alone. The significance threshold for ranked Bayes factors was set to 2.0, and the environmental variables considered were latitude, longitude and ten climate variables. All climate variables were standardized before analysis. In this analysis, we included the geographical information such as latitude and longitude to find out the false positives because they are sometimes correlated with some environmental variables.
Putative functions for the detected outlier loci were identified by performing BLASTx searches of the NCBI database using Blast2GO (Conesa et al., 2005, Supplementary Table 1).
Mapping the outlier loci on the linkage map, and LD
The YI pedigree used to determine the positions of the outlier loci of C. japonica had previously been used to construct a linkage map, in which a total of 438 markers were assigned to 11 large linkage groups (LGs) and some small or nonintegrated LGs, giving a total map length of 1372.2 cM (Tani et al., 2003). The mapped population was genotyped using Illumina's 1536-plex GoldenGate array, as discussed above. All linkage analyses and map estimations were performed using the JoinMap v3.0 software package (Van Ooijen and Voorrips, 2001). During map construction, markers were assigned to tentative LGs by comparing the linkages formed at logarithm of odds thresholds ranging from 3.0 to 9.0, increasing in increments of 1.0. Finally, the markers were ordered at a logarithm of odds threshold of 8.0. Map distances were calculated using the Kosambi mapping function (Kosambi, 1944).
Coefficients of LD were calculated as described by Weir (1996), using squared allele–frequency correlations (r2) for pairs of loci on the basis of genotype data from the investigated populations. The differences from equilibrium were verified by the χ2 tests (Weir, 1996). These analyses were performed using the GDA software package (Lewis and Zaykin, 2002).
Assuming a random distribution of markers, if the genome were divided into N intervals, the number of markers per interval would follow a Poisson distribution. To determine whether outlier loci were randomly distributed, every LG was divided into 15, 20, 25 and 50-cM intervals. We used the χ2 test to compare the actual distribution of outlier loci to that expected for a Poisson distribution, as described by Kang et al. (2010).
Results
SNP genotyping
Of the 1536 SNPs, 1031 (67.1%) yielded data that met our quality thresholds according to the GoldenGate genotyping system (Uchiyama et al., 2012). The median GC50 score across all usable SNPs was 0.81, with an average call rate of 98.6%. A minimum GenCall50 (GC50) score of 0.30 was chosen for inclusion of SNP loci in the final data set and genotypic clusters were edited manually as needed. Of those 1031 SNPs, 5 were monomorphic and were therefore discarded. The remaining 1026 SNPs were used in subsequent analyses.
Genetic structure
The Pl values for the SNPs ranged from 0.786 to 0.951 with an average of 0.901. The Rs values also varied, from 1.569 to 1.794 with an average of 1.721, and the He values ranged from 0.292 to 0.320 with an average of 0.311 (Table 2). In contrast to findings from previous studies, these parameters do not reveal any clear geographical trends (Takahashi et al., 2005; Tsumura et al., 2007). The average FIS value for all loci was not significantly different from expectations under the Hardy–Weinberg equilibrium except for one locus, SNPg04215.
Table 2. Genetic diveristy of 14 investigated populations in C. japonica based on 1023 SNPs.
| Population a | Number of investigated individuals | Ho | Unbiased He | Pl | RS | FIS |
|---|---|---|---|---|---|---|
| AJG | 14 | 0.304 | 0.304 | 0.890 | 1.739 | 0.000 |
| NBT | 15 | 0.305 | 0.314 | 0.925 | 1.766 | 0.029 |
| ISN | 14 | 0.312 | 0.318 | 0.916 | 1.738 | 0.020 |
| DND | 16 | 0.309 | 0.312 | 0.930 | 1.776 | 0.008 |
| BJD | 18 | 0.314 | 0.314 | 0.951 | 1.794 | −0.001 |
| AST | 9 | 0.311 | 0.311 | 0.865 | 1.684 | −0.001 |
| KWZ | 16 | 0.311 | 0.314 | 0.928 | 1.731 | 0.012 |
| ASH | 13 | 0.311 | 0.318 | 0.942 | 1.768 | 0.025 |
| OKI | 17 | 0.301 | 0.308 | 0.919 | 1.764 | 0.022 |
| AZJ | 8 | 0.312 | 0.309 | 0.850 | 1.657 | −0.009 |
| SNG | 16 | 0.306 | 0.313 | 0.940 | 1.767 | 0.023 |
| YNS | 8 | 0.312 | 0.320 | 0.875 | 1.602 | 0.026 |
| ONN | 6 | 0.298 | 0.304 | 0.786 | 1.569 | 0.021 |
| YKU |
16 |
0.284 |
0.292 |
0.900 |
1.736 |
0.027 |
| Mean | 13.3 | 0.306 | 0.311 | 0.901 | 1.721 | 0.015 |
Abbreviation: SNPs, single-nucleotide polymorphisms.
See Table 1.
The overall genetic differentiation among populations at the 1026 loci was low (FST=0.0391). We also conducted an analysis of molecular variance to determine the variation within and among groups (the two varieties) and populations, and to test the significance of the among-population variation. The variation among populations was 7.72% and was highly significant (P<0.001), while the proportions of variance among varieties and among populations within varieties were 2.74% and 4.98%, respectively.
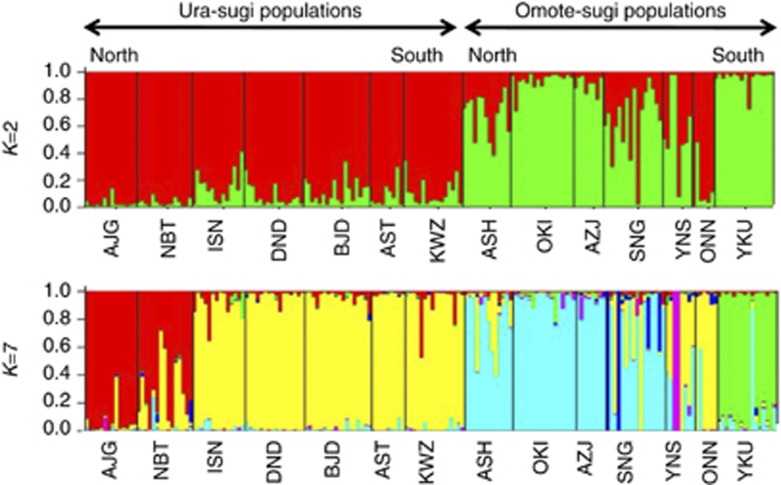
Bayesian clustering of the information from the 743 loci after excluding the outlier loci demonstrated that the models with K=2 and K=7 both provided satisfactory explanations of the observed data, as judged by the delta K and the highest log-likelihood values, respectively (Supplementary Figure 1). Both of the bar plots for K=2 and K=7 revealed clear differences between populations of the two varieties with the exception of the Oninome population (Figure 2). In addition, the bar plot for K=7 separated the populations for each variety. The ura-sugi variety populations were divided into two groups, with the first consisting of two northern populations and the second containing all the others. Conversely, the omote-sugi populations were divided into three major groups. The first comprised four northern populations, the southernmost population from Yakushima and two others. The two other omote-sugi population groups carried a higher proportion of ura-sugi-type alleles.
Figure 2.
Genetic relationships among the 14 populations surveyed using STRUCTURE (Pritchard et al., 2000) based on 1023 SNPs. The model with K=2 and K=7 showed the delta K value and the highest log-likelihood value, respectively.
Outlier loci
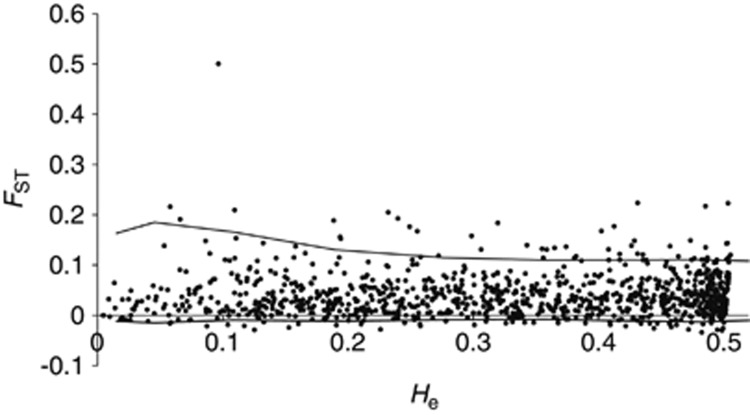
Using the FST-based method (Fdist2), we identified 63 loci lying outside the 99% confidence interval (upper), and an additional 40 loci that were also outside (below) the CI (Figure 3). The method of considering the hierarchical structure, such as ura-sugi and omote-sugi varieties, using Arlequin was identified for yet another 34 loci (Table 3). BLASTx searches identified putative functions for 41 of these loci (65% Table 3).
Figure 3.
Distribution of FST values as a function of the within-population heterozygosity (He) based on 1026 loci in C. japonica. The envelope of values (99% confidential interval) corresponds to neutral expectations in the infinite-allele model constructed according to the method of Beaumont and Nichols (1996).
Table 3. Outlier loci detected by Fdist2 (P<0.99), Arlequin (P<0.99) and BayeScan (Bayes factor >2) analyses, and their linkage group and putative function by BLASTx search (E-value cutoff ⩽1 × 10−3).
| Locus |
Fdist2
|
Arlequin FST |
BayeScan
|
Linkage group | Putative function | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
All populations
|
Omote | Ura | P value | All populations | Omote | Ura | |||||
| He | FST | P | P | ||||||||
| SNPg03850 | 0.4314 | 0.2237 | 0.0000 | * | * | 0.0010 | 0.0000 | * | — | No hit | |
| SNPg04215 | 0.0965 | 0.5006 | 0.0000 | * | 0.0000 | 0.0062 | 7 | Endo-1,4-beta-glucanase | |||
| SNPg01385 | 0.5036 | 0.2231 | 0.0000 | 0.0004 | 0.0000 | — | No hit | ||||
| SNPg04024 | 0.4856 | 0.2173 | 0.0000 | * | 0.0004 | 0.0000 | 11 | MYBPA1 protein | |||
| SNPg01330 | 0.2321 | 0.2050 | 0.0000 | * | 0.0002 | 0.0000 | 11 | Adenosine 3′-phospho 5′-phosphosulfate transporter | |||
| SNPg04607 | 0.2399 | 0.1927 | 0.0000 | * | 0.0017 | 0.0016 | — | No hit | |||
| SNPg04808 | 0.3198 | 0.1837 | 0.0001 | * | 0.0026 | 3 | Leucine-rich repeat receptor protein kinase EXS precursor | ||||
| SNPg02882 | 0.4127 | 0.1772 | 0.0001 | 0.0032 | 2 | Putrescine aminopropyltransferase-related protein | |||||
| SNPg05302 | 0.4025 | 0.1679 | 0.0001 | * | — | Dead box ATP-dependent RNA helicase | |||||
| SNPg02447 | 0.2492 | 0.1766 | 0.0002 | * | 0.0060 | — | Amidophosphoribosyltransferase | ||||
| SNPg00860 | 0.2554 | 0.1675 | 0.0004 | 0.0030 | 0.0022 | 6 | No hit | ||||
| SNPg00921 | 0.2987 | 0.1582 | 0.0005 | * | * | — | GDSL-motif lipase hydrolase family protein | ||||
| SNPg04111 | 0.4737 | 0.1490 | 0.0005 | * | 2 | No hit | |||||
| SNPg03578 | 0.4289 | 0.1486 | 0.0005 | * | — | 2-Hydroxyacid dehydrongenase | |||||
| SNPg04480 | 0.1938 | 0.1560 | 0.0010 | * | 0.0008 | 0.0022 | 11 | Glucan endo-1,3-beta-glucosidase precursor | |||
| SNPg01805 | 0.4264 | 0.1386 | 0.0012 | 0.0022 | 11 | Glucan endo-1,3-beta-glucosidase precursor | |||||
| SNPg03910 | 0.1887 | 0.1887 | 0.0012 | * | 0.0012 | 3 | Endo-1,4-beta-glucanase | ||||
| SNPg02822 | 0.1943 | 0.1525 | 0.0013 | 0.0011 | 0.0002 | 11 | Methyltransferase-like protein 13-like | ||||
| SNPg03082 | 0.4573 | 0.1376 | 0.0013 | 0.0008 | 0.0100 | 2 | No hit | ||||
| SNPg03744 | 0.5032 | 0.1437 | 0.0014 | 0.0074 | 0.0034 | — | No hit | ||||
| SNPg03195 | 0.5025 | 0.1429 | 0.0015 | 0.0019 | 0.0016 | — | Protein-binding protein | ||||
| SNPg01042 | 0.3742 | 0.1366 | 0.0019 | 0.0038 | 0.0028 | 10 | Harpin-induced protein-like | ||||
| SNPg03273 | 0.3429 | 0.1397 | 0.0019 | * | 11 | Putative TIR/NBS/LRR disease resistance protein | |||||
| SNPg01079 | 0.5026 | 0.1117 | 0.0022 | * | 0.0098 | * | — | No hit | |||
| SNPg04166 | 0.3650 | 0.1342 | 0.0023 | * | 0.0082 | — | No hit | ||||
| SNPg00581 | 0.4857 | 0.1353 | 0.0025 | 9 | No hit | ||||||
| SNPg04244 | 0.4947 | 0.1104 | 0.0025 | 2 | Major facilitator protein | ||||||
| SNPg02884 | 0.3551 | 0.1324 | 0.0026 | — | LPXTG-motif cell wall anchor domain protein | ||||||
| SNPg04589 | 0.4082 | 0.1313 | 0.0028 | 2 | Extracellular ligand-gated ion channel | ||||||
| SNPg00517 | 0.3589 | 0.1310 | 0.0029 | * | — | lon protease homolog peroxisomal-like | |||||
| SNPg04896 | 0.4868 | 0.1085 | 0.0029 | 2 | TIR-NBS class disease resistance protein | ||||||
| SNPg02875 | 0.3063 | 0.1311 | 0.0036 | 8 | No hit | ||||||
| SNPg00066 | 0.0580 | 0.2161 | 0.0036 | * | 0.0021 | 2 | VTC2-like protein | ||||
| SNPg03555 | 0.5042 | 0.1058 | 0.0037 | 10 | Strictosidine synthase family protein | ||||||
| SNPg05041 | 0.4941 | 0.1051 | 0.0039 | — | No hit | ||||||
| SNPg01589 | 0.4926 | 0.1047 | 0.0040 | — | S-receptor kinase | ||||||
| SNPg05257 | 0.2724 | 0.1186 | 0.0040 | * | 4 | 20-s Proteasome subunit paf1 | |||||
| SNPg01300 | 0.4314 | 0.1225 | 0.0041 | * | — | Light-regulated zinc finger protein 1 | |||||
| SNPg01839 | 0.1095 | 0.2095 | 0.0043 | * | * | 0.0009 | — | No hit | |||
| SNPg02637 | 0.4356 | 0.1210 | 0.0046 | 0.0014 | 0.0062 | — | p-Aminobenzoate synthase | ||||
| SNPg02373 | 0.4827 | 0.1109 | 0.0047 | 5 | Unknown | ||||||
| SNPg05351 | 0.4849 | 0.1108 | 0.0047 | 5 | No hit | ||||||
| SNPg00531 | 0.1106 | 0.1537 | 0.0048 | 0.0026 | 3 | Chaperone protein DnaJ chloroplast | |||||
| SNPg02059 | 0.2579 | 0.1161 | 0.0048 | * | — | Aldo keto reductase | |||||
| SNPg04667 | 0.4172 | 0.1104 | 0.0049 | 1 | No hit | ||||||
| SNPg05574 | 0.4359 | 0.1101 | 0.0050 | 0.0077 | 10 | No hit | |||||
| SNPg04724 | 0.4084 | 0.1100 | 0.0050 | * | 8 | Hexokinase 1 | |||||
| SNPg02675 | 0.4385 | 0.1100 | 0.0050 | 3 | Glycoside hydrolase, family 17 | ||||||
| SNPg01003 | 0.1929 | 0.1151 | 0.0052 | 7 | Late embryogenesis-abundant hydroxyproline-rich glycoprotein | ||||||
| SNPg03769 | 0.4170 | 0.1220 | 0.0055 | 0.0019 | 7 | Brassinosteroid insensitive 1-associated receptor kinase 1precursor | |||||
| SNPg02786 | 0.3866 | 0.1085 | 0.0056 | 5 | 2-Hydroxyacid dehydrongenase | ||||||
| SNPg01044 | 0.4640 | 0.1174 | 0.0060 | 7 | Lactoylglutathione lyase | ||||||
| SNPg03110 | 0.4545 | 0.1067 | 0.0064 | 10 | No hit | ||||||
| SNPg00114 | 0.5056 | 0.1203 | 0.0066 | 2 | No hit | ||||||
| SNPg02495 | 0.0659 | 0.1912 | 0.0070 | * | 0.0051 | 2 | Early-responsive to dehydration 7 | ||||
| SNPg00236 | 0.2515 | 0.1106 | 0.0070 | — | F-box and wd40 domain protein | ||||||
| SNPg02706 | 0.3823 | 0.1176 | 0.0076 | 0.0078 | 8 | ATP-dependent RNA helicase DHX8 | |||||
| SNPg03565 | 0.4769 | 0.1133 | 0.0081 | 0.0080 | — | Histone h4 | |||||
| SNPg01835 | 0.5041 | 0.1153 | 0.0091 | 10 | No hit | ||||||
| SNPg00660 | 0.4872 | 0.1147 | 0.0094 | 8 | Nitrite reductase | ||||||
| SNPg01833 | 0.3560 | 0.1142 | 0.0096 | * | 1 | No hit | |||||
| SNPg05180 | 0.3810 | 0.1141 | 0.0097 | 1 | F-box-like protein | ||||||
| SNPg03808 | 0.4746 | 0.1106 | 0.0098 | 6 | ARC6h-like protein | ||||||
| SNPg00006 | 0.0004 | — | Pinus taeda anonymous locus umn_2311_01 genomic sequence | ||||||||
| SNPg04004 | * | 0.0009 | — | No hit | |||||||
| SNPg01566 | * | 0.0010 | 3 | No hit | |||||||
| SNPg04934 | 0.0012 | 5 | Ankyrin partial | ||||||||
| SNPg01953 | 0.0014 | 6 | PREDICTED: uncharacterized protein LOC100253581 | ||||||||
| SNPg05269 | 0.0019 | 7 | No hit | ||||||||
| SNPg04802 | 0.0020 | 9 | Galactose oxidase-like | ||||||||
| SNPg01166 | 0.0022 | 4 | P. taeda anonymous locus 0_6264_01 genomic sequence | ||||||||
| SNPg03800 | 0.0027 | — | LRR-kinase protein | ||||||||
| SNPg03964 | 0.0030 | 11 | GCK domain-containing protein | ||||||||
| SNPg02633 | 0.0031 | 9 | Leucine-rich repeat transmembrane protein kinase | ||||||||
| SNPg04798 | * | 0.0032 | — | No hit | |||||||
| SNPg03049 | 0.0038 | — | Pentatricopeptide repeat-containing | ||||||||
| SNPg04439 | 0.0039 | 4 | No hit | ||||||||
| SNPg01041 | * | 0.0041 | 7 | Cell number regulator 1 | |||||||
| SNPg00017 | * | 0.0044 | — | No hit | |||||||
| SNPg02303 | 0.0045 | — | No hit | ||||||||
| SNPg00923 | * | 0.0050 | — | Acidic endochitinase-like | |||||||
| SNPg01152 | 0.0055 | 8 | Transmembrane receptors ATP-binding protein | ||||||||
| SNPg01574 | 0.0055 | 9 | UDP-glycosyltransferase | ||||||||
| SNPg03804 | 0.0057 | 7 | No hit | ||||||||
| SNPg00315 | 0.0060 | — | Predicted protein (Micromonas sp. RCC299) | ||||||||
| SNPg00024 | 0.0064 | — | Pyruvate kinase | ||||||||
| SNPg05588 | 0.0065 | 4 | Vesicle-associated protein 4-2-like | ||||||||
| SNPg05020 | 0.0067 | — | No hit | ||||||||
| SNPg02220 | * | 0.0072 | 6 | DNA-binding protein | |||||||
| SNPg00438 | 0.0073 | — | No hit | ||||||||
| SNPg02428 | 0.0080 | 3 | Maternal effect embryo arrest 12 protein | ||||||||
| SNPg00094 | 0.0082 | — | No hit | ||||||||
| SNPg01108 | 0.0082 | 3 | Unknown | ||||||||
| SNPg00054 | 0.0091 | — | PI-PLC × domain-containing protein at5g67130-like | ||||||||
| SNPg00531 | 0.0093 | — | Chaperone protein DnaJ | ||||||||
| SNPg02256 | 0.0096 | 11 | Microtubule-associated protein map65-1a | ||||||||
| SNPg03226 | 0.0096 | 11 | Vps51 Vps67 family (components of vesicular transport) protein | ||||||||
| SNPg00161 | * | — | Pseudo-response receiver | ||||||||
| SNPg01491 | * | 0.0084 | 1 | Glycoside hydrolase family 47 protein | |||||||
| SNPg02982 | * | — | Glycosylphosphatidylinositol anchor biosynthesis protein 11 | ||||||||
| SNPg04411 | * | 2 | Resistance gene region between conserved kinase-2 and p-loop domains | ||||||||
| SNPg05191 | 0.0052 | 2 | E3 ubiquitin-protein ligase ring1-like | ||||||||
| SNPg00033 | * | 3 | Yippee-like protein | ||||||||
| SNPg00140 | * | * | 3 | Nramp transporter | |||||||
| SNPg02378 | * | 3 | No hit | ||||||||
| SNPg00718 | * | 5 | No hit | ||||||||
| SNPg01667 | * | — | FAD/NAD(P)-binding oxidoreductase family protein | ||||||||
| SNPg02442 | * | 5 | Condensation domain-containing protein | ||||||||
| SNPg05366 | * | 5 | Phenylalanine ammonia lyase | ||||||||
| SNPg00892 | * | — | Nucleotide-sugar transporter family protein | ||||||||
| SNPg01169 | * | 7 | EREBP transcription factor | ||||||||
| SNPg02235 | * | 1 | No hit | ||||||||
| SNPg02348 | * | 7 | No hit | ||||||||
| SNPg02489 | * | 10 | Unknown | ||||||||
| SNPg04760 | * | 10 | No hit | ||||||||
| SNPg02148 | * | * | 11 | Transmembrane protein 45B-like | |||||||
| SNPg02671 | * | 11 | No hit | ||||||||
| SNPg03025 | * | 11 | Lysyl-tRNA synthetase | ||||||||
| SNPg01727 | * | — | FAD NAD-binding oxidoreductase family protein | ||||||||
| SNPg01899 | * | — | Unknown | ||||||||
| SNPg03715 | * | — | No hit | ||||||||
| SNPg04159 | * | — | opc-8:0 ligase1 | ||||||||
| SNPg04392 | * | — | Leucine-rich repeat-containing protein | ||||||||
| SNPg04510 | * | — | No hit | ||||||||
| SNPg04924 | * | — | No hit | ||||||||
| SNPg05020 | * | — | No hit | ||||||||
| SNPg05148 | * | — | Ilectin receptor-like kinase | ||||||||
Abbreviation: SNP, single-nucleotide polymorphism.
Statistically siginificant level. *<0.01.
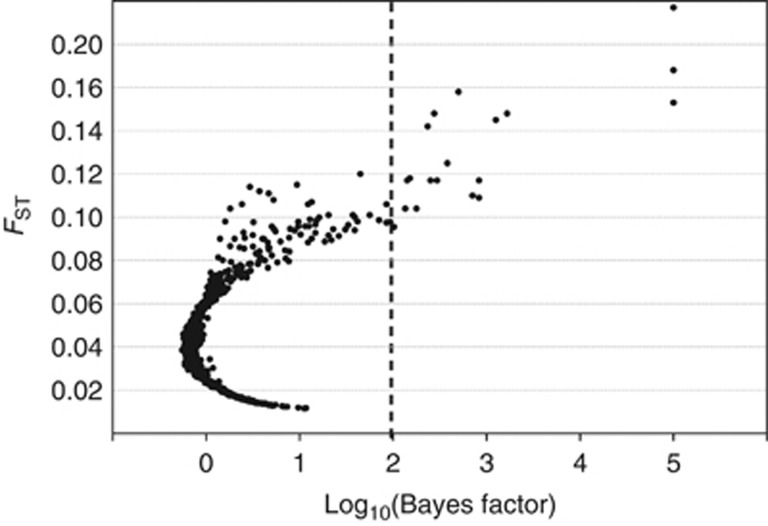
Using the Bayesian method implemented in BayeScan, we identified 52 loci as outliers with a log10 Bayes factor above 1, all of which were also detected by the FST-based method except for SNPg05191. Of these outlier loci, 20 had log10 Bayes factor values above 2 (Figure 4); 19 of these 20 were also identified as outliers by the FST-based method (Table 3). Five of the 20 loci had log10 Bayes factors above 3, corresponding to a posterior probability of locus effects above 0.999; four had a log10 Bayes factor of 5, corresponding to a posterior probability of 1.000. BLASTx searches identified putative functions for 11 of the 20 loci with log10 Bayes factors above 2 (55% Table 3).
Figure 4.
Plot of FST values and Bayes factors (log10) for 1026 loci obtained using the BayeScan outlier test. Dashed lines indicate the Bayes factor threshold of 2 (log10) that provides ‘decisive' evidence for selection corresponding to a posterior probability of 0.99.
Using within-variety FST analysis, 54 outlier loci were identified. All but three of the identified outlier loci were detected only in either omote-sugi or ura-sugi variety populations; the three exceptions were SNPg03850, SNPg01839 and SNPg00140 (Table 3). The 25 outlier loci were also detected when all populations were included in the analysis. Using the Bayesian method, only four loci were identified in either the omote-sugi or ura-sugi populations, two of which were present in all populations.
Relationships between specific loci and environmental variables
Eighteen loci were associated with environmental variables (Table 4). The environmental variable with the greatest number of associations was the daily maximum temperature, which was related to six loci; the maximum snow depth and latitude were associated with five and three, respectively. All environmental variables were associated with at least one locus. The 12 out of 18 loci found to be related to an environmental variable were also identified as outliers in either the Fdist2, Arlequin or BayeScan analyses. BLASTx searches identified putative functions for 10 of the 18 environment-associated loci.
Table 4. Significant associations between loci and environmnental variables (P<0.01), and their linkage group and the putative function by BLAST search.
|
Environmental variable
|
Fdist2 a | BayeScan * | Arlequin * | Linkage group | Putative function | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Longitude | Latitude | Altitude | Mean annual temperature | Daily maximum temperature | Daily minimum temperature | Annual precipitation | Annual sunshine hours | Amount of global solar radiation | Maximum snow depth | ||||||
| SNPg00563 | ** | * | * | 3 | No hit | ||||||||||
| SNPg00660 | ** | ** | ** | 8 | Nitrite reductase | ||||||||||
| SNPg01042 | ** | ** | ** | ** | 10 | Harpin-induced protein-like | |||||||||
| SNPg01330 | ** | ** | *** | ** | ** | *** | 11 | Adenosine 3-phospho 5-phosphosulfate | |||||||
| SNPg01385 | ** | ** | *** | ** | ** | *** | *** | — | No hit | ||||||
| SNPg01589 | ** | ** | ** | *** | * | 6 | S-receptor kinase | ||||||||
| SNPg01835 | ** | ** | * | * | 10 | No hit | |||||||||
| SNPg02192 | ** | — | Calcium-dependent protein | ||||||||||||
| SNPg02235 | ** | ** | 7 | No hit | |||||||||||
| SNPg02446 | ** | 5 | No hit | ||||||||||||
| SNPg02685 | ** | 11 | Hydroxyproline-rich glycoprotein family protein-like | ||||||||||||
| SNPg02929 | ** | * | — | No hit | |||||||||||
| SNPg03082 | ** | ** | ** | ** | *** | 2 | No hit | ||||||||
| SNPg03259 | ** | 4 | Subtilisin-like protease | ||||||||||||
| SNPg04024 | ** | ** | ** | *** | *** | *** | 11 | MYBPA1 protein | |||||||
| SNPg04308 | ** | — | Uncharacterized protein | ||||||||||||
| SNPg04607 | ** | *** | ** | ** | — | No hit | |||||||||
| SNPg05593 | ** | ** | — | O-acyltransferase wsd1-like | |||||||||||
Abbreviation: SNP, single-nucleotide polymorphism.
Statistically siginificant level. *<0.05, **<0.01 and ***<0.001.
Distribution of outlying loci on the linkage map, and LD
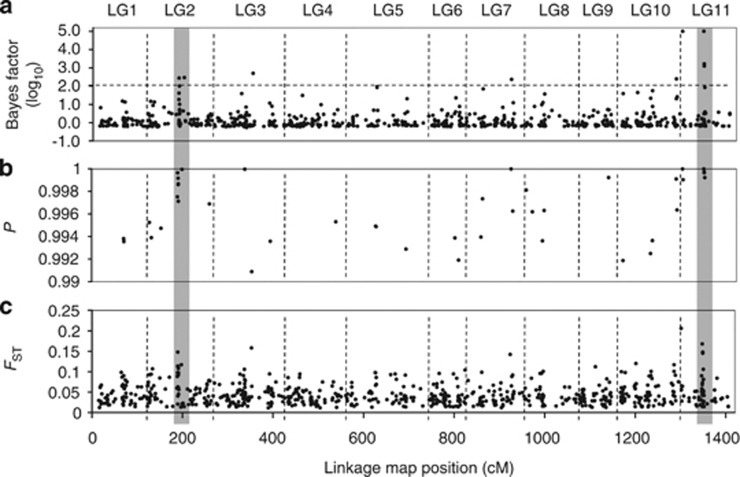
In total, 100 loci were identified as outlier loci or loci associated with environmental variables for all populations. Sixty-four of these loci were successfully mapped on the C. japonica linkage map, and their distribution was studied (Figure 5, Moriguchi et al., 2012). The numbers of mapped outlier loci in each LG varied substantially. For example, LG 4 contained only three loci, while LG2 contained 11. The observed distribution of loci deviated significantly from the Poisson distribution for all marker intervals considered (15, 20, 25 and 50 cM) and was therefore not random (P<0.001). Eight of the 14 loci that were detected by all of the Fdist2, Arlequin and BayeScan methods were mapped on the linkage map; 4 on LG11, and one on each of LG2, LG6, LG7 and LG10. The outlier loci thus formed clumps on specific LGs, particularly LG2 and LG11 (Figure 5). The outlier distribution was related to the FST distribution; regions with high FST values had high numbers of outlier loci.
Figure 5.
The distribution of markers identified as being potentially involved in local adaptation across the genome of C. japonica. The figure shows plots of marker position (based on 1255 mapped SNPs or the other DNA markers; Moriguchi et al., 2012) in cumulative centiMorgans (cM) versus (a) Bayes factor (log10), as determined by BayeScan analysis, (b) the probability that the locus is an outlier as judged by the FST-based test (Beaumont and Nichols, 1996) and (c) F-statistic value, FST. Vertical dashed lines demarcate the 11 LGs. The two gray zones indicate candidate regions associated with local adaptation.
LD was detected for only five of the 2145 possible combinations of the 66 loci. Three pairs of loci were closely linked to one another on LG2, LG5 and LG11, respectively, two of which corresponded to the clumped regions of outlier loci discussed above. Locus SNPg04215 was mapped to LG7 but exhibited high LD (more than r2=0.270) with SNPg00066 and SNPg02495 on LG2.
Discussion
Genetic structure
The genetic differentiation between the studied C. japonica populations was low (FST=0.0391). Hierarchical analysis indicated that genetic variation among varieties and populations accounted for 2.74% and 4.98% of the total variation, respectively. These results are similar to those from our previous study (Tsumura et al., 2007). While there was little genetic differentiation between variety groups, what differentiation there was pertained to genes related for local adaptation. A K value of 7 yielded the highest log-likelihood value in the structure analysis and provided a much clearer picture of the species' genetic structure than had been obtained in our previous study (Tsumura et al., 2007). Although the extent of the genetic differentiation between varieties was relatively modest, they were clearly distinguished from one another in the structure analysis excluding ONN population in omote-sugi populations. This population might be established immigration from Chugoku populations such as Azouji and others after extinction of natural forest in Kyushu Island (Takahara, 1998). We therefore examined outlier loci both for all populations and within populations of individual varieties after excluding ONN population from omote-sugi variety populations. However, the obtained result is not largely different in case of inclusion of ONN population.
Detection of outlier loci
The number of outlier loci identified with Fdist2 and Arlequin was roughly three times greater than the number identified with BayeScan for all populations, even when using the same significance threshold (0.99) in both cases and considering the hierarchical structure. Fdist2 may be more prone to detecting false positives than other methods (Pérez-Figueroa et al., 2010), but all of the loci identified by BayeScan were also detected by Fdist2 with the exception of six loci (Table 3). This illustrates the necessity of performing multiple tests to detect outlier loci. It may be that BayeScan is more efficient than Fdist2 at identifying true positives. Pérez-Figueroa et al. (2010) conducted a simulation to compare the efficiency of three different programs (DFDIST, DETSELD and BayeScan) for detecting loci under directional selection using dominant markers. They concluded that BayeScan is more efficient than the alternatives because it automatically optimizes the values of its parameters (Pérez-Figueroa et al., 2010). However, some false positives are inevitable even when multitest corrections and multiple methods are used.
Tests for detecting outlier loci that deviate from neutral expectations are unable to identify false positives (type I errors). We therefore conducted all Fdist2, Alrequin and BayeScan tests, imposing a 99% P lower limit for the identification of genes under selective pressure; the expected number of false positives was thus 1026 × 0.01=10.26. As 14 outlier loci were identified by both tests, it appears that at least some are unlikely to be false positives (owing to type I errors). The 14 loci identified by all Fdist2, Alrequin and BayeScan are thus strong candidates as genes involved in adaptation to local environments.
1.4% of the 1026 loci investigated were identified as candidate loci. This proportion of candidate loci is similar to that found in a previous study (2.7%) in which 148 cleaved amplified polymorphic sequence loci were examined (Tsumura et al., 2007). Eckert et al. (2010) identified 22 candidate loci (BF log10>2.0) associated with climate variables from 1730 loci in 682 loblolly pine tree samples covering the full range of the species. Prunier et al. (2011) also identified 10 candidate loci above 99% confidence interval in black spruce. The proportion of detected loci (1.3% and 1.7%, respectively) associated to the climate variables was very similar with our results. The average FST value of the 14 candidate loci was 0.1943 (the values for individual candidate loci ranged from 0.1210 to 0.5006), which is substantially higher than the average FST value for all loci (0.0391). This means that different populations have substantially different allelic compositions at these 14 loci, presumably because of the different selective pressures acting upon them.
When we examined the status of outlier loci within groups representing the two varieties, two-thirds of the loci detected in all populations were not identified as outliers in either variety group by Fdist2. With BayeScan, only three of the loci detected in all populations were identified as outliers in one of the variety groups or the other. These genes might be important in the two varieties' adaptation to their different environments. In Japan, the coast of the Japan Sea experiences heavy snowfall but the Pacific Ocean side is quite dry in the winter. The ura-sugi variety is found on the Japan Sea side and has short needles with narrow angles, which are thought to confer selective advantages in areas with heavy snowfall in the winter. The outlier genes were detected in all populations and also in each variety group, and are therefore likely to be important in local adaptation.
Relationships with environmental variables
Correlation analyses were performed for several environmental variables, bearing in mind the possibility that multiple loci might be associated with any one of these variables. Some of the environmental variables were correlated with one another—specifically, the latitude and the longitude (r=0.81), and the latitude and the annual precipitation (r=−0.86). This reflects the fact that the Japanese archipelago extends in a rough line from the southwest to the northeast and the environmental gradient examined in this study mirrors that of the archipelago; the natural C. japonica forest has a wide-ranging distribution, from 30°N, 130°E to 40°N, 140°E (Table 1, Figure 1). There is heavy snowfall on the Japan Sea side in the winter season but the Pacific Ocean side is quite dry in the winter, and the annual precipitation is much higher in the southern part of Japan than elsewhere in the country. There are thus significant differences in the environmental factors affecting the different regions examined in this work, especially in terms of the annual precipitation and the maximum depth of fallen snow (Table 1). There is a cline of environmental variables such as precipitation, temperature and snowfall along the Japanese archipelago from the northeast to southwest. The formation of the Japanese archipelago began around 17 million years ago, with the earliest incarnation of the Japan Sea forming around 7 million years ago (Taira, 2001). Ever since then, there have been significant climatic differences between the Japan Sea side and Pacific Ocean sides of Japan. A C. anglica fossil dated to the later phase of the Miocene has been found in Japan, and is a likely ancestor of C. japonica. Moreover, C. japonica fossils have been found that date back to the Pleiocene, and so post-date the formation of the Japanese archipelago (Uemura, 1981). C. japonica has thus adapted climatic differences over extended periods of time while expanding its distribution, in the face of global climate changes including several glacial and interglacial periods.
Eight of the 14 candidate genes identified by all Fdist2, Arlequin and BayeScan were associated with at least one of the latitude, the mean annual temperature, the daily minimum or maximum temperature, the maximum snow depth and the annual sunshine hours (Table 3). Three of them were associated with two or more environmental variables that correlated with one another, suggesting that the genes might have responded to the same selection pressures.
Putative functions for four of these eight candidate genes were revealed by BLASTx searches. For example, SNPg04024 is a putative MYBPA1 protein and would thus activate the promoters of several of the general flavonoid pathway genes (Bogs et al., 2007). This kind of function is important for defense against diseases and predators, and for the survival of the individual and/or population. Individuals with genotypes that confer advantages in their specific environments would be selected for, changing the allele frequency at the candidate loci in that population. However, it is important to properly account for hitchhiking effects when making suggestions of this kind, because it is possible that the genes identified may simply be closely linked to the true adaptive gene.
In addition to the location and climate data, other environmental variables such as soil type and the species composition of the forest will also have important effects on selection. Phenotypic differences of the two varieties such as needle, branchlets and wood characteristics that are the result of selection will also be important for association study.
Such information should be collected and used in future association analyses.
The clumpy distribution of the outlier loci, and LD
Significant deviations from the Poisson distribution of outlier loci were observed for all marker intervals (P<0.001), indicating that the 64 mapped outlier loci and loci associated with environmental variables were not randomly distributed on the linkage map; instead, they tended to cluster, especially on LG2 and LG11 (Figure 5), in a manner reminiscent of the ‘genomic islands of divergence' discussed by Nosil et al. (2009). These two genomic regions, which are likely to have a major role in local adaptation, cover around 1.7 cM on LG2 and about 2.1 cM on LG11, corresponding to about 10.2 and 12.6 Mb, respectively, in this species. Three of the outlier loci in the LG11 region (SNPg01330, SNPg02685 and SNPg04024) were associated with some environment variables. Two of the loci in the LG11 region had Bayes factors above 3 (log10), which is generally interpreted as a ‘decisive' level of significance. The SNPg04024 locus has a Bayes factor of 5, giving a posterior probability of 1. We are thus convinced that the LG11 region is affected by directional selection along the environmental gradient, presumably by factors such as mean annual temperature, daily maximum temperature, hours of sunshine and/or maximum snow depth. Scotti-Saintagne et al. (2004) identified some important regions of their genomes associated with adaptation using both methods of outlier detection and quantitative trait locus mapping.
Surprisingly, the LD among the loci in each of the region is also high, affecting one pair in each of LG2 and LG11. Although LD in forest trees decays rapidly within several thousand basepairs (Neale and Savolainen, 2004), Eckert et al. (2010) have observed high LD in the loblolly pine, affecting the set of five SNPs located on LG 8. The LD of forest trees has to date only been estimated in coding regions; it is possible that the LD values of non-coding regions might be higher than those for coding regions, as is the case in angiosperms (Gaut et al., 2007; Moritsuka et al., 2012). The SNPg04215 locus on LG7 exhibited substantial LD with two loci on LG2, which is unusual; the high LD might reflect the effects of directional selection, resulting in the same selection pressure. Evolutionary mechanisms such as co-adaptation of gene complexes might be affected by this LD (Dobzhansky, 1970).
The outlier loci detected in the other regions with high significance levels are also strong candidates for involvement in local adaptation. It will be necessary to confirm whether these loci correspond to genuine adaptive genes, to compare the sequences of the full-length regions containing these genes in populations from different environments, and to determine the LD associated with these loci. The identification of outlier loci with high significance levels is essential for conservation efforts and will be required for future work on molecular breeding.
Data Archiving
Data have been deposited at Dryad: doi:10.5061/dryad.gt178.
Acknowledgments
We thank H Tachida for insightful comments and discussions concerning earlier versions of this manuscript. We also thank Y Taguchi for excellent technical assistance. This study was supported by the Program for the Promotion of Basic and Applied Research for Innovations in Bio-oriented Industry.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond Ser B. 1996;263:1619–1626. [Google Scholar]
- Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007;143:1347–1361. doi: 10.1104/pp.106.093203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis infunctional genomics research. Bioinformatics. 2005;21:3674. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of the Evolutionary Process. Columbia University Press: New York; 1970. [Google Scholar]
- Eckert AJ, Bower AD, González-Martínez SC, Wegrzyn JL, Coop G, Neale DB. Back to nature: ecological genomics of loblolly pine (Pinus taeda, Pinaceae) Mol Ecol. 2010;19:3789–3805. doi: 10.1111/j.1365-294X.2010.04698.x. [DOI] [PubMed] [Google Scholar]
- El Mousadik A, Petit RJ. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor Appl Genet. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Eveno E, Collada C, Guevara MA, Léger V, Soto A, Díaz L, et al. Contrasting patterns of selection at Pinus pinaster Ait. drought stress candidate genes as revealed by genetic differentiation analyses. Mol Biol Evol. 2008;25:417–437. doi: 10.1093/molbev/msm272. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–995. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Wright SI, Rizzon C, Dvorak J, Anderson LK. Recombination: an underappreciated factor in the evolution of plant genomes. Nat Rev Genet. 2007;8:77–84. doi: 10.1038/nrg1970. [DOI] [PubMed] [Google Scholar]
- González-Martínez SC, Krutovsky KV, Neale DB. Forest tree population genomics and adaptive evolution. New Phytol. 2006;170:227–238. doi: 10.1111/j.1469-8137.2006.01686.x. [DOI] [PubMed] [Google Scholar]
- Goudet J.2000FSTAT: a program to estimate and test gene diversities and fixation indicesVer. 2.9.1. Available at http://www2.unil.ch/popgen/softwares/fstat.htm .
- Grattapaglia D, Plomion C, Kirst M, Sederoff RR. Genomics of growth traits in forest trees. Curr Opin Plant Biol. 2009;12:148–156. doi: 10.1016/j.pbi.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Hayashi Y.1960Taxonomical and Phytogeographical Study of Japanese Conifers Norin-Shuppan: Tokyo; in Japanese. [Google Scholar]
- Hoffmann AA, Willi Y. Detecting genetic responses to environmental change. Nat Rev Genet. 2008;9:421–432. doi: 10.1038/nrg2339. [DOI] [PubMed] [Google Scholar]
- Holliday JA, Ritland K, Aitken SN. Widespread, ecologically relevant genetic markers developed from association mapping of climate-related traits in Sitka spruce (Picea sitchensis) New Phytol. 2010;188:501–514. doi: 10.1111/j.1469-8137.2010.03380.x. [DOI] [PubMed] [Google Scholar]
- Howe GT, Aitken SN, Neale DB, Jermstad KD, Wheeler NC, Chen THH. From genotype to phenotype: unraveling the complexities of cold adaptation in forest trees. Can J Bot. 2003;81:1247–1266. [Google Scholar]
- Japan Meteorological Agency . Mesh Climatic Data of Japan. Japan Meteorological Agency: Japan Meteorological Business Support Center, Tokyo; 2002. [Google Scholar]
- Kang BY, Mann IK, Major JE, Rajora OP. Near-saturated and complete genetic linkage map of black spruce (Picea mariana) BMC Genomics. 2010;11:515. doi: 10.1186/1471-2164-11-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer M, Dieringer D, Schlötterer C. A microsatellite variability screen for positive selection associated with the ‘out of Africa' habitat expansion of Drosophila melanogaster. Genetics. 2003;165:1137–1148. doi: 10.1093/genetics/165.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Lewis PO, Zaykin D.2002GDAAvailable via http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php .
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- Moriguchi Y, Ujino-Ihara T, Uchiyama K, Futamura N, Saito M, Ueno S, et al. The construction of a high-density linkage map for identifying SNP markers that are tightly linked to a nuclear-recessive major gene for male sterility in Cryptomeria japonica D. Don. BMC Genomics. 2012;13:95. doi: 10.1186/1471-2164-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritsuka E, Hisataka Y, Tamura M, Uchiyama K, Watanabe A, Tsumura Y, et al. Extended linkage disequilibrium in non-coding regions in a conifer, Cryptomeria japonica. Genetics. 2012;190:1145–1148. doi: 10.1534/genetics.111.136697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai S.1947Major forestry tree species in the Tohoku region and their varietal problemsIn: Kokudo Saiken Zourin Gijutsu Kouenshu, Aomori-rinyukai (eds). pp131–151.in Japanese.
- Neale DB. Genomics to tree breeding and forest health. Curr Opin Genet Dev. 2007;17:1–6. doi: 10.1016/j.gde.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Neale DB, Ingvarsson PK. Population, quantitative and comparative genomics of adaptation in forest trees. Curr Opin Plant Biol. 2008;11:1–7. doi: 10.1016/j.pbi.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Neale DB, Kremer A. Forest tree genomics: growing resources and applications. Nat Rev Genet. 2011;12:111–122. doi: 10.1038/nrg2931. [DOI] [PubMed] [Google Scholar]
- Neale DB, Savolainen O. Association genetics of complex traits in conifers. Trends Plant Sci. 2004;9:325–330. doi: 10.1016/j.tplants.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Nei M. F-statistics and analysis of gene diversity in subdivided populations. Ann Hum Genet. 1977;41:225–233. doi: 10.1111/j.1469-1809.1977.tb01918.x. [DOI] [PubMed] [Google Scholar]
- Nei M, Chesser RK. Estimation of fixation indices and gene diversities. Ann Hum Genet. 1983;47:253–259. doi: 10.1111/j.1469-1809.1983.tb00993.x. [DOI] [PubMed] [Google Scholar]
- Namroud MC, Beaulieu J, Juge N, Laroche J, Bousquet J. Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce. Mol Ecol. 2008;17:3599–3616. doi: 10.1111/j.1365-294X.2008.03840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Ohba K.1993Clonal forestry with sugi (Cryptomeria japonica)In: Ahuja MR, Libby WJ, (eds).Clonal Forestry II. Conservation and Application Springer-Verlag: Berlin; 66–90. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelgas B, Bousquet J, Meirmans PG, Ritland K, Isabel N. QTL mapping in white spruce: gene maps and genomic regions underlying adaptive traits across pedigrees, years and environments. BMC Genomics. 2011;12:145. doi: 10.1186/1471-2164-12-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Figueroa A, García-Pereira MJ, Saura M, Rolán-Alvarez E, Caballero A. Comparing three different methods to detect selective loci using dominant markers. J Evol Biol. 2010;23:2267–2276. doi: 10.1111/j.1420-9101.2010.02093.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier J, Laroche J, Beaulieu J, Bousquet J. Scanning the genome for gene SNPs related to climate adaptation and estimating selection at the molecular level in boreal black spruce. Mol Ecol. 2011;20:1702–1716. doi: 10.1111/j.1365-294X.2011.05045.x. [DOI] [PubMed] [Google Scholar]
- Savolainen O, Pyhäjärvi T. Genomic diversity in forest trees. Curr Opinion Plant Biol. 2007;10:162–167. doi: 10.1016/j.pbi.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Savolainen O, Pyhäjärvi T, Knürr T. Gene flow and local adaptation in forest trees. Ann Rev Ecol Evol Syst. 2007;38:595–619. [Google Scholar]
- Scotti-Saintagne C, Mariette S, Porth I, Goicoechea PG, Barreneche T, Bodénès C, et al. Genome scanning for interspecific differentiation between two closely related oak species Quercus robur L. & Q. petraea (Matt.) Liebl. Genetics. 2004;168:1615–1626. doi: 10.1534/genetics.104.026849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J, Reger J, Feulner PGD, Smadja C, Galindo J, Ekblom R, et al. Adaptation genomics: the next generation. Trends Ecol Evol. 2010;25:705–712. doi: 10.1016/j.tree.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Taira A. Tectonic evolution of the Japanese island arc system. Ann Rev Earth Planet Sci. 2001;29:109–134. [Google Scholar]
- Takahashi T, Tani N, Taira H, Tsumura Y. Microsatellite markers reveal high allelic variation in natural populations of Cryptomeria japonica near refugial areas of the last glacial period. J Plant Res. 2005;118:83–90. doi: 10.1007/s10265-005-0198-2. [DOI] [PubMed] [Google Scholar]
- Takahara H.1998Distribution history of Cryptomeria forest. Pp 207–223In Yasuda Y, Miyoushi N, (eds)Vegetation history of the Japanese Archipelago Asakura-Shoten: Tokyo; 207–223.in Japanese. [Google Scholar]
- Tani N, Takahashi T, Iwata H, Yuzuru M, Ujino-Ihara T, Matsumoto A, et al. A consensus linkage map for sugi (Cryptomeria japonica) from two pedigrees, based on microsatellites and expressed sequence taqs. Genetics. 2003;165:1551–1568. doi: 10.1093/genetics/165.3.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M. Altitudinal and latitudinal migration of Cryptomeria japonica for the past 20,000 years in Japan. Quat Res. 1986;26:135–152. [Google Scholar]
- Tsumura Y, Kado T, Takahashi T, Tani N, Ujino-Ihara T, Iwata H. Genome-scan to detect genetic structure and adaptive genes of natural populations of Cryptomeria japonica. Genetics. 2007;176:2393–2403. doi: 10.1534/genetics.107.072652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Ujino-Ihara T, Ueno S, Taguchi Y, Futamura N, Shinohara K, et al. 2012Single nucleotide polymorphisms in Cryptomeria japonica: their discovery and validation for genome mapping and diversity studies Tree Genet Genomee-pub ahead of print 4 May 2012; doi: 10.1007/s11295-012-0508-5 [DOI]
- Uemura K.1981Ancestor and change of distribution in Cryptomeria japonica Iden 3574–79.in Japanese. [Google Scholar]
- Van Ooijen JW, Voorrips RE.2001. JoinMap: version 3.0, software for the calculation of genetic linkage mapsPlant Research International: Wageningen, The Netherlands [Google Scholar]
- Vasemägi A, Primmer CR. Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Mol Ecol. 2005;14:3623–3642. doi: 10.1111/j.1365-294X.2005.02690.x. [DOI] [PubMed] [Google Scholar]
- Vitalis R, Dawson K, Boursot P. Interpretation of variation across marker loci as evidence of selection. Genetics. 2001;158:1811–1823. doi: 10.1093/genetics/158.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis R, Dawson K, Boursot P, Belkhir K. DetSel 1.0: a computer program to detect markers responding to selection. J Hered. 2003;94:429–431. doi: 10.1093/jhered/esg083. [DOI] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic Data Analysis II. Sinauer Associates: Sunderland, MA; 1996. [Google Scholar]
- Wright S. Coefficients of inbreeding and relationship. Am Nat. 1922;56:330–338. [Google Scholar]
- Wright S. Evolution and the Genetics of Populations. Variability within and among natural populations. vol. 4. The University of Chicago Press: Chicago; 1978. [Google Scholar]
- Yamazaki T.1995CryptomeriaceaeIn: Iwatsuki K, Yamazaki T, Boufford DE, Ohba H, (eds).Flora of Japan. Vol I. Pteridophyta and Gymnospermae Kodansha: Tokyo; p.264 [Google Scholar]
- Yasue M, Ogiyama K, Suto S, Tsukahara H, Miyahara F, Ohba K. Geographical differentiation of natural Cryptomeria stands analyzed by diterpene hydrocarbon constituents of individual trees. J Jpn For Soc. 1987;69:152–156. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.