Summary
A landmark in cell biology, the discovery of the JAK-STAT pathway provided a simple mechanism for gene regulation that dramatically advanced our understanding of the action of hormones, interferons, colony stimulating factors, and interleukins. As we learn more about the complexities of immune responses, new insights into the functions of this pathway continue to be revealed, aided by technology that permits genomewide views. As we celebrate the 20th anniversary of the discovery of this paradigm in cell signaling, it is particularly edifying to see how this knowledge has rapidly been translated to human immune disease. Not only have genomewide association studies demonstrated that this pathway is highly relevant to human autoimmunity but targeting JAKs is now a reality in immune-mediated disease.
The importance of interferons (IFNs) and hormones such as erythropoietin, growth hormone and prolactin has been recognized for more than half a century. With the advent of molecular biology era came the discovery of a plethora of other cytokines, which we now know regulate all aspects of cell development and differentiation. Cytokines, though, represent a collection of structurally distinct ligands that bind to different classes of receptors. A major subgroup of cytokines, comprising roughly 60 factors, bind to receptors termed Type I/II cytokine receptors. Cytokines that bind these receptors include Type I IFNs, IFN-γ, many interleukins and colony stimulating factors. From an immunology perspective, these cytokines are important for initiating innate immunity, orchestrating adaptive immune mechanisms and constraining immune and inflammatory responses.
As discussed by Darnell and Stark in this issue, the discovery of JAKs and Stats stemmed from attempts to understand how IFNs exerted their effect. However, we now know that all Type I/II cytokine receptors selectively associate with JAKs (JAK1, JAK2, JAK3 or TYK2). For these receptors, activation of the receptor-bound JAKs is critical for initiating phosphorylation of the cytokine receptor and subsequent recruitment of one or more STATs. Over the past two decades, multiple lines of evidence have clearly established the roles of different JAKs and STATs in mediating the effect of cytokines that use Type I/II cytokine receptors in immunoregulation, host-defense and immunopathology (Darnell et al., 1994; Leonard and O’Shea, 1998; O’Shea and Murray, 2008).
As our understanding of these processes have become more sophisticated, additional roles for this pathway have been recognized. For instance, with the identification of “newer” helper subsets comes the appreciation of important roles of STATs in these subsets as well as new roles for STATs in recognized subsets. As our understanding of the mechanisms involved in innate immunity expands, new roles of STATs in these processes become evident. In addition, new technologies also allow comprehensive views of STAT action whereas insights from genomewide association studies clearly implicate JAKs and STATs in human autoimmunity. Finally, the possibility of targeting the JAK-STAT pathway in autoimmune disease has now become a reality. In this review, we will try to briefly discuss these exciting advances. We recognize that this is a challenging task given the immense amount of exciting work in this field. In the interest of brevity, we have been forced to limit our discussion and we apologize in advance in advance for any omissions.
New insights into the immunoregulatory roles of JAKs and STATs
When the STATs were first discovered, the palette of helper T cells was simple - Th1 and Th2 cells. TYK2, JAK2 and STAT4 were found to be critical for IL-12 signals and Th1 differentiation whereas JAK1, JAK3 and STAT6 were key for IL-4 signaling (Darnell et al., 1994; Leonard and O’Shea, 1998; O’Shea and Murray, 2008). In various models of infectious disease and immune-mediated disease, deficiency of STAT4 and STAT6 had the expected outcomes [Goenka, 2011 #3629; Wurster, 2000 #3633; Murphy, 2000 #3636; [Oestreich, 2012 #3692; Paternoster, 2011 #3436].
New roles for STATs in “old” helper T cell subsets
It is now appreciated, however, that Th2 responses can occur in the absence of STAT6(van Panhuys et al., 2008). In fact, early Th2 differentiation can by driven by IL-2, which upregulates GATA3 and enhances IL-4 receptor expression(Paul, 2010). Activated by IL-2, STAT5A/B can directly bind the Il4r gene and promote its expression (Liao et al., 2008); however, STAT5A/B can also enhance Th1 responses by regulating Tbx21 and Il12rb2 (Liao et al., 2011b). Interestingly, STAT3 is also a contributor to Th2 differentiation and binds Th2-associated gene loci (Liao et al., 2008; Stritesky et al., 2011). Thus, in contrast to the previous views equating STAT6 with Th2 differentiation, it appears that this process involves more subtle and complex interactions of STAT3, STAT5 and STAT6 with the relevant genetic loci.
Role of STATs in Treg cell function
Along with TGFβ, IL-2 is a key regulator of differentiation of Treg cells in the thymus and the periphery. As mediators of IL-2 signaling, STAT5A/B are critical for the differentiation of Treg cells. Their effect is very direct in that STAT5/A directly bind the Foxp3 gene and drive expression of this key gene (Burchill et al., 2007; Yao et al., 2006; Yao et al., 2007; Zorn et al., 2006). In addition, STAT5A/B regulate IL2ra, expression of which is also a critical for Treg cells. Surprisingly, STAT3 also has an important role in Treg cell function (Chaudhry et al., 2009). Deletion of STAT3 in Treg cells results in lethal gastrointestinal disease, but the effect is selective and does not globally impair Treg cell function. Treg cells retain the ability to limit T cell proliferation but have impaired ability to block Th17-mediated pathology. Of interest, STAT3 physically associates with Foxp3.
Roles of STATs in “new” helper cell subsets
With the recognition of a multiplicity of fates for T cells, it has become clear that STATs are also key elements for these “new” subsets. We now know that STAT3 is critical for Th17 differentiation both in mouse and humans, mediating signals by IL-23 and IL-6(Chen et al., 2006; Mathur et al., 2007; Milner et al., 2008; Yang et al., 2007). STAT3 regulates Th17 differentiation by directly binding Il17a/f, Rorc and Il23r, as well as other genes involved in Th17 differentiation (Durant et al., 2010).
Interestingly, IL-2, acting via STAT5A/B, is an important negative regulator of Th17 differentiation (Laurence et al., 2007). In this case, the action of STAT5A/B action is very direct – they compete with STAT3 binding to the Il17a/f locus (Yang et al., 2011). Intriguingly, by sequestering IL-2, regulatory T cells promote Th17 differentiation (Chen et al., 2011b; Pandiyan et al., 2011).
One of the newest “lineages” of CD4 T cells is the follicular helper T cell, which provides help to B cells in germinal centers. Cytokines like IL-6 and IL-21 act on STAT3 and promote expression of Bcl6 and other molecules that contribute to the phenotype and function of this subset (Batten et al., 2010; Eddahri et al., 2009; Nurieva et al., 2008). However, IL-12 and STAT4 also turn out to be drivers of Tfh cells (Nakayamada et al., 2011; Schmitt et al., 2009). STAT4 directly binds many genes involved in Tfh differentiation, including Bcl6 and Il21. Conversely, IL-2 inhibits Tfh differentiation and once again, the action of STAT5 appears to be very direct. It competes with STAT3 binding to the Bcl6 locus and also promotes expression of Prdm1, which encodes Blimp1 (Johnston et al., 2012; Nurieva et al., 2012; Weinmann, 2012).
Perhaps less surprising given its role in transmitting IL-4 signals, STAT6 is an important regulator of Th9 cells (Goswami et al., 2011).
STATs and CD8 memory
IL-7 and IL-15 are important for CD8 memory and accordingly STAT5A/B are also important (Hand et al., 2010; Tripathi et al., 2010). STAT5A/B are essential for the survival of viral-specific CD8 T cells and expression of Bcl-2. In contrast though, in the setting of viral infection, the numbers of CD4 effector T cells are unaffected by the absence of STAT5A/B. However, STAT5A/B are not the only family members important for CD8 cell function; STAT3 is also important, mediating signals by IL-10 and IL-21 (Cui et al., 2011). Expression of such key molecules as Eomes, Bcl-6, Blimp-1, and Socs-3 are all reduced in STAT3-deficient CD8 T cells. A similar defect in CD8 T cell memory was seen in patients with hyperimmunoglobulin E syndrome and dominant-negative STAT3 mutations(Siegel et al., 2011).
STAT5 in B cells
IL-7, acting via STAT5A/B, is important in B lymphopoiesis, controlling survival and development (Malin et al., 2010). Conversely, the B cell adapter, BLNK, antagonizes IL-7 signaling via inhibition of JAK3, and absence of BLNK leads to constitutive JAK-Stat activation and leukomogenesis(Nakayama et al., 2009).
STATs and innate immunity
STATs also have numerous functions in innate immunity – too many to review in detail in a short review(Murray, 2007; O’Shea and Murray, 2008). The importance of STAT1 in mediating IFN effects has long been recognized as has the role of STAT3 in IL-6 signaling and the acute phase response. CSFs and cytokines like GM-CSF, G-CSF and IL-5, which regulate myeloid development, also signal via STATs. Consequently, STATs have key functions for neutrophils and macrophages (Croker et al., 2004; Lee et al., 2002; Nguyen-Jackson et al., 2010; Panopoulos et al., 2006; Zhang et al., 2010a). GM-CSF inhibits Flt3L-mediated plasmacytoid DC production and conventional DC growth and STAT5 is important in this process(Esashi et al., 2008). In contrast, STAT3 is important for the expansion of DC progenitors.
The importance of IL-22, acting via STAT3, in regulating the barrier function of epithelial cells and wound repair is a topic of considerable interest (Sonnenberg et al., 2011). Like IL-10, IL-22 is produced by and acts on innate immune cells, and has critical anti-inflammatory properties (Sonnenberg et al., 2011; Zenewicz and Flavell, 2011). Precisely how STAT3 promotes inflammation in some circumstances and inhibits in others is an important, but challenging question (El Kasmi et al., 2006). STAT3 can negatively regulate IFN responses and has been proposed to inhibit TLR signaling either by inducing anti-inflammatory molecules or by a direct suppression of NF-κB (Wang et al., 2011). Nonetheless, a clear understanding of the pro- and anti-inflammatory actions of STAT3 remains elusive.
Recently, the role of innate immune cells in promoting Th2 responses has become increasingly apparent (Oliphant et al., 2011; Saenz et al., 2010). Thymic stromal lymphopoetin (TSLP) in particular is an important Type I cytokine that promotes allergic responses. It acts on multiple cells, but a critical effect is on basophils, which are major producers of IL-4 (Siracusa et al., 2011; van Panhuys et al., 2011)The identity of the JAKs responsible for signaling had been enigmatic, but we now know that TSLP signals via JAK1 and JAK2 to activate STAT5 (Rochman et al., 2010).
In addition to the classical mode of activating macrophages via IFN-γ, the appreciation of the importance of Th2 cytokines to generate alternatively activated macrophages (AAM) is now recognized (Gordon and Martinez, 2010). AAM appear to be important in a range of processes including host defense, fibrosis, metabolic regulation, obesity and cancer. As IL-4 and IL-13 are major drivers of the AAM, STAT6 is a key player for these cells. STAT6 is important in regulating insulin action, lipid metabolism and expression of proliferation-activated receptor isoforms (Ricardo-Gonzalez et al., 2010; Szanto et al., 2010). Very recently, AAM and STAT6 have been implicated in the mammalian stress response, the response to cold (Nguyen et al., 2011). Intriguingly, AAM secrete catacholamines in a STAT6-dependent manner and induce thermogenic gene expression in brown adipose tissue and lipolysis in white adipose tissue. Beyond their role as transcription factors, a direct role of STATs in mitochondrial function makes the argument for key roles in metabolism even more compelling (Gough et al., 2009; Potla et al., 2006; Wegrzyn et al., 2009).
While it has long been recognized that viruses can disrupt IFN signaling by disrupting STAT signaling (Ramachandran and Horvath, 2009), recent work shows that T. gondii alters host response by injecting a kinase, ROP16 that activates both STAT3 and STAT6 (Butcher et al., 2011; Saeij et al., 2007). In macrophages, the effect is down-regulation of proinflammatory cytokine signaling and deviation to an alternatively activated phenotype. Viruses can also activate STAT6 and can do so apparently in a JAK-independent manner (Chen et al., 2011a). In this case though, Stat6 activation is protective in terms of host response.
Towards a genomic view of STAT action: transcriptional and epigenetic roles
The advent of chromatin precipitation and massive parallel sequencing (ChIP-Seq) has permitted the understanding of STAT action on a global scale. Analysis of the genomewide targets of STATs via Chip-seq analysis for all the STATs has now been obtained, albeit in a limited number of tissues with relatively few stimuli and time points (reviewed in(Kanno et al., 2011; O’Shea et al., 2011). Gene expression is dramatically influenced by chromatin organization and until recently, the importance of STATs in regulating epigenetics has only been implicated by analysis of selected regions of certain genes. However, new technologies in in measuring cell-specific transcriptome and epigenome, coupled with the use of knockout mice, allows assessments of the global impact of STAT-dependent signaling. What emerges is that STATs have thousands of genomic targets, and have major effects on transcription and epigenetic modifications on a substantial portion of these genes (Durant et al., 2010; Elo et al., 2010; Good et al., 2009; Kanno et al., 2011; Liao et al., 2011a; Wei et al., 2010) In the case of STAT6, about half of its target genes are affected in terms gene expression, epigenetic modifications or both when STAT6 is lacking in polarized Th2 cells (Wei et al., 2010). The impact of STAT4 in Th1 cells is less, but this is expected as both STAT4 and STAT1 contribute to Th1 differentiation (Schulz et al., 2009).
In addition to their roles in driving transcription, it is also clear from genomic studies that a major function of STATs is their role as functional repressors (Mandal et al., 2011a; Wei et al., 2010; Yang et al., 2011). In B cells, IL-7-mediated activation of STAT5 maintains proliferation and represses Igk germline transcription. Recently it has been shown that STAT5 binds the Igk intronic enhancer as a tetramer. This results in the recruitment of the histone methyltransferase Ezh2, which in turn induces histone H3 lysine 27 trimethylation, a repressive mark(Mandal et al., 2011a). Genome-wide analyses showed a STAT5 tetrameric binding motif is frequently associated with transcriptional repression. As indicated above, in T cells STAT5 displaces STAT3 and inhibits IL-17 expression (Yang et al., 2011). In Th1 and Th2 cells, STAT4 and STAT6 binding is frequently associated with repression. However, the mechanism of inhibition is not necessarily mediated by competition; in a large number of cases they bind distinct sites (Wei et al., 2010). Thus, it is clear STATs can both enhance and repress gene expression depending upon the complexes they recruit.
Equally intriguing is evidence that aside from phosphorylating STATs, JAK can have a direct role in regulating chromatin (Dawson et al., 2009). JAK2 has been found in the nuclei of haematopoietic cells, where it phosphorylates histone H3 tyrosine 41. Phosphorylation of this residue prevents heterochromatin protein 1alpha binding, and thereby counteracts gene silencing (Li, 2008; Shi et al., 2006).
Evidence for genetic links between cytokines and cytokine signaling and human autoimmune disease
While data from numerous animal models have implicated Type I/II cytokine receptors and the JAK/STAT pathway in autoimmune disease, the limitations is that they are just models. However, human genetics provides the ability to directly link genes to human disease. The field has moved rapidly from candidate gene to genome-wide investigation of single nucleotide polymorphisms (SNPs); systematic interrogation of the entire genome through next-generation sequencing is also now feasible (Mardis, 2011). Genome-wide association studies (GWAS) have led to an explosion of loci associated with risk of immune-mediated diseases. Importantly, these data show that inherited variation in genes encoding cytokines, Type I/II cytokines, JAKs and STATs are associated with these disorders.
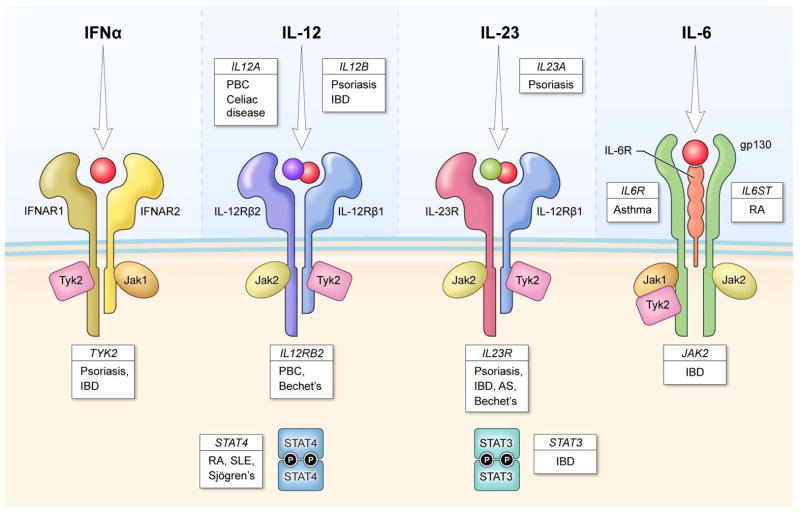
Among the strongest evidence is work showing that multiple genes in the IL-23 signaling pathway are involved in human autoimmunity. One of the first variants to be identified was a non-synonymous variant of the IL-23R (Arg381Gln)(Duerr et al., 2006), which is associated with reduced risk of of IBD, psoriasis (Cargill et al., 2007; Nair et al., 2009) and ankylosing spondylitis (Burton et al., 2007). More recently, additional coding variants have been found to influence disease susceptibility to Crohn’s and Behcet’s disease (Momozawa et al., 2011; Remmers et al., 2010b). Subsequently, polymorphisms of the genes encoding both subunits of IL-23 (p19/IL23A and p40/IL12B), JAK2, TYK2, and STAT3 have all been linked to autoimmunity (Bowes et al., 2011; Chu et al., 2011; Franke et al., 2010; Jakkula et al., 2010).
STAT3 is also activated by IL-6 and its receptors, IL6R and gp130 (latter encoded by IL6ST), have also been implicated in immune-mediated disease (Alloza et al., 2011; Ferreira et al., 2011; Stahl et al., 2010). IL6R may also be associated with cardiovascular disease (Elliott et al., 2009) and a disease-associated missense allele correlates with CRP levels (Dehghan et al., 2011; Melzer et al., 2008).
Multiple genes in the IL-12 pathway have also been implicated by GWAS. IL12A and IL12RB2, which are unique to IL-12 and not shared by IL-23, and STAT4 are associated with multiple autoimmune diseases (Hirschfield et al., 2009; Mells et al., 2011; Radstake et al., 2010; Remmers et al., 2010a; Remmers et al., 2007; Trynka et al., 2011; Zhernakova et al., 2011). It needs to be borne in mind that STAT4 is not only activated by IL-12, but can be activated by IL-23 and Type I IFNs.
With respect to allergic disease, polymorphisms of STAT6, and IL13 are associated with IgE levels and atopic dermatitis (Granada et al., 2011; Paternoster et al., 2011).
Despite these exciting leads, there are challenges of interpreting the biological function of genetic association data. Most disease-associated SNPs fall outside of protein-coding regions, and several genes may be in the region of linkage disequilibrium (LD) surrounding the SNP. The best biological candidate gene in the region is assumed to be the causal gene, but this may not be the correct assumption. For instance, there is an association of RA and multiple sclerosis with a SNP near the IL6ST gene (Alloza et al., 2011; Stahl et al., 2010); there is no direct evidence that the disease-associated variant disrupts IL6ST function. Similarly, IL12RB2 and IL23R are adjacent to each other, and it is not clear whether the associated Behcet’s risk allele influences one gene or the other.
Another challenge is inferring function when genes can be involved in multiple pathways. STAT4 is one example, but Tyk2 is another – both are involved in signaling by IL-12, IL-23 and Type I IFNs. Exactly who is the bad actor? Bioinformatic methods have been developed to search for relationships across genetic risk loci in order to find patterns that might otherwise be difficult to decipher (Hu et al., 2011; Raychaudhuri et al., 2009; Rossin et al., 2011; Segre et al., 2010). Future studies aimed at functional integration of genetic risk loci are a major effort to follow-up GWAS findings. Regardless though, the data clearly implicate the JAK-STAT pathway and cognate cytokines in the human immune-mediated disease.
Targeting Cytokine Signaling
The role of cytokine and cytokine signaling in mediating immune-mediated disease, now supported by GWAS data, has made these attractive pharmacological targets (Plenge, 2010). In fact, monoclonal antibodies directed against cytokines and cytokine receptors (e.g. ustekinumab, tocilizumab, mepolizumab, lebrikinumab, and daclizumab) have already shown efficacy in a variety of clinical settings. Additionally, the prospect of targeting intracellular signaling by these cytokines is also now a reality.
Janus kinases Inhibitors (JAKinibs)
As discussed by Notarangelo/Holland/Casanova (cite review in this issue), the unequivocal in vivo importance of the JAK/STAT pathways was first established by the identification of patients with severe combined immunodeficiency with JAK3 mutations. The profound, but selective phenotype associated with JAK3-deficiency led to the proposition that targeting JAKs would represent a new class of immunomodulatory drugs (Ghoreschi et al., 2009; O’Shea et al., 2004; Russell et al., 1995).
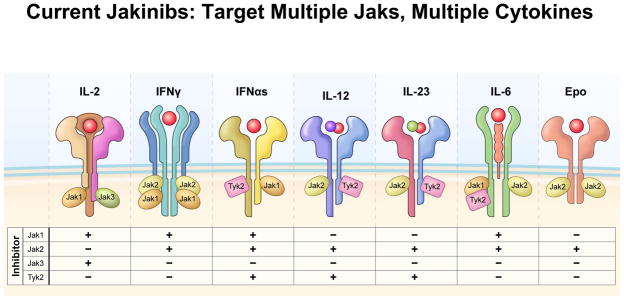
Tofacitinib, formerly designated CP-690,550, was the first JAK inhibitor to be studied in humans. It inhibits JAK3 and JAK1 and to a lesser extent JAK2. Consequently, tofacitinib potently inhibits common γ chain cytokines but also blocks IFN-γ, IL-6 and to a lesser extent IL-12 and IL-23 (Ghoreschi et al., 2011). Functionally, tofacitinib affects both innate and adaptive immune responses (Ghoreschi et al., 2011). Remarkably, tofacitinib has little activity on kinases other than JAKs (Karaman et al., 2008).
Tofacitinib was effective in preclinical models (Changelian et al., 2003)and has shown efficacy in a variety of Phase II and III trials in rheumatoid arthritis, as monotherapy and in combination with other drugs (Fleischmann et al., 2012; Kremer et al., 2009; Kremer et al., 2011; Tanaka et al., 2011). Importantly, Tofacitinib is effective in patients who have failed one or more biologic and also prevents structural damage. Tofacitinib is under investigation for psoriasis, inflammatory bowel disease, sicca syndrome and prevention of transplant rejection.
Other JAK inhibitors are also rapidly moving ahead in preclinical assessment and clinical trials (Table I) (Fridman et al., 2010; Lin et al., 2010; Lu et al., 2011; Stump et al., 2011). As discussed by Green and Staudt in this issue, the JAK1 and JAK2 inhibitor, Ruxolitinib, is efficacious in polycythemia/myelofibrosis, a disorder due to gain-of-function JAK2 mutations. As might be expected, based on its ability to block cytokines that use JAK1 and JAK2, this drug is also efficacious in arthritis (Fridman et al., 2010). Conversely, drugs that have relative selectivity for individual JAKs (JAK1, JAK2 and JAK3), also appear to have utility in preclinical and early clinical trials (Table I)
Table 1.
Selected JAKinibs
| Agent | Targets | Indication/Phase |
|---|---|---|
| Tofacitinib | JAK3/JAK1/JAK2 | RA/Phase III Psoriasis/Phase II IBD/Phase II |
| VX-509 | JAK3 | RA/Phase II |
| R-348 | JAK3 | RA/Phase I |
| Ruxolitinib | JAK1/JAK2 | Approved – MF/PV |
| INCB-28050 | JAK1/JAK2 | RA/Phase II |
| GLPG-0634 | JAK1 | RA/Phase II |
| AC-430 | JAK2 | RA/Phase I Lymphoma/Phase I |
| Lestaurtinib | FLT3/TrkA/JAK2 | AML/Phase III Psoriasis/Phase II Pancreatic cancer/Phase II |
| CEP-33779 | JAK2 | Preclinical |
The adverse effects associated with JAKinibs appear to be largely related to its mode of action. Infections are among the common adverse effects, but opportunistic infections are uncommon. Anemia and neutropenia, presumably related to JAK2 inhibition and interference with signaling by erythropoietin and other colony-stimulating factors, can also occur. Increases in serum LDL also occur, as has been seen with the IL-6 blocker, tocilizumab (Kawashiri et al., 2011). Little reduction in CD4+ T cells has been noted in nonhuman primates treated with tofacitinib, but more significant reduction in NK cells and CD8+ T cells can occur (Conklyn et al., 2004; Paniagua et al., 2005). Whether this is will be pertinent and clinically relevant in humans remains to be determined. A decline in functional Treg cells has not been noted in human subjects in a renal transplant study (Sewgobind et al., 2010).
Given the profound role of cytokines in disorders ranging from malignancy to autoimmunity, JAKinibs have enormous potential utility. The extent to which JAK inhibitors will be used as steroid-sparing agents or even supplant the use of steroids in diseases like the vasculitides or systemic lupus erythematosus remains to be seen. A surprise in the field is that targeting multiple kinases is not necessarily detrimental, especially in circumstances in which multiple cytokines drive pathogenesis. Conversely though, it is conceivable that more selective JAK inhibitors (e.g. selective JAK1 and JAK3 inhibitors) might have efficacy with reduced adverse effects related to JAK2 inhibition. It is likely that we will soon see if this is the case given the intense interest in JAKinibs.
The prospect of targeting STATs?
Given their importance and circumscribed functions, it would also seem logical to target STATs – especially if different STATs could be selectively targeted. A number of STAT inhibitors have been described (Nelson et al., 2011; Yue and Turkson, 2009); however, to date, there is no STAT inhibitor that is near clinical development. Conceptually, one might target STATs by: 1.) blocking STAT phosphorylation, 2.) disrupting STAT binding to phosphorylated receptors or dimerization (both of which are mediated by the STAT src homology (SH)2 domain; or 3.) interfering with DNA binding. Phosphopepitidomimetics continue to be designed that interrupt phosphotyrosine-SH2 binding (Mandal et al., 2011b; Zhang et al., 2010b; Zhao et al., 2010); however, the challenge will be to generated compounds with in vivo efficacy and selectivity. Targeting of the N-terminal domain has also been proposed as a strategy (Timofeeva et al., 2007). Screening of libraries has also revealed that small molecules like pimozide, nifruroxaide and pyrimethamine may be useful STAT inhibitors (Nelson et al., 2011). Whether any of these strategies ultimately generate orally available drugs that have efficacy with acceptable safety remains to be determined. However, given the prominent role of STATs in cancer, it is like that work will continue in this area.
Conclusions
The elegance of the JAK-STAT pathway is that it provides a simple, membrane to nucleus mechanism for rapidly inducing gene expression. As complexities of immune cell function continue to be unraveled, JAKs and STATs remain central players in all of the key cells, ranging from the “newest” CD4 helper cell subset to alternatively activated macrophages. Curiously though, there is still a paucity of information on conditional JAK and STAT knockouts. While some were quickly generated and studies in other cases we are still relatively ignorant about tissue specific functions of others (e.g. JAK1, JAK2, JAK3, TYK2, STAT1, STAT4 and STAT6).
In addition, although the simplicity of the pathway is appealing, some subtleties have become apparent. For instance, in contrast to the simplistic linear view, most cytokines activate more than one STAT. Precisely what this means in terms of the molecular basis of cytokine action is still being unraveled. However, technologic advances have certainly facilitated a broader understanding of the function of STAT proteins. It is now clear that STATs activate and repress gene expression and serve to organize the epigenetic landscape of immune cells. Nonetheless, our understanding how this occurs is still in its infancy. Despite the gaps in our knowledge, it is clear that this pathway is directly relevant to human disease and the pathway can be successfully targeted. For all these reasons, the next twenty years are likely to be just as exciting as the first.
Figure 1.
Genetics links of cytokine signaling with human autoimmune disease. Although various animal models have implicated cytokines, their receptors, JAKs and STATs with autoimmune disease, genomewide association studies (GWAS) now show that these factors are truly relevant to human disease. This work shows that pathways that lead to STAT3 and STAT4 activation lie at the heart of many common autoimmune diseases Adapted from (Cho and Gregersen, 2011). AS – ankylosing spondylitis, IBD – inflammatory bowel disease; PBC –primary biliary cirrhosis, SLE – systemic lupus erythematosus.
Figure 2.
Consequence of Jak inhibition on signaling by key immunoregulatory cytokines. A variety of JAKinibs have been developed with varying degrees of specificity for the different Jaks. The consequences of inhibiting each of the Jaks on these selected cytokines is depicted. Most inhibitors in clinical use inhibit more than one Jak; however, increasingly selective JAKinibs are in development. A selective Tyk2 inhibitor has yet to be reported.
Footnotes
Competing interests:
JO’S and National Institutes of Health (NIH) hold patents related to targeting JAKs as targets for immunomodulatory agents and have a Collaborative Research Agreement and Development Award with Pfizer.
References
- Alloza I, Otaegui D, de Lapuente AL, Antiguedad A, Varade J, Nunez C, Arroyo R, Urcelay E, Fernandez O, Leyva L, et al. ANKRD55 and DHCR7 are novel multiple sclerosis risk loci. Genes and Immunity. 2011 doi: 10.1038/gene.2011.81. [DOI] [PubMed] [Google Scholar]
- Batten M, Ramamoorthi N, Kljavin NM, Ma CS, Cox JH, Dengler HS, Danilenko DM, Caplazi P, Wong M, Fulcher DA, et al. IL-27 supports germinal center function by enhancing IL-21 production and the function of T follicular helper cells. The Journal of experimental medicine. 2010;207:2895–2906. doi: 10.1084/jem.20100064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes J, Orozco G, Flynn E, Ho P, Brier R, Marzo-Ortega H, Coates L, McManus R, Ryan AW, Kane D, et al. Confirmation of TNIP1 and IL23A as susceptibility loci for psoriatic arthritis. Annals of the rheumatic diseases. 2011;70:1641–1644. doi: 10.1136/ard.2011.150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. Journal of Immunology. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher BA, Fox BA, Rommereim LM, Kim SG, Maurer KJ, Yarovinsky F, Herbert DR, Bzik DJ, Denkers EY. Toxoplasma gondii rhoptry kinase ROP16 activates STAT3 and STAT6 resulting in cytokine inhibition and arginase-1-dependent growth control. PLoS pathogens. 2011;7:e1002236. doi: 10.1371/journal.ppat.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP, Matsunami N, Ardlie KG, Civello D, Catanese JJ, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changelian PS, Flanagan ME, Ball DJ, Kent CR, Magnuson KS, Martin WH, Rizzuti BJ, Sawyer PS, Perry BD, Brissette WH, et al. Prevention of organ allograft rejection by a specific Janus kinase 3 inhibitor. Science. 2003;302:875–878. doi: 10.1126/science.1087061. [DOI] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell. 2011a;147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- Chen Y, Haines CJ, Gutcher I, Hochweller K, Blumenschein WM, McClanahan T, Hammerling G, Li MO, Cua DJ, McGeachy MJ. Foxp3(+) regulatory T cells promote T helper 17 cell development in vivo through regulation of interleukin-2. Immunity. 2011b;34:409–421. doi: 10.1016/j.immuni.2011.02.011. [DOI] [PubMed] [Google Scholar]
- Chen Z, Laurence A, Kanno Y, Pacher-Zavisin M, Zhu BM, Tato C, Yoshimura A, Hennighausen L, O’Shea JJ. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci U S A. 2006;103:8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Gregersen PK. Genomics and the multifactorial nature of human autoimmune disease. The New England journal of medicine. 2011;365:1612–1623. doi: 10.1056/NEJMra1100030. [DOI] [PubMed] [Google Scholar]
- Chu X, Pan CM, Zhao SX, Liang J, Gao GQ, Zhang XM, Yuan GY, Li CG, Xue LQ, Shen M, et al. A genome-wide association study identifies two new risk loci for Graves’ disease. Nature Genetics. 2011;43:897–901. doi: 10.1038/ng.898. [DOI] [PubMed] [Google Scholar]
- Conklyn M, Andresen C, Changelian P, Kudlacz E. The JAK3 inhibitor CP-690550 selectively reduces NK and CD8+ cell numbers in cynomolgus monkey blood following chronic oral dosing. Journal of leukocyte biology. 2004;76:1248–1255. doi: 10.1189/jlb.0504282. [DOI] [PubMed] [Google Scholar]
- Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, Williams E, et al. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An Interleukin-21- Interleukin-10-STAT3 Pathway Is Critical for Functional Maturation of Memory CD8(+) T Cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, Pellikka N, Wallaschofski H, Kettunen J, Henneman P, et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–738. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddahri F, Denanglaire S, Bureau F, Spolski R, Leonard WJ, Leo O, Andris F. Interleukin-6/STAT3 signaling regulates the ability of naive T cells to acquire B-cell help capacities. Blood. 2009;113:2426–2433. doi: 10.1182/blood-2008-04-154682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi KC, Holst J, Coffre M, Mielke L, de Pauw A, Lhocine N, Smith AM, Rutschman R, Kaushal D, Shen Y, et al. General nature of the STAT3-activated anti-inflammatory response. Journal of Immunology. 2006;177:7880–7888. doi: 10.4049/jimmunol.177.11.7880. [DOI] [PubMed] [Google Scholar]
- Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, Peden JF, Erdmann J, Braund P, Engert JC, Bennett D, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. Jama. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elo LL, Jarvenpaa H, Tuomela S, Raghav S, Ahlfors H, Laurila K, Gupta B, Lund RJ, Tahvanainen J, Hawkins RD, et al. Genome-wide profiling of interleukin-4 and STAT6 transcription factor regulation of human Th2 cell programming. Immunity. 2010;32:852–862. doi: 10.1016/j.immuni.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Esashi E, Wang YH, Perng O, Qin XF, Liu YJ, Watowich SS. The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity. 2008;28:509–520. doi: 10.1016/j.immuni.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, Danoy P, Baltic S, Nyholt DR, Jenkins M, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann R, Cutolo M, Genovese MC, Lee EB, Kanik KS, Sadis S, Connell CA, Gruben D, Krishnaswami S, Wallenstein G, et al. Phase IIb dose-ranging study of the oral JAK inhibitor tofacitinib (CP-690,550) or adalimumab monotherapy versus placebo in patients with active rheumatoid arthritis with an inadequate response to disease-modifying antirheumatic drugs. Arthritis and rheumatism. 2012;64:617–629. doi: 10.1002/art.33383. [DOI] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nature Genetics. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman JS, Scherle PA, Collins R, Burn TC, Li Y, Li J, Covington MB, Thomas B, Collier P, Favata MF, et al. Selective inhibition of JAK1 and JAK2 is efficacious in rodent models of arthritis: preclinical characterization of INCB028050. Journal of Immunology. 2010;184:5298–5307. doi: 10.4049/jimmunol.0902819. [DOI] [PubMed] [Google Scholar]
- Ghoreschi K, Jesson MI, Li X, Lee JL, Ghosh S, Alsup JW, Warner JD, Tanaka M, Steward-Tharp SM, Gadina M, et al. Modulation of innate and adaptive immune responses by tofacitinib (CP-690,550) Journal of Immunology. 2011;186:4234–4243. doi: 10.4049/jimmunol.1003668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K, Laurence A, O’Shea JJ. Selectivity and therapeutic inhibition of kinases: to be or not to be? Nature Immunology. 2009;10:356–360. doi: 10.1038/ni.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good SR, Thieu VT, Mathur AN, Yu Q, Stritesky GL, Yeh N, O’Malley JT, Perumal NB, Kaplan MH. Temporal induction pattern of STAT4 target genes defines potential for Th1 lineage-specific programming. Journal of Immunology. 2009;183:3839–3847. doi: 10.4049/jimmunol.0901411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Goswami R, Jabeen R, Yagi R, Pham D, Zhu J, Goenka S, Kaplan MH. STAT6-Dependent Regulation of Th9 Development. Journal of Immunology. 2011 doi: 10.4049/jimmunol.1102840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granada M, Wilk JB, Tuzova M, Strachan DP, Weidinger S, Albrecht E, Gieger C, Heinrich J, Himes BE, Hunninghake GM, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. The Journal of allergy and clinical immunology. 2011 doi: 10.1016/j.jaci.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand TW, Cui W, Jung YW, Sefik E, Joshi NS, Chandele A, Liu Y, Kaech SM. Differential effects of STAT5 and PI3K/AKT signaling on effector and memory CD8 T-cell survival. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16601–16606. doi: 10.1073/pnas.1003457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Gu X, Walker EJ, Jing K, Juran BD, Mason AL, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. The New England journal of medicine. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Kim H, Stahl E, Plenge R, Daly M, Raychaudhuri S. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am J Hum Genet. 2011;89:496–506. doi: 10.1016/j.ajhg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakkula E, Leppa V, Sulonen AM, Varilo T, Kallio S, Kemppinen A, Purcell S, Koivisto K, Tienari P, Sumelahti ML, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010;86:285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. The Journal of experimental medicine. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and Epigenetic Control of T Helper Cell Specification: Molecular Mechanisms Underlying Commitment and Plasticity. Annual Review of Immunology. 2012 doi: 10.1146/annurev-immunol-020711-075058. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nature Biotechnology. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Kawashiri SY, Kawakami A, Yamasaki S, Imazato T, Iwamoto N, Fujikawa K, Aramaki T, Tamai M, Nakamura H, Ida H, et al. Effects of the anti-interleukin-6 receptor antibody, tocilizumab, on serum lipid levels in patients with rheumatoid arthritis. Rheumatol Int. 2011;31:451–456. doi: 10.1007/s00296-009-1303-y. [DOI] [PubMed] [Google Scholar]
- Kremer JM, Bloom BJ, Breedveld FC, Coombs JH, Fletcher MP, Gruben D, Krishnaswami S, Burgos-Vargas R, Wilkinson B, Zerbini CA, et al. The safety and efficacy of a JAK inhibitor in patients with active rheumatoid arthritis: Results of a double-blind, placebo-controlled phase IIa trial of three dosage levels of CP-690,550 versus placebo. Arthritis and rheumatism. 2009;60:1895–1905. doi: 10.1002/art.24567. [DOI] [PubMed] [Google Scholar]
- Kremer JM, Cohen S, Wilkinson BE, Connell CA, French JL, Gomez-Reino J, Gruben D, Kanik KS, Krishnaswami S, Pascual-Ramos V, et al. A Phase 2B dose-ranging study of the oral JAK inhibitor tofacitinib (CP-690,550) versus placebo in combination with background methotrexate in patients with active rheumatoid arthritis and inadequate response to methotrexate alone. Arthritis and rheumatism. 2011 doi: 10.1002/art.33419. [DOI] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Lee CK, Raz R, Gimeno R, Gertner R, Wistinghausen B, Takeshita K, DePinho RA, Levy DE. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17:63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annual Review of Immunology. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- Li WX. Canonical and non-canonical JAK-STAT signaling. Trends in Cell Biology. 2008;18:545–551. doi: 10.1016/j.tcb.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol. 2011a;23:598–604. doi: 10.1016/j.coi.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011b;12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, Leonard WJ. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor γ±-chain expression. Nature Immunology. 2008;9:1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TH, Hegen M, Quadros E, Nickerson-Nutter CL, Appell KC, Cole AG, Shao Y, Tam S, Ohlmeyer M, Wang B, et al. Selective functional inhibition of JAK-3 is sufficient for efficacy in collagen-induced arthritis in mice. Arthritis and rheumatism. 2010;62:2283–2293. doi: 10.1002/art.27536. [DOI] [PubMed] [Google Scholar]
- Lu LD, Stump KL, Wallace NH, Dobrzanski P, Serdikoff C, Gingrich DE, Dugan BJ, Angeles TS, Albom MS, Mason JL, et al. Depletion of autoreactive plasma cells and treatment of lupus nephritis in mice using CEP-33779, a novel, orally active, selective inhibitor of JAK2. Journal of Immunology. 2011;187:3840–3853. doi: 10.4049/jimmunol.1101228. [DOI] [PubMed] [Google Scholar]
- Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nature Immunology. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Powers SE, Maienschein-Cline M, Bartom ET, Hamel KM, Kee BL, Dinner AR, Clark MR. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nature Immunology. 2011a;12:1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Gao F, Lu Z, Ren Z, Ramesh R, Birtwistle JS, Kaluarachchi KK, Chen X, Bast RC, Jr, Liao WS, et al. Potent and selective phosphopeptide mimetic prodrugs targeted to the Src homology 2 (SH2) domain of signal transducer and activator of transcription 3. Journal of medicinal chemistry. 2011b;54:3549–3563. doi: 10.1021/jm2000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. A decade’s perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O’Malley JT, Kapur R, Levy DE, Kansas GS, Kaplan MH. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178:4901–4907. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, Heneghan MA, Neuberger JM, Donaldson PT, Day DB, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nature Genetics. 2011;43:329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, Rafferty I, Lauretani F, Murray A, Gibbs JR, Paolisso G, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS genetics. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, Kanno Y, Spalding C, Elloumi HZ, Paulson ML, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momozawa Y, Mni M, Nakamura K, Coppieters W, Almer S, Amininejad L, Cleynen I, Colombel JF, de Rijk P, Dewit O, et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nature Genetics. 2011;43:43–47. doi: 10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- Murray PJ. The JAK-STAT signaling pathway: input and output integration. Journal of Immunology. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nature Genetics. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Yamamoto M, Hayashi K, Satoh H, Bundo K, Kubo M, Goitsuka R, Farrar MA, Kitamura D. BLNK suppresses pre-B-cell leukemogenesis through inhibition of JAK3. Blood. 2009;113:1483–1492. doi: 10.1182/blood-2008-07-166355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, et al. Early Th1 Cell Differentiation Is Marked by a Tfh Cell-like Transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EA, Sharma SV, Settleman J, Frank DA. A chemical biology approach to developing STAT inhibitors: molecular strategies for accelerating clinical translation. Oncotarget. 2011;2:518–524. doi: 10.18632/oncotarget.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KD, Qiu Y, Cui X, Goh YP, Mwangi J, David T, Mukundan L, Brombacher F, Locksley RM, Chawla A. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480:104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Jackson H, Panopoulos AD, Zhang H, Li HS, Watowich SS. STAT3 controls the neutrophil migratory response to CXCR2 ligands by direct activation of G-CSF-induced CXCR2 expression and via modulation of CXCR2 signal transduction. Blood. 2010;115:3354–3363. doi: 10.1182/blood-2009-08-240317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, Huang H, Wen R, Wang J, Li HS, et al. STAT5 negatively regulates T follicular helper (Tfh) cell generation and function. The Journal of biological chemistry. 2012 doi: 10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nature reviews Immunology. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Pesu M, Borie DC, Changelian PS. A new modality for immunosuppression: targeting the JAK/STAT pathway. Nature reviews Drug discovery. 2004;3:555–564. doi: 10.1038/nrd1441. [DOI] [PubMed] [Google Scholar]
- Oliphant CJ, Barlow JL, McKenzie AN. Insights into the initiation of type 2 immune responses. Immunology. 2011;134:378–385. doi: 10.1111/j.1365-2567.2011.03499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandiyan P, Conti HR, Zheng L, Peterson AC, Mathern DR, Hernandez-Santos N, Edgerton M, Gaffen SL, Lenardo MJ. CD4(+)CD25(+)Foxp3(+) regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity. 2011;34:422–434. doi: 10.1016/j.immuni.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua R, Si MS, Flores MG, Rousvoal G, Zhang S, Aalami O, Campbell A, Changelian PS, Reitz BA, Borie DC. Effects of JAK3 inhibition with CP-690,550 on immune cell populations and their functions in nonhuman primate recipients of kidney allografts. Transplantation. 2005;80:1283–1292. doi: 10.1097/01.tp.0000177643.05739.cd. [DOI] [PubMed] [Google Scholar]
- Panopoulos AD, Zhang L, Snow JW, Jones DM, Smith AM, El Kasmi KC, Liu F, Goldsmith MA, Link DC, Murray PJ, et al. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood. 2006;108:3682–3690. doi: 10.1182/blood-2006-02-003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternoster L, Standl M, Chen CM, Ramasamy A, Bonnelykke K, Duijts L, Ferreira MA, Alves AC, Thyssen JP, Albrecht E, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nature Genetics. 2011 doi: 10.1038/ng.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul WE. What determines Th2 differentiation, in vitro and in vivo? Immunology and Cell Biology. 2010;88:236–239. doi: 10.1038/icb.2010.2. [DOI] [PubMed] [Google Scholar]
- Plenge R. GWASs and the age of human as the model organism for autoimmune genetic research. Genome Biology. 2010;11:212. doi: 10.1186/gb-2010-11-5-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potla R, Koeck T, Wegrzyn J, Cherukuri S, Shimoda K, Baker DP, Wolfman J, Planchon SM, Esposito C, Hoit B, et al. Tyk2 tyrosine kinase expression is required for the maintenance of mitochondrial respiration in primary pro-B lymphocytes. Molecular and Cellular Biology. 2006;26:8562–8571. doi: 10.1128/MCB.00497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radstake TR, Gorlova O, Rueda B, Martin JE, Alizadeh BZ, Palomino-Morales R, Coenen MJ, Vonk MC, Voskuyl AE, Schuerwegh AJ, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nature Genetics. 2010;42:426–429. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A, Horvath CM. Paramyxovirus disruption of interferon signal transduction: STATus report. J Interferon Cytokine Res. 2009;29:531–537. doi: 10.1089/jir.2009.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS genetics. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers EF, Cosan F, Kirino Y, Ombrello MJ, Abaci N, Satorius C, Le JM, Yang B, Korman BD, Cakiris A, et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet’s disease. Nat Genet. 2010a;42:698–702. doi: 10.1038/ng.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers EF, Cosan F, Kirino Y, Ombrello MJ, Abaci N, Satorius C, Le JM, Yang B, Korman BD, Cakiris A, et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet’s disease. Nature Genetics. 2010b;42:698–702. doi: 10.1038/ng.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, de Bakker PI, Le JM, Lee HS, Batliwalla F, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo-Gonzalez RR, Red Eagle A, Odegaard JI, Jouihan H, Morel CR, Heredia JE, Mukundan L, Wu D, Locksley RM, Chawla A. IL-4/STAT6 immune axis regulates peripheral nutrient metabolism and insulin sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:22617–22622. doi: 10.1073/pnas.1009152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman Y, Kashyap M, Robinson GW, Sakamoto K, Gomez-Rodriguez J, Wagner KU, Leonard WJ. Thymic stromal lymphopoietin-mediated STAT5 phosphorylation via kinases JAK1 and JAK2 reveals a key difference from IL-7-induced signaling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19455–19460. doi: 10.1073/pnas.1008271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Cotsapas C, Daly MJ. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS genetics. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, Aman MJ, Migone TS, Noguchi M, Markert ML, Buckley RH, et al. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- Saeij JP, Coller S, Boyle JP, Jerome ME, White MW, Boothroyd JC. Toxoplasma coopts host gene expression by injection of a polymorphic kinase homologue. Nature. 2007;445:324–327. doi: 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz SA, Noti M, Artis D. Innate immune cell populations function as initiators and effectors in Th2 cytokine responses. Trends in Immunology. 2010;31:407–413. doi: 10.1016/j.it.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Schmitt N, Morita R, Bourdery L, Bentebibel SE, Zurawski SM, Banchereau J, Ueno H. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 2009;31:158–169. doi: 10.1016/j.immuni.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz EG, Mariani L, Radbruch A, Hofer T. Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-gamma and interleukin-12. Immunity. 2009;30:673–683. doi: 10.1016/j.immuni.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS genetics. 2010:6. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewgobind VD, Quaedackers ME, van der Laan LJ, Kraaijeveld R, Korevaar SS, Chan G, Weimar W, Baan CC. The Jak inhibitor CP-690,550 preserves the function of CD4CD25FoxP3 regulatory T cells and inhibits effector T cells. Am J Transplant. 2010;10:1785–1795. doi: 10.1111/j.1600-6143.2010.03200.x. [DOI] [PubMed] [Google Scholar]
- Shi S, Calhoun HC, Xia F, Li J, Le L, Li WX. JAK signaling globally counteracts heterochromatic gene silencing. Nature Genetics. 2006;38:1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel AM, Heimall J, Freeman AF, Hsu AP, Brittain E, Brenchley JM, Douek DC, Fahle GH, Cohen JI, Holland SM, et al. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 2011;35:806–818. doi: 10.1016/j.immuni.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siracusa MC, Saenz SA, Hill DA, Kim BS, Headley MB, Doering TA, Wherry EJ, Jessup HK, Siegel LA, Kambayashi T, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 2011;477:229–233. doi: 10.1038/nature10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nature Genetics. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D, Travers J, Nguyen ET, Levy DE, Kaplan MH. The Transcription Factor STAT3 Is Required for T Helper 2 Cell Development. Immunity. 2011;34:39–49. doi: 10.1016/j.immuni.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump KL, Lu LD, Dobrzanski P, Serdikoff C, Gingrich DE, Dugan BJ, Angeles TS, Albom MS, Ator MA, Dorsey BD, et al. A highly selective, orally active inhibitor of Janus kinase 2, CEP-33779, ablates disease in two mouse models of rheumatoid arthritis. Arthritis research & therapy. 2011;13:R68. doi: 10.1186/ar3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, Szeles L, Poliska S, Oros M, Evans RM, et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARg-regulated gene expression in macrophages and dendritic cells. Immunity. 2010;33:699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Suzuki M, Nakamura H, Toyoizumi S, Zwillich SH. Phase II study of tofacitinib (CP-690,550) combined with methotrexate in patients with rheumatoid arthritis and an inadequate response to methotrexate. Arthritis Care Res (Hoboken) 2011;63:1150–1158. doi: 10.1002/acr.20494. [DOI] [PubMed] [Google Scholar]
- Timofeeva OA, Gaponenko V, Lockett SJ, Tarasov SG, Jiang S, Michejda CJ, Perantoni AO, Tarasova NI. Rationally designed inhibitors identify STAT3 N-domain as a promising anticancer drug target. ACS chemical biology. 2007;2:799–809. doi: 10.1021/cb700186x. [DOI] [PubMed] [Google Scholar]
- Tripathi P, Kurtulus S, Wojciechowski S, Sholl A, Hoebe K, Morris SC, Finkelman FD, Grimes HL, Hildeman DA. STAT5 is critical to maintain effector CD8+ T cell responses. Journal of Immunology. 2010;185:2116–2124. doi: 10.4049/jimmunol.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, Bakker SF, Bardella MT, Bhaw-Rosun L, Castillejo G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nature Genetics. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Panhuys N, Prout M, Forbes E, Min B, Paul WE, Le Gros G. Basophils are the major producers of IL-4 during primary helminth infection. Journal of Immunology. 2011;186:2719–2728. doi: 10.4049/jimmunol.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Panhuys N, Tang SC, Prout M, Camberis M, Scarlett D, Roberts J, Hu-Li J, Paul WE, Le Gros G. In vivo studies fail to reveal a role for IL-4 or STAT6 signaling in Th2 lymphocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12423–12428. doi: 10.1073/pnas.0806372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WB, Levy DE, Lee CK. STAT3 negatively regulates type I IFN-mediated antiviral response. Journal of Immunology. 2011;187:2578–2585. doi: 10.4049/jimmunol.1004128. [DOI] [PubMed] [Google Scholar]
- Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann ASea. STAT5 negative regulation of Tfh. Nature Immunology. 2012 in pres. [Google Scholar]
- Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenewicz LA, Flavell RA. Recent advances in IL-22 biology. Int Immunol. 2011;23:159–163. doi: 10.1093/intimm/dxr001. [DOI] [PubMed] [Google Scholar]
- Zhang H, Nguyen-Jackson H, Panopoulos AD, Li HS, Murray PJ, Watowich SS. STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood. 2010a;116:2462–2471. doi: 10.1182/blood-2009-12-259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yue P, Fletcher S, Zhao W, Gunning PT, Turkson J. A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochemical pharmacology. 2010b;79:1398–1409. doi: 10.1016/j.bcp.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Jaganathan S, Turkson J. A cell-permeable Stat3 SH2 domain mimetic inhibits Stat3 activation and induces antitumor cell effects in vitro. The Journal of biological chemistry. 2010;285:35855–35865. doi: 10.1074/jbc.M110.154088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, Westra HJ, Fehrmann RS, Kurreeman FA, Thomson B, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS genetics. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, Bellucci R, Raderschall E, Canning C, Soiffer RJ, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108:1571–1579. doi: 10.1182/blood-2006-02-004747. [DOI] [PMC free article] [PubMed] [Google Scholar]