Abstract
Context:
Mild traumatic brain injury (mTBI) represents a major health problem in civilian populations as well as among the military service members due to (1) lack of effective treatments, and (2) our incomplete understanding about the progression of secondary cell injury cascades resulting in neuronal cell death due to deficient cellular energy metabolism and damaged mitochondria.
Aims:
The aim of this study was to identify and delineate the mitochondrial targeted genes responsible for altered brain energy metabolism in the injured brain.
Settings and Design:
Rats were either grouped into naïve controls or received lateral fluid percussion brain injury (2–2.5 atm) and followed up for 7 days.
Materials and Methods:
Rats were either grouped into naïve controls or received lateral fluid percussion brain injury (2–2.5 atm) and followed for 7 days. The severity of brain injury was evaluated by the neurological severity scale—revised (NSS-R) at 3 and 5 days post TBI and immunohistochemical analyses at 7 days post TBI. The expression profiles of mitochondrial-targeted genes across the hippocampus from TBI and naïe rats were also examined by oligo-DNA microarrays.
Results:
NSS-R scores of TBI rats (5.4 ± 0.5) in comparison to naïe rats (3.9 ± 0.5) and H and E staining of brain sections suggested a mild brain injury. Bioinformatics and systems biology analyses showed 31 dysregulated genes, 10 affected canonical molecular pathways including a number of genes involved in mitochondrial enzymes for oxidative phosphorylation, mitogen-activated protein Kinase (MAP), peroxisome proliferator-activated protein (PPAP), apoptosis signaling, and genes responsible for long-term potentiation of Alzheimer's and Parkinson's diseases.
Conclusions:
Our results suggest that dysregulated mitochondrial-focused genes in injured brains may have a clinical utility for the development of future therapeutic strategies aimed at the treatment of TBI.
Keywords: Brain trauma, fluid percussion, gene expression, hippocampus, mitochondria, neurological severity scale
INTRODUCTION
Traumatic brain injury (TBI) has recently been recognized as the silent, signature wound of the wars in Iraq and Afghanistan.[1] From 2000 to 2010, approximately 1.6 million service members have been deployed to date in Iraq and Afghanistan. During this time period almost 179,000 service members sustained a TBI with the vast majority, about 137,000 of those being classified as mild in terms of severity.[2] In the United States, TBIs are also common in the civilian population, with an estimated 1.7 million civilians sustaining a mild to moderate TBI annually where approximately 75% of these injuries are classified as mild TBI (mTBI).[3] Therefore, even a slight improvement in the diagnostic and neuroprotective strategies will have significant health implications.
The mild–moderate closed head trauma is initiated by the impact of the brain against the inner side of the calvarium. Subsequently, there begins a series of complex biochemical, cellular, metabolic, and physiological injury cascade that may take longer to complete complete.[4,5] These mild–moderate brain injuries are invisible from the outside, but the cognitive deficits, including memory impairment, are common neurological consequences of TBI and the underlying mechanisms are poorly defined.[6] The hippocampus is one of the key regions of the brain responsible for learning and long-term memories. Both clinical and experimental evidence suggest that long-term effects of mTBI cause hippocampal neuronal cell death, which is associated with cognitive impairment.[7] Lateral fluid percussion injury (FPI) in rodents replicates several clinically relevant features of human TBI. It has been widely used in the investigation of biochemical and pathological responses observed in human closed head injuries.[8]
The growing evidence suggests the important role of mitochondria in the progression of TBI and a subcellular target for neuroprotective strategies.[9] Following TBI, excessive release of the neurotransmitter glutamate can result in mitochondrial dysfunctions by causing a toxic influx of calcium into brain cells. Mitochondrion has its own DNA (mtDNA), which encodes only 37 genes: 13 for various subunits of electron transport chain complexes I, II, III, IV, and V; 22 mt genes for mitochondrial tRNA and remaining 2 genes encodes rRNA. Most mitochondrial proteins including those for electron transport chain enzymes to produce cellular ATP, cell signaling, and apoptosis are encoded and/or regulated by nuclear DNA. In comparison to nuclear DNA, mtDNA is very sensitive to oxidative damage due to lack of histones.[10–16] Therefore, any damage to mtDNA will have grave consequences on the depletion of cellular ATP, increase free radicals, DNA damage, and neuronal cell death, which is observed in clinical and experimental TBI.[1,9] To our knowledge, neuronal-targeted expression profiles of mitochondria-focused genes have not been examined as a potential therapeutic target for mTBI. To date, methods employing a systems biology approach to identify the mitochondria-focused genes in the hippocampus underlying the pathogenesis of TBI have also not been documented.
In this study, we have examined the neurobehavioral and histochemical response to the brain injury, and analyzed the mitochondria-focused oligo-DNA microarray to determine the expression profile of mitochondria-focused genes in the hippocampus of rats with TBI induced by lateral fluid percussion injury.
MATERIALS AND METHODS
Adult, male Sprague–Dawley rats (225–275 G, Harlan, Frederick, MD, USA) were pair housed in standard polycarbonate shoebox cages (42.5 × 20.5 × 20 cm) with hardwood chip bedding (Pine-Dri) in an environmentally controlled animal facility under a 12-h reverse light-dark cycle. Food (Harlan Teklad 4% Mouse/Rat Diet 7001) and water were available ad libitum. Rats were left to acclimate for 1 week to the vivarium before use. All procedures were performed to National Institutes of Health (NIH) guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC). A total of 12 animals/group were included in this study. All animals had neurobehavioral evaluation. Hippocampi from three animals/group were used for hematoxylin and eosin stain (H and E staining) (total six animals), and three animals from each group had mitochondrial genetic profile mapped (total six animals). Remaining tissues were stored for further analysis.
Instrumentation and induction of fluid percussion injury
Mild injury was induced in rats according to our published procedure.[17] In brief, animals were anesthetized with 1–3% isoflourane in oxygen. Under sterile conditions, a 3 cm sagittal incision was made along the midline to expose the cranium. A 5 mm burr hole was placed 2 mm to the right of the sagittal suture halfway between bregma and lamda using a 5 mm trephine drill bit exposing the dura. A Luer-Loc needle hub was placed into the burr hole and cemented to the cranium using cyanoacrilate. The glue was allowed to completely dry, and the empty Luer-Loc hub was filled with normal saline before being connected to the TBI device. A fluid percussion pulse of 2.0–2.5 atm was administered by an injury cannula positioned parasagittally over the right cerebral cortex. The fluid percussion pulse was administered by a pendulum modulated fluid percussion biomechanical device (Richmond, VA, USA), the Luer-Loc hub was removed and defects in the cranium were repaired with bone wax. The skin was then closed with a surgical skin stapler. Animals were allowed to stabilize in the warm blanket and then returned to their cages and moved to the animal facility at the Uniformed Services University of the Health Sciences. At 7 days post TBI, animals were anesthetized, and brains were removed followed by the dissection of hippocampus. Hippocampi were dissected according to the method described by Jacobowitz and modified by Hefner et al.[18,19] The consistency of the hippocampus region of the brain was confirmed by measuring the regional specific protein levels, which were in the range of 10.8-11.2 mg/ml in all the animals examined. Simultaneously, animals in the naïve control group were also housed in the same environmental conditions. These naïve animals were time matched and did not receive any brain injury. Seven days postinjury (day 7), TBI and naïve animals were sacrificed. The hippocampi from the right side of the brain of three animals per group were used for RNA isolation and subsequent gene profiling.
Animal behavioral measures
Behavior was observed during the animals’ dark cycle. Behaviors were measured prior to injury (baseline) and at two time points during the week after injury (3 days post TBI = T1; 5 days post TBI = T2). Behavioral measures included the neurological severity scale–revised (NSS-R) to measure neurobehavioral responses. The NSS-R is a series of 10 specific behavioral measures[20] based on several neurological severity scales (NSS) designed for rats.[21–24] The NSS-R measures motor, sensory, and reflex responses. The NSS-R was scored by an experienced rater who was previously trained until reaching high inter-rater reliability with other independent raters. Each response was scored from 0–2, where 0 = normal response, 1 = partial and/or compromised impairment, and 2 = lack of ability to perform the task. A composite score was calculated (0–20), where the higher the score indicates more neurobehavioral impairment. The testing was conducted using two empty polycarbonate cages (46 × 36 × 20 cm) with no bedding or lid. The equipment was cleaned between each animal.
Immunohistochemistry
One week postinjury, a subset of three animals per group were subjected to formalin perfusion and immunohistochemistry as described previously.[17] Briefly, rats were deeply anesthetized with isoflurane inhalational anesthesia perfused transcardially with 400 ml of 4% fresh depolymerized paraformaldehyde in phosphate buffered saline (Fisher Scientific) using a peristaltic pump at a flow rate of approximately 60–80 ml/min. Brains were carefully dissected, and postfixed in 4% paraformaldehyde for 1 day. The tissues were cryoprotected by sequential passage through 10%, 20%, and 30% sucrose in phosphate buffered solution (PBS) for 1 day each. Tissues were kept in 30% sucrose at 4°C until used. The brains were rapidly frozen and 10–20 μm coronal sections were cut in a cryostat set at a temperature of-20°C. Sections collected on treated slides were stained with H and E to determine a broad range of cytoplasmic, nuclear, and extracellular matrix features. Hematoxylin has a deep blue-purple color and stains nucleic acids, while eosin is pink and stains proteins nonspecifically. In a typical tissue, nuclei are stained blue, whereas the cytoplasm and extracellular matrix have varying degrees of pink staining.
RMNchip
High quality rat mitochondrion-neuron focused microarrays (rMNChip) were designed and fabricated as described previously.[15] Briefly, rMNChip contains 1,500 genes including 37 mitochondrial DNA (mtDNA)-encoded genes, 1,098 nuclear DNA (nDNA)-encoded and mitochondria-focused genes, and 365 neuron-related genes. The oligonucleotides for genes and positive (80 housekeeping genes) and negative (no rat DNA) controls on rMNChip were printed, each in triplicates, onto the N-hydroxysuccinimide ester reactive groups-coated glass slides. rMNChip microarrays were fabricated in the Class 100 super-clean environment as described previously.[15]
RNA isolation and purification
Each sample of total RNA was purified from hippocampi collected in a RNA tube and processed using the RNA Kit (PAXgene RNA Kit, 762164, QIAGEN, USA) following the manufacturer's instructions.
Microarray hybridization
One microgram RNA per sample was used for Cy5-dUTP labeling of cDNA by use of the express array detection kit (3DNA Array 900, Genisphere, Hatfield, PA, USA) following the manufacturer's instructions. Slides were scanned using 5 micron resolution and LOWESS method with Scan Array Microarray Scanner (PerkinElmer). Triplicate microarray experiments were performed for each and every RNA sample. Thus, the RNA level of each and every gene was measured as many as nine times. The background-subtracted mean values of the measurements were used for microarray data analysis. All microarray experiments were performed in the same laboratory of Gen Pro Markers, Inc.[15]
Microarray database, bioinformatics and systems biology
Gene expression database was constructed using FileMaker software (FileMaker Pro, Inc., Santa Clara, CA, USA). Database construction, data filtering, and selection were performed as described previously.[10,15] The quantile normalization method[25] in software R version 2.7.1 (the R Foundation for Statistical Computing) was used to normalize microarray data. The normalized data was used to cluster and visualize genes and hippocampus tissue samples by using software Cluster version 3.0 and the resulting heat map was visualized by using MapleTree software (http://rana.lbl.gov/EisenSoftware.htm). Gene information including identification numbers, official symbols and full names were updated using the National Center for Biotechnology Information (NCBI) database (www.ncbi.nlm.nih.gov/gene). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were computed using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov).
Statistics
The quantile normalization method in software R/Bioconductor version 2.7.1 (The R Foundation for Statistical Computing) was used to normalize data. Statistical calculations were performed on triplicate spots per gene and triplicate array experiments using XLSTAT 2006 (XLSTAT, New York, NY, USA). The moderated P-values and false discovery rate (FDR) for multiple statistical testing with Benjamini and Hochberg methods were calculated with DAVID Bioinformatics Resources version 6.7 (http://david.abcc.ncifcrf.gov). Differentially expressed genes were identified arbitrarily by ≥1.4-fold change in the average expression of the background-subtracted mean intensity ratios of a gene between TBI and naïve rats, with P-value < 0.05. Repeated measures analysis of variance (rANOVA) were conducted for the NSS-R (N = 24). All tests were two tailed using alpha = 0.05.
RESULTS
Effects of TBI on neurological severity scale - revised
Figure 1 presents the NSS-R scores. There were no differences in NSS-R scores between experimental groups at baseline. Post TBI, the TBI rats (5.58 ± 0.48) had significantly greater NSS-R scores compared with the naïve rats (3.92 ± 0.48), F(1, 22) = 5.93, P = 0.023, η2 = 0.212. Additionally, the TBI rats (5.42 ± 0.50) had significantly greater scores than the naïve rats (3.25 ± 0.50) at T2, F(1, 22) = 9.23, P = 0.006, η2 = 0.295. This finding suggests that TBI rats suffered more neurological impairment than the naïve rats.
Figure 1.
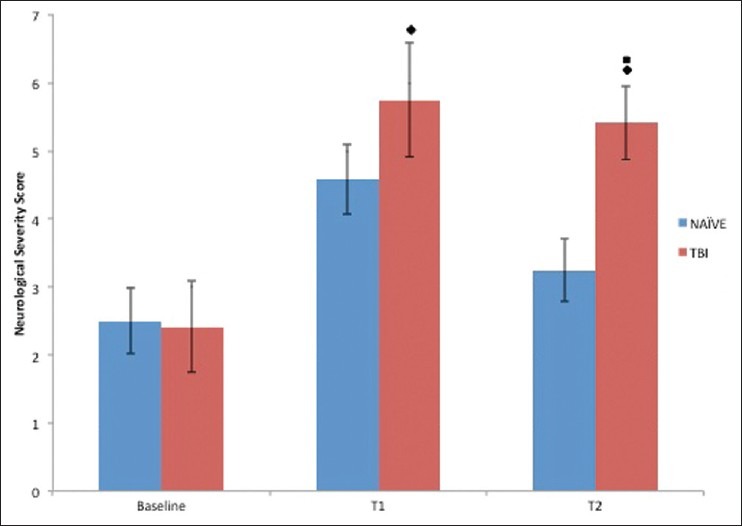
Effect of TBI on neurological severity score– revised (NSS-R): Neurological severity is determined using a 10-item test with a score of 0, 1, or 2, given on each task. A greater score for a rat indicates more neurobehavioral impairment. All animals were evaluated at baseline (BL), day 3 (T1) and day 5 (T2) postinjury. Time matched naïve animals were also evaluated at BL, T1 and T2 time period. ■ P < 0.05, naïve compared with TBI at T2, ♦ P < 0.05, differed from BL (either T1 or T2)
Hematoxylin and eosin staining
Figure 2 depicts the H and E immunosatined brain section from TBI and naïve rats. In comparison to naïve rats [Figure 2a], the injured brain section [Figure 2b] is showing the hippocampal commissure, which is the anterior part of the white matter of the hippocampus, and is inverted v shaped fiber bundle at the center of the tissue. Obvious histological injury is only on the cortex, which is at the right upper part of the ‘TBI’ section on the cortex. A breach in the surface of the cortex and an increase in hematoxylin stained (blue) nuclei ‘newer’ cells in the lesion suggest a minor injury on the surface and possibly the healing process through the generation of new cells.
Figure 2.
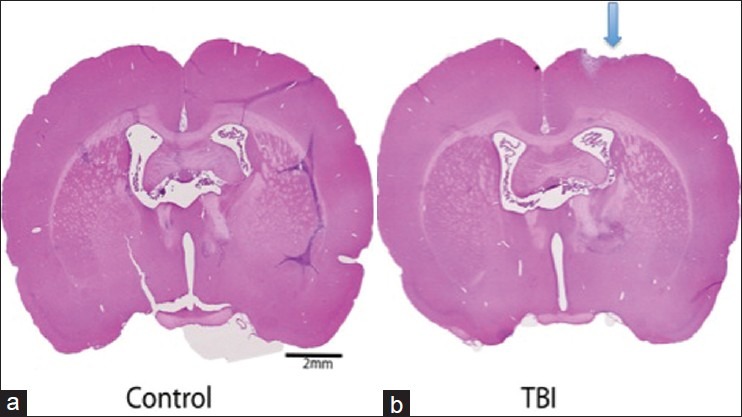
(a,b) Histochemical examination of TBI: Comparison of hematoxylin and eosin staining in brain sections obtained from naïve and post TBI. In comparison to naïve, the histological injury is only seen as a breach in the surface of the cortex, which is at the right upper part of the TBI
Expression levels of 1,448 of 1,500 mitochondrion-neuron focused genes are informative
Expression clusters of mitochondria-focused genes were obtained from both TBI and naïve groups of animals. Total RNA samples were extracted from the hippocampus of rats with or without TBI and labeled for triplicate microarray experiments using our recently developed rMNChip. To avoid misclassification, each of 1,500 genes on rMNChip was measured nine times (three identical probes per microarray and three microarray experiments per specimen), which generated reliable expression data for further analysis. The microarray data of spots across all gene chips used for RNA samples were filtered by uniform statistic and bioinformatic criteria described previously,[10] which generated 1,448 genes with informative expression data. Figure 3 shows the box plots of RNA levels of 1,448 genes before and after the data normalization. The normalized data were used for unsupervised clustering analysis and identification of differentially expressed genes.
Figure 3.
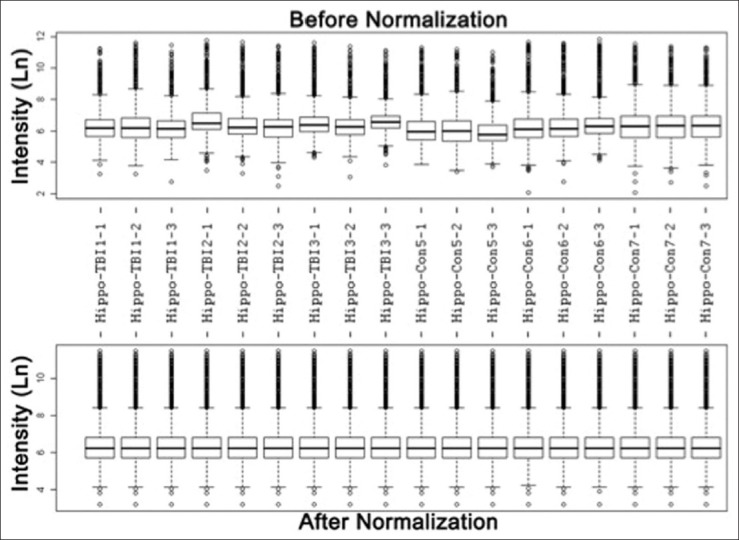
Box plots of expression data before and after normalization: The quantile normalization algorithms were used to adjust the values of the background-subtracted mean pixel intensities of triplicate measurements per microarray for each and every set of 1,500 genes. The expression levels of each of 1,500 genes in most of these hippocampus samples of six rats with or without TBI were measured by microarray experiments with both technical triplicates and experimental triplicates. In contrast to the prenormalization boxplots (top panel), the postnormalized box plots distribute in the same intervals with the same density center, indicating successful adjustment of data. The postnormalized data were used for further analysis
Expression patterns of 235 and 105 genes in hippocampus differentiate mTBI from naïve rats
By using ≥1.25-fold change in the average expression of the background-subtracted mean intensity ratios of a gene between TBI and naïve as criteria, 235 of 1,448 informative genes in the hippocampus are able to differentiate mTBI from the naïve rats based on the unsupervised cluster analysis [Figure 4a and b]. By increasing the stringency to ≥1.60-fold changes, the patterns of 105 genes differentiate mTBI from the naïve rats with much clear cluster results [Figure 4c].
Figure 4.
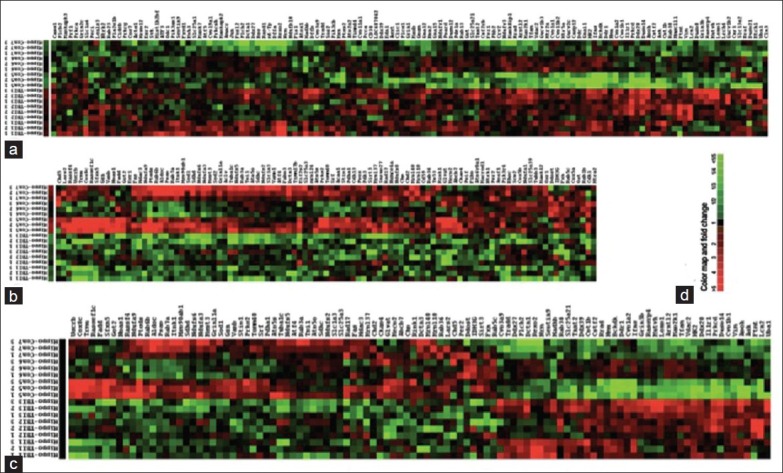
Dendrogram and heat maps of 235 gene expression in hippocampus of rat model of traumatic brain injury (TBI) and naïve. The dendrogram of unsupervised cluster of three TBI and three naïve (a) and (b) based on expression similarities of 235 rat mitochondria-neuron genes. (c) The dendrogram of upsupervised cluster of the same samples based on expression similarities of 105 genes. These heat maps classifies the hippocampus tissue samples into two groups. (d) Color map and fold changes indicated. Green: Down-regulation; red: Up-regulation; black: No change
Identification of 10 canonical molecular pathways with a significant number of dysregulated genes
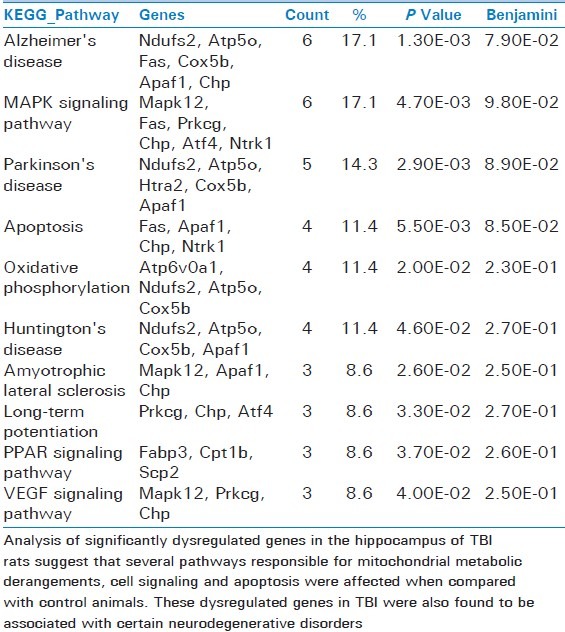
By applying differentially expressed genes to the NCBI DAVID Bioinformatics Resources, we identified 10 canonical molecular pathways including Alzheimer's disease, mitogen-activated protein kinase (MAPK) signaling, Parkinson's disease, apoptosis, oxidative phosphorylation, Huntington's disease, amyotrophic lateral sclerosis, long-term potentiation, peroxisome proliferator-activated receptors (PPARs) signaling, and vascular endothelial growth factor (VEGF) signaling pathways [Table 1].
Table 1.
Pathways with a significant number of dysregulated genes in rat traumatic brain injury hippocampus
DISCUSSION
The aim of this translational study was to examine the expression profiling of mitochondria-focused genes in the hippocampus of a rodent TBI model: To identify the gene clusters, the pathways related to either diseases or to related signal transduction pathways. These pathways are indicated as possible therapeutic targets for TBI. In this study we also searched for changes at individual gene expression level, which potentially may serve as a therapeutic target for the treatment of TBI and associated neurodegenerative diseases.
The rat model of lateral fluid percussion injury is one of the most accepted animal model to replicate human TBI in terms of biochemical and metabolic pathways.[26] However, in this rat model, it is difficult to diagnose the severity of brain injury because animals are anesthetized at the time of injury. Therefore, the level of consciousness cannot be used as the measurement of brain injury severity. However, we have compared the NSS-R scores prior to injury and then 3 and 5 days post TBI. Baseline scores were similar between groups, but by 3 and 5 days post TBI, the TBI animals had almost a 2-fold increase in their NSS-R score that remained elevated over both time points. This suggests an impaired neurobehavioral function similar to mild-moderate TBI in humans. Our H and E stained brain sections did not show any gross and/or histological damage in the hippocampus except a slight tissue distortion at the point of impact suggesting a mild TBI. FPI induces both focal and diffuse axonal injury, which often results in the failure of cellular metabolism and neuronal cell death. Therefore, we have compared the neuronal focused mitochondrial genes in the hippocampus of injured and naïve animals due to its key role in cognitive functions.
The advent of high-throughput genomics, such as gene microarrays, has led to an unprecedented surge of cross-disciplinary work between biology and statistics. However, quality assessment of microarray data is an important and often challenging aspect of gene expression analysis. The application of machine-learning techniques (also known as pattern-recognition techniques) to large biological data sets has been used to identify groups of functionally related genes,[11] to predict broader biological phenotypes and genetic interactions,[27] and to diagnosis post-traumatic stress disorder (PTSD).[14] Since the algorithmic basis of clustering, the application of unsupervised machine-learning techniques to identify the patterns inherent in a data set, is well established, we have used this approach in this current study. Unsupervised techniques are exploratory. It lets the data organize itself, and then we try to find biological meaning. This approach is to understand whole data and produce a visualization of the data. Briefly, like the methods used in other research areas, we first quantified the scanned image into numeric data amenable to statistical analysis. We filtered the raw data set, all spots across all gene chips of 16 rats (TBI and naïve) by removing the high-noise and low-signal spots. After that, we normalized the data to remove spatial variability, channel imbalances and inter array heterogeneity [Figure 3]. Then, we were able to use the cluster approaches to identify broad patterns. Using this approach we were able to produce a visualization of the data, hierarchical clustering. Our hierarchical clustering generated dendrograms demonstrated up- and downregulated gene clusters in all rats, including TBI and naïve [Figure 4]. These unsupervised analysis data indicated grouping of genes having similar expression patterns or clustering. The approaches are based on the assumption that the whole set of microarray data is a finite mixture of a certain type of distributions with different parameters. Application of the model-based algorithms to unsupervised clustering has been reported in PTSD study.[14] We hypothesize that unknown gene X is similar in expression to known genes A and B, or they are involved in the same/related pathway.
In a complex disorder such as mTBI, clustering a variety of risk factors can improve predictive value. Integrative biomarkers can be developed through superposition of mRNA. Clustering can prioritize functionally plausible candidate genes. Clustering aggregates information from multiple loci into association scores for pathways[28] and informs drug development by predicting therapeutic gene targets. Clustering can also identify pathways affected by an existing drug, either to predict the mode of action or suggest drug repurposing in which drugs developed for known disorders can serve as a new compound for TBI. Based on the unsupervised cluster results, 10 networks of dysregulated genes involved in neuron function and neurological disorders were identified and presented in Table 1. These pathways are involved in several networks related to neuronal disorders, such as Alzheimer′s, Huntington, and Parkinson's diseases. Our results also provide an important direction to monitor the TBI patients for these neurodegenerative disorders as a consequence of delayed traumatic brain injury due to progressive mitochondrial damage.[4,7,29]
In addition to the identification of mitochondrial genes for the neurodegenerative disorders, we have also documented several genes responsible for altered metabolic pathways, signaling and apoptotic pathways in TBI rats compared with naïve animals. Therefore, an orchestrated mitochondrial genes network and pathways mechanisms identified in TBI [Table 1] may also serve as future blood-based mitogenetic biomarkers for TBI.
Limitations of the study
The data obtained from three samples in each group is the bare minimum number allowing calculation of standard deviation and statistical significant difference permitting the comparison between TBI and control group. Although a larger sample size would strengthen this paper significantly, this small sample size is also the starting point for a pilot study to determine the reliability and validity of our mitochondrial focused gene-profiling data in response to TBI. This study will also allow us to estimate the sample size for a larger-scale study.
In this study, we have used naïve animals without craniotomy or anesthesia as control group. Our study also raises the question of the appropriate use of control animals for TBI induced by FPI that requires the craniotomy. In a recent study by Cole et al. (2011), craniotomy alone results in traumatic brain injury with morphological, biochemical, and behavioral correlates.[30] In addition, the significantly increased expression in cytokines after craniotomy alone can initiate numerous secondary cascades similar to the posttraumatic changes in brain physiology. Therefore, due to the diverse nature of injury caused by craniotomy procedures, it may be ben eficial to avoid this procedure as a control group and use naïve animals as controls.
CONCLUSION
Our data suggest that mitochondria-mediated cellular and molecular response to TBI is essential for the cell survival and in prevention of long-term neurodegenerative disorders associated with brain injury. Our findings also suggest that mitochondria-focused and TBI-responsive genes may play a key role in the pathogenesis of TBI. Meanwhile, the identification and validation of the dysregulated genes enhances our understanding of the cellular and molecular mechanisms underlying the pathophysiology of TBI. We expect more studies to test the clinical utility of mitochondria-focused genes as therapeutic targets for the treatment of TBI.
Analysis of significantly dysregulated genes in the hippocampus of TBI rats suggest that several pathways responsible for mitochondrial metabolic derangements, cell signaling and apoptosis were affected when compared with control animals. These dysregulated genes in TBI were also found to be associated with certain neurodegenerative disorders.
ACKNOWLEDGEMENT
We thank Dr. Xianzhang Hu and Dr. Xiaoxia Li for RNA isolation and purification. Dr. Fei Meng and Mr. Michael Tang for their services of microarray contract, and Mrs. Brandi Benford for her excellent surgical skills in performing TBI.
Footnotes
Source of Support: DARPA to PS. Award No. W911NF-11-1-0005
Conflict of Interest: No
REFERENCES
- 1.Hoge CW, Castro CA, Messer SC, McGurk D, Cotting DI, Koffman RL. Combat duty in Iraq and Afghanistan, mental health problems, and barriers to care. N Engl J Med. 2004;351:13–22. doi: 10.1056/NEJMoa040603. [DOI] [PubMed] [Google Scholar]
- 2.Warden D. Military TBI during the Iraq and Afghanistan wars. J Head Trauma Rehabil. 2006;21:398–402. doi: 10.1097/00001199-200609000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Center for Disease Control, NIH. Traumatic Brain Injury in the United States: A Report to Congress. 2001 [Google Scholar]
- 4.Giza CC, Hovda DA. The neurometabolic cascade of concussion. J Athl Train. 2001;36:228–35. [PMC free article] [PubMed] [Google Scholar]
- 5.Verweij BH, Muizelaar JP, Vinas FC, Peterson PL, Xiong Y, Lee CP. Impaired cerebral mitochondrial function after traumatic brain injury in humans. J Neurosurg. 2000;93:815–20. doi: 10.3171/jns.2000.93.5.0815. [DOI] [PubMed] [Google Scholar]
- 6.Sanders MJ, Sick TJ, Perez-Pinzon MA, Dietrich WD, Green EJ. Chronic failure in the maintenance of long-term potentiation following fluid percussion injury in the rat. Brain Res. 2000;861:69–76. doi: 10.1016/s0006-8993(00)01986-7. [DOI] [PubMed] [Google Scholar]
- 7.Royo NC, Conte V, Saatman KE, Shimizu S, Belfield CM, Soltesz KM, et al. Hippocampal vulnerability following traumatic brain injury: A potential role for neurotrophin-4/5 in pyramidal cell neuroprotection. Eur J Neurosci. 2006;23:1089–102. doi: 10.1111/j.1460-9568.2006.04642.x. [DOI] [PubMed] [Google Scholar]
- 8.Thompson HJ, Lifshitz J, Marklund N, Grady MS, Graham DI, Hovda DA, et al. Lateral fluid percussion brain injury: A 15-year review and evaluation. J Neurotrauma. 2005;22:42–75. doi: 10.1089/neu.2005.22.42. [DOI] [PubMed] [Google Scholar]
- 9.Cernak I, Wang Z, Jiang J, Bian X, Savic J. Ultrastructural and functional characteristics of blast injury-induced neurotrauma. J Trauma. 2001;50:695–706. doi: 10.1097/00005373-200104000-00017. [DOI] [PubMed] [Google Scholar]
- 10.Bai X, Wu J, Zhang Q, Alesci S, Manoli I, Blackman MR, et al. Third-generation human mitochondria-focused cDNA microarray and its bioinformatic tools for analysis of gene expression. Biotechniques. 2007;42:365–75. doi: 10.2144/000112388. [DOI] [PubMed] [Google Scholar]
- 11.Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–7. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 12.Madrigal JL, Olivenza R, Moro MA, Lizasoain I, Lorenzo P, Rodrigo J, et al. Glutathione depletion, lipid peroxidation and mitochondrial dysfunction are induced by chronic stress in rat brain. Neuropsychopharmacology. 2001;24:420–9. doi: 10.1016/S0893-133X(00)00208-6. [DOI] [PubMed] [Google Scholar]
- 13.Manoli I, Alesci S, Blackman MR, Su YA, Rennert OM, Chrousos GP. Mitochondria as key components of the stress response. Trends Endocrinol Metab. 2007;18:190–8. doi: 10.1016/j.tem.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Su YA, Wu J, Zhang L, Zhang Q, Su DM, He P, et al. Dysregulated mitochondrial genes and networks with drug targets in postmortem brain of patients with posttraumatic stress disorder (PTSD) revealed by human mitochondria-focused cDNA microarrays. Int J Biol Sci. 2008;4:223–35. doi: 10.7150/ijbs.4.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su YA, Zhang Q, Su DM, Tang MX. Rat mitochondrion-neuron focused microarray (rMNChip) and bioinformatics tools for rapid identification of differential pathways in brain tissues. Int J Biol Sci. 2011;7:308–22. doi: 10.7150/ijbs.7.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Zhou R, Li X, Ursano RJ, Li H. Stress-induced change of mitochondria membrane potential regulated by genomic and non-genomic GR signaling: A possible mechanism for hippocampus atrophy in PTSD. Med Hypotheses. 2006;66:1205–8. doi: 10.1016/j.mehy.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 17.Sharma P, Benford B, Li ZZ, Ling GS. Role of pyruvate dehydrogenase complex in traumatic brain injury and Measurement of pyruvate dehydrogenase enzyme by dipstick test. J Emerg Trauma Shock. 2009;2:67–72. doi: 10.4103/0974-2700.50739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heffner TG, Hartman JA, Seiden LS. A rapid method for the regional dissection of the rat brain. Pharmacol Biochem Behav. 1980;13:453–6. doi: 10.1016/0091-3057(80)90254-3. [DOI] [PubMed] [Google Scholar]
- 19.Jacobowitz DM. Removal of discrete fresh regions of the rat brain. Brain Res. 1974;80:111–5. doi: 10.1016/0006-8993(74)90726-4. [DOI] [PubMed] [Google Scholar]
- 20.Kamnaksh A, Kovesdi E, Kwon SK, Wingo D, Ahmed F, Grunberg NE, et al. Factors affecting blast traumatic brain injury. J Neurotrauma. 2011;28:2145–53. doi: 10.1089/neu.2011.1983. [DOI] [PubMed] [Google Scholar]
- 21.Hamm RJ. Neurobehavioral assessment of outcome following traumatic brain injury in rats: An evaluation of selected measures. J Neurotrauma. 2001;18:1207–16. doi: 10.1089/089771501317095241. [DOI] [PubMed] [Google Scholar]
- 22.Mahmood A, Lu D, Wang L, Li Y, Lu M, Chopp M. Treatment of traumatic brain injury in female rats with intravenous administration of bone marrow stromal cells. Neurosurgery. 2001;49:1196–203. [PubMed] [Google Scholar]
- 23.Marti M, Mela F, Fantin M, Zucchini S, Brown JM, Witta J, et al. Blockade of nociceptin/orphanin FQ transmission attenuates symptoms and neurodegeneration associated with Parkinson's disease. J Neurosci. 2005;25:9591–601. doi: 10.1523/JNEUROSCI.2546-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shohami E, Novikov M, Bass R. Long-term effect of HU-211, a novel non-competitive NMDA antagonist, on motor and memory functions after closed head injury in the rat. Brain Res. 1995;674:55–62. doi: 10.1016/0006-8993(94)01433-i. [DOI] [PubMed] [Google Scholar]
- 25.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 26.McIntosh TK, Vink R, Noble L, Yamakami I, Fernyak S, Soares H, et al. Traumatic brain injury in the rat: Characterization of a lateral fluid-percussion model. Neuroscience. 1989;28:233–44. doi: 10.1016/0306-4522(89)90247-9. [DOI] [PubMed] [Google Scholar]
- 27.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–13. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Wang C, Eberly LE, Caffo BS, Schwartz BS. Adaptive control of the false discovery rate in voxel-based morphometry. Hum Brain Mapp. 2009;30:2304–11. doi: 10.1002/hbm.20669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parihar MS, Brewer GJ. Mitoenergetic failure in Alzheimer disease. Am J Physiol Cell Physiol. 2007;292:C8–23. doi: 10.1152/ajpcell.00232.2006. [DOI] [PubMed] [Google Scholar]
- 30.Cole JT, Yarnell A, Kean WS, Gold E, Lewis B, Ren M, et al. Craniotomy: True sham for traumatic brain injury, or a sham of a sham? J Neurotrauma. 2011;28:359–69. doi: 10.1089/neu.2010.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]


