Abstract
Cryptococcus gattii, a pathogenic fungus historically appreciated to be endemic to tropical regions, was recognized to emerge in a more temperate zone of North America in the 1990s. Early reports focused on an outbreak that was first apparent on Vancouver Island (BC, Canada), involving both the veterinary and human population. More recently, it has been recognized that this organism is endemic to a wider geography in western North America, with recognized disease caused by unique molecular subtypes in both healthy and immunosuppressed human hosts and a variety of domestic and wild animals. A number of cases of disease caused by C. gattii isolates that are unrelated to the Vancouver Island–Pacific Northwest outbreak strains have also been recognized in different parts of the USA. As microbiology laboratories have historically not identified these organisms to the species level, our current understanding of the scope of this infection is probably an underestimate. Ongoing public health epidemiologic efforts will be facilitated by increased attention towards culture-confirmed diagnosis and species identification in clinical microbiology laboratories. Early experience presents a strong rationale for increasing diagnostic attention, with multiple clinical features that are unique to this infection, including variability in antifungal susceptibilities and a heightened need for aggressive management of inflammatory responses. Larger prospective studies to evaluate and optimize clinical management are needed.
Keywords: cryptococcosis, Cryptococcus gattii, North America
Cryptococcal disease is a serious threat to the health of both humans and animals, with the bulk of infections caused by the cosmopolitan member of the genus, Cryptococcus neoformans. This organism is particularly prevalent as a cause of severe CNS disease in HIV-infected people who reside in resource-poor areas of the world. A recent report that estimated the global burden of cryptococcosis using published studies found that there are over 600,000 attributable deaths each year, with the highest case rate in sub-Saharan Africa, which approximates 1 million per year [1]. As such, cryptococcosis is defined as one of the most important worldwide opportunistic fungal infections.
Although C. neoformans has the widest geographic distribution, increased attention has been drawn to another species, Cryptococcus gattii, which holds the distinction of more frequently causing infection in people who have apparently healthy immune systems. Both of these yeasts are free-living in nature, with some variable environmental prevalence. Pigeon guano or other avian fecal matter is considered to provide a habitat conducive for C. neoformans growth, although the organism can be found in other soil and decaying wood [2,3]. C. gattii has been considered to be more geographically restricted, with associations typically reported from tree and plant materials in tropical regions that are rich in Eucalypts [4]. The tropical restriction and epidemiologic link to Eucalyptus trees has been challenged more recently with the emergence of a unique C. gattii subtype in the more temperate climates of western North America, and recovery of these organisms in other regions with more diverse vegetation [2,5,6]. Emergence of a multispecies outbreak of C. gattii infections in British Columbia (BC) and the Pacific Northwest (PNW) region of the USA defines it as a recently recognized ‘endemic mycosis’ in North America.
Microbiology
Pathogenic cryptococci are categorized into one of four subtypes (A–D) based on capsular antigens. Serotype A, the most common worldwide pathogen, is C. neoformans var grubi and D is C. neoformans var neoformans. Serotypes B and C are considered the distinct species, C. gattii. This species is further divided into molecular ‘VG’ types, with isolates falling into one of four different VG groups (VGI–IV), or, less commonly, intraspecies or intravarietal hybrids [7,8].
Understanding the epidemiology of cryptococcosis is made difficult by the fact that diagnoses are not routinely established with complete identification of the species involved. In some cases, diagnosis is only supported by antigen results, lacking culture. When the isolate is recovered from the site of disease, many clinical microbiology laboratories do not typically distinguish between C. neoformans and C. gattii. Some laboratories have started to use specialized screening methods to distinguish between C. gattii and C. neoformans, but this is not common despite commercial availability of L-canavanine glycine bromothymol blue medium. Growth on this medium allows for easy identification of C. gattii by virtue of colony color; one recent study showed that 25 out of 27 (93%) of C. gattii strains induced a blue color change, in contrast to none out of 86 of C. neoformans [9]. Hence, our current understanding of the distribution between C. neoformans and C. gattii is probably incomplete.
It is also important to note that there is some controversy over genotyping. In general, diagnostic laboratories use DNA finger-printing methods (e.g., M13 fingerprinting) that result in the so-called ‘VG’ fingerprints. However, this method of genotyping has a relatively low resolution; for instance, the VGIV genotype was actually found to contain two very distinct AFLP genotypes called AFLP7 and AFLP10 (and serotype C and B isolates, respectively) [10]. It is clear that our understanding is limited by the methods used in prior investigations, and much needs to be learned.
The Vancouver Island outbreak
A unique C. gattii isolate emerged as a cause of disease on Vancouver Island (BC, Canada) in humans and animals, with the first cases of this outbreak recognized in 1999. Very quickly after the first cases were reported on Vancouver Island, the Canadian public health department enabled reporting and tracking that would prove critical in capturing the scope and nature of development of this new multispecies outbreak. Investigators documented that the outbreak, which is currently ongoing and geographically spreading, caused disease with an unusually high incidence. It was noted that the incidence of C. gattii infection among people of Vancouver Island from 1999 to 2003 was between 8.5 and 37 cases per million residents per year, compared with a reported 0.94 cases per million residents in Australia [11,12].
Environmental scientists recovered C. gattii isolates from tree sources, soil, air, and fresh- and sea-water, with ‘hot spots’ noted in areas around the southern, coastal regions of Vancouver Island, especially Parksville [5]. The bulk of isolates type as VGII; analysis of isolates recovered from humans, animals and the environment demonstrated a ‘major’ VGII genotype (VGIIa) and a ‘minor’ genotype (VGIIb), with the VGIIa isolate molecularly unique compared with prior isolates worldwide [11,13,14]. VGI isolates were also recovered, albeit at a lower frequency [5].
By 2004, the infection was noted to have emerged on mainland BC as well, with cases documented in people who had no prior exposure to Vancouver Island, and the isolate was recovered from multiple environmental sources [5,12]. By 2010, 281 human cases had been recognized by the British Columbia Centres of Disease Control. The multispecies nature of the outbreak has also been notable. Infection and colonization has been documented in aquatic mammals, small wild and large animals, and domestic dogs and cats.
Why, and how, this infection emerged in this area of the world has been a matter of debate. Some investigators postulated that the VGIIb/minor genotype, which is similar to those previously recognized in Australia, may have been transported into the region in association with typical vegetation, with same-sex recombination yielding the unique VGIIa/major variant [11,13,14]. These isolates then found themselves in a temperate region that was made more hospitable to a ‘tropical’ fungus, possibly as a result of a warmed environment. It was also shown that these isolates may disperse geographically by transportation of people and objects, potentially on feet and tires of travelers in the region, and that forestry practices probably affect airborne concentrations and spread [5].
When did this outbreak first become appreciated in the USA? Despite documented ‘spread’ of the Vancouver Island isolate to mainland BC, early efforts to recover the isolate in the USA, by sampling on the Gulf islands as well as inland regions around the Puget Sound (WA) and northern Washington, failed to recover C. gattii from the environment [Marr KA, Personal Observations] [15]. Canadian investigators did recover the isolate from the environment in northern Washington, just south of the USA–Canadian border [12]. Some people first believed that this infection was limited to Canada. However, our understanding of the epidemiology of disease in the USA has been limited by diagnostic bias; although some local clinicians were aware of the outbreak in Canada, microbiology laboratories in US states had not routinely identified isolates to the species level, instead reporting ‘C. neoformans’ without serotype/species typing (to this day, this is the usual practice in most clinical microbiology laboratories). In response, we initiated efforts to identify cases in Seattle in 2005 [16]. We screened cases that were available from local hospitals to identify isolates that could be further evaluated to species level, with attention placed on evaluating isolates from people that were not infected with HIV; results of these efforts recovered one case of focal pulmonary disease, documented to be caused by C. gattii VGIIa, from a patient with a myelodysplastic syndrome who resided on Orcas Island [16]. Despite surveillance, we failed to recover the isolate from his home or surroundings on the north part of the island [Marr KA, Unpublished data]. Two cases of human disease also became apparent to the Oregon state department of health; these cases, dating back to 2004 and 2005, were subsequently shown to be C. gattii [12].
Although these cases were only first recognized in the USA in the mid-2000s, there is evidence that these isolates have been in our environment in western USA for a longer period of time. The outbreak strain of VGIIa is genetically similar to a C. gattii isolate that was submitted to the American-type culture collection (ATCC) from the CDC several decades ago, and another highly related isolate recovered from environmental sampling in the San Francisco Bay area in California in the 1990s (NIH444/CBS6956) [13]. The origin of the ATCC isolate is not clear. In way of anecdote, I was contacted (along with K Bartlett) several years ago by a patient from the Seattle area, who claimed to have been treated for C. gattii disease in the 1970s by physicians who were practicing at a local private hospital. Although he claimed to have contributed this isolate to the CDC, and hence ATCC, we were unable to confirm this history. Either way, I, and others, believe that C. gattii may have been ‘emerging’ for a longer period of time in a broader geography of the PNW. The detailed studies that our Canadian colleagues performed in BC were a critical step that served to initiate the awareness that has now generated an understanding of a more complex emerging infection involving western North America.
Current epidemiology of cryptococcal disease in North America
What we know about the current epidemiology of cryptococcal disease is largely the result of voluntary reporting of cases and submission of isolates. In 2008, we were asked to convene a group to write a ‘white paper’ summarizing the state of disease emergence in the PNW [17]. This group of academics and clinicians from the USA and Canada (including public health officials from the British Columbia Centres of Disease Control) were joined by representatives from the CDC and Washington and Oregon state public health systems to generate the C. gattii Public Health Working Group [18]. In 2009, this group took the critical step of formalizing surveillance of infections and developing an isolate repository housed at the CDC. In my opinion, the critical pieces that allowed for generation of an increased understanding of this outbreak included: establishment of reference laboratories in US states to distinguish C. gattii from C. neoformans; and multidisciplinary collaboration between medical and veterinary clinicians and public health officials. However, the limitations to our methods and data are obvious; most laboratories are still not routinely identifying these isolates to the species level, and all surveillance has been contingent on voluntary reporting. Hence, most of what we know still comes from case recognition from interested and aware clinicians, and communication to academic and public health reference laboratories. For this reason our understanding of the scope and distributions of this outbreak – and our understanding of the epidemiology of disease in the USA – is still critically incomplete.
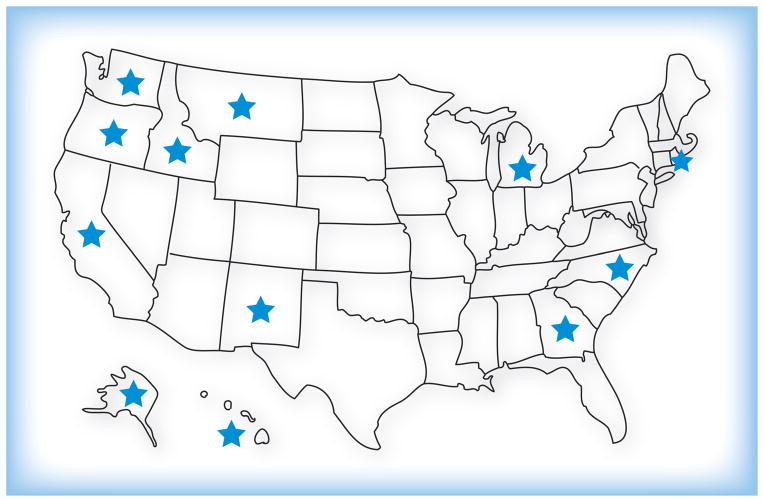
Despite limitations in our knowledge and methods, many human and animal cases have now been documented in a broad geography of the PNW, and detailed molecular typing, including whole genome sequencing, has documented that there are two unique VGII genotype isolates that have demonstrated clonal expansion, as well as multiple other C. gattii types observed to cause disease in the region [14,19]. The VGIIa (Vancouver Island)/major genotype causes the bulk of human and veterinary disease and has been recovered from the environment in BC, Washington and Oregon. Other cases have been reported in states in which there has been some question regarding the origin of the infection. Another unique VGII isolate, dubbed VGIIc, has been recovered from humans and the environment in Oregon; this isolate also appears to be expanding clonally [11,20]. Human infections have now been documented in people residing in other states, although the molecular type of the isolates recovered have been variable and unrelated to the outbreak strain, spanning VG-I, -II and -III genotypes (Figure 1) [21–24]. Some of these cases have had travel exposure, some have not. Hence, the outbreak is the complex result of multiple variables. There has been clonal expansion of the original VGIIa/major isolate from Vancouver Island, and establishment of another unique VGII strain, VGIIc, as well as identification of other C. gattii isolates having ‘non-outbreak’ strain types from distant states.
Figure 1. States in which human cases of Cryptococcus gattii infection have been recently documented.
Some of these may indicate travel-related exposure.
Adapted with permission from [45].
Closer examination of culture collections has now documented that C. gattii disease has been underappreciated as a cause of disease in HIV-infected people as well. A series of isolates recovered from AIDS patients in southern California was studied in detail to identify species; although the isolates were first identified as C. neoformans, a relatively large proportion were actually C. gattii [25,26]. These isolates were then recently examined by multilocus sequence typing and found to be VGIII, which is likely to be endemic to the region. Recent cases of CNS disease caused by C. gattii VGIII have been observed in people with HIV infection as well as in people with no recognized immunosuppression [Filler S, Pers. Comm.].
Our understanding of the outbreak is further enhanced by veterinary evaluations; detailed case reporting and surveillance studies have led to a critical understanding of the endemicity of the isolates. Early studies performed in the Vancouver Island outbreak region found C. gattii as a cause of disease and nasal colonization in both domestic cats and dogs, horses and wild animals (e.g., gray squirrels) [27]. Outbreaks and cases have been reported among domestic cat and dog populations in Washington in Oregon, llamas, horses and numerous aquatic mammals [5,12,27–30]. One interesting case of horizontal transmission in a pregnant porpoise was recently described [31].
Environmental scientists have now recovered C. gattii in both soil and water in tropical and temperate regions. In western North America, much of the distribution appears to overlap with the Coastal Douglas Fir and Coastal Western Hemlock biogeo-climatic zones, and C. gattii isolates have been recovered from decayed tree products representing over 50 different species [2,5,26]. Thus, it is likely that this infection is more globally distributed than previously appreciated.
Finally, a very interesting report described a case of C. gattii infection in a Japanese man who developed infection caused by a strain identical to the Vancouver Island VGIIa outbreak strain [32]. This occurred despite lack of travel to the endemic region in North America, raising the possibility that this isolate has spread to other regions of the world. Indeed, other investigators have isolated C. gattii from the environment in The Netherlands [33] and in a patient in southern Italy [34]. It appears evident that there is much more to learn about the epidemiology of this pathogenic fungus.
C. gattii causes a unique clinical syndrome
There is a large body of evidence that suggests that the disease caused by C. gattii is unique relative to that caused by C. neo-formans, with consideration of the hosts at risk, clinical presentation, antifungal susceptibilities and optimal management. Historically, this ‘tropical fungus’ was noted to cause disease proportionately more frequently in people who have no apparent cause of immunosuppression. Recent studies describing the clinical conditions in people who have developed C. gattii disease in North America suggests that roughly half do have some degree of immunosuppression, albeit different types and less often involving HIV compared with C. neoformans. In one Canadian study, risks involved receipt of oral corticosteroids, lung disease, older age, smoking, as well as underlying HIV and cancer [35]. The case patients identified in the USA have had variable underlying conditions, including receipt of solid organ transplants, hematological malignancies and rheumatologic conditions [22]. There are people who have developed this infection despite no apparent defect in immunity, both young and old. Some believe that there may be underlying subclinical immunological defects and/or ‘differences’ leading to an enhanced risk. Antibody responses to Cryptococcus were studied in one otherwise healthy 38-year-old woman. Although her total anti-Cryptococcus antibody levels increased months after treatment of disease, she failed to develop potentially protective Cryptococcus-specific IgG2 antibodies [36]. This observation evokes the hypothesis that some otherwise healthy people may have more subtle immunologic differences, such as specific antibody deficiencies, as an underlying risk for invasive disease.
While C. neoformans most frequently presents as a symptomatic meningoencephalitis, there appears to be much more diversity in the clinical presentation of C. gattii disease, in both immunocompromised and immunosuppressed hosts. While presentation after CNS disease development is common, C. gattii also frequently presents with underlying pulmonary disease, with concurrent pneumonia and focal lung masses, as well as pulmonary nodules [22,36,37].
There may be clinically significant differences in anti-fungal susceptibility of Cryptococcus species, with more apparent variability in azole in vitro susceptibilities among C. gattii isolates, especially those of the VGII lineage. Specifically, minimal inhibitory concentrations to fluconazole can be high (≥32 ug/ml) in VGII (a–c) isolates that have been recovered from humans and animals [10,38–41]. Whether this causes true microbial ‘resistance’ has not yet been clarified, although my anecdotal observation has been of frequent failure with C. gattii infection treated with fluconazole alone. There is also evidence that the clinical syndrome caused by C. gattii is unique with consideration of pressure management needs and neurologic outcomes. Cases of C. gattii infection presenting as fulminent intracranial hypertension have been reported [42]. Also, many of these patients present needs for longer-term CNS drainage and administration of corticosteroids in order to control recurrent symptoms and sterile arachnoiditis [43,44]. Anecdotally, I have found that persistent elevated CNS pressures are common, and people do well with short-term courses of steroids, especially in the setting of prolonged CNS inflammation. The reported outcomes in clinical series support this observation, with high numbers of people developing neurologic morbidity [22]. More systematic studies need to be performed to evaluate the course of disease and optimize management strategies.
Expert commentary
Several academic and public health efforts are ongoing to enable an understanding of the scope of this infection, both worldwide and in North America. Many critical questions have arisen regarding where these organisms have come from, how prevalent they are, overall risks, as well as optimal management of disease (and prevention). Although steps have been taken to enhance identification and reporting of cases, I believe that there are critical limitations in our approach to diagnosis, especially with regards to our widespread failure to identify this organism to the species level. It is important for clinicians to understand that most laboratories do not distinguish between C. neoformans/C. gattii species complex. As evidence accumulates that these species cause unique syndromes, with potential differences in antifungal and pressure management, there arises an increased need to be more precise with consideration of Cryptococcal diagnosis.
Five-year view
The Canadian public health department responded swiftly with consideration of enabling case identification and reporting, and the CDC is coordinating critical efforts to understand the microbial epidemiology of this disease. In the next 5 years, there needs to be more efforts established to capture accurate diagnoses, track cases, and enable prospective clinical cases to evaluate specific immunologic risks and management needs, especially with consideration of optimizing antifungal management and other therapeutic approaches. I believe that this infection will become more widely known as not only increasingly important in North America, but also in other regions of the world.
Key issues.
A unique variant of Cryptococcus gattii has emerged as an important cause of pulmonary and CNS disease in humans and animals in western North America.
Our epidemiologic understanding is limited by current (and historic) lack of routine testing to distinguish Cryptococcus species in clinical microbiology laboratories.
Disease caused by this species appears to require a unique approach, with observation of variable susceptibility to azole antifungals and excessive inflammatory responses associated with neurologic morbidity in human hosts.
Effective prevention and treatment of disease caused by Cryptococcus species requires incorporation of diagnostic testing in clinical microbiology laboratories, interaction with public health agencies, and prospective studies focused on specific host risks and outcomes.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure
KA Marr receives support from NIH K24 AI085118. She has served as an advisor for Astellas, Merck, Optimer and Pfizer, and receives research funding from Astellas, Merck, Pfizer and Schering Plough. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23(4):525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhary A, Rhandhawa SH, Prakash A, Meis JF. Environmental prevalence of Cryptococcus neoformans and Cryptococcus gattii in India: an update. Crit Rev Microbiol. 2012;38(1):1–16. doi: 10.3109/1040841X.2011.606426. [DOI] [PubMed] [Google Scholar]
- 3.Ellis DH, Pfeiffer TJ. Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet. 1990;336(8720):923–925. doi: 10.1016/0140-6736(90)92283-n. [DOI] [PubMed] [Google Scholar]
- 4.Ellis DH, Pfeiffer TJ. Natural habitat of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1990;28(7):1642–1644. doi: 10.1128/jcm.28.7.1642-1644.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Kidd SE, Chow Y, Mak S, et al. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific northwest of the United States. Appl Environ Microbiol. 2007;73(5):1433–1443. doi: 10.1128/AEM.01330-06. A comprehensive environmental sampling was performed. Results confirmed hot spots and characterized the type of soil in which Cryptococcus gattii was found. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kidd SE, Bach PJ, Hingston AO, et al. Cryptococcus gattii dispersal mechanisms, British Columbia, Canada. Emerg Infect Dis. 2007;13(1):51–57. doi: 10.3201/eid1301.060823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aminnejad M, Diaz M, Arabatzis M, et al. Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia. 2011;173(5–6):337–346. doi: 10.1007/s11046-011-9491-x. [DOI] [PubMed] [Google Scholar]
- 8.Bovers M, Hagen F, Kuramae EE, et al. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6(4):599–607. doi: 10.1111/j.1567-1364.2006.00082.x. [DOI] [PubMed] [Google Scholar]
- 9.McTaggart L, Richardson SE, Seah C, Hoang L, Fothergill A, Zhang SX. Rapid identification of Cryptococcus neoformans var. grubii, C neoformans var neoformans, and C gattii by use of rapid biochemical tests, differential media, and DNA sequencing. J Clin Microbiol. 2011;49(7):2522–2527. doi: 10.1128/JCM.00502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagen F, Illnait-Zaragozi MT, Bartlett KH, et al. In vitro antifungal susceptibilities and amplified fragment length polymorphism genotyping of a worldwide collection of 350 clinical, veterinary, and environmental Cryptococcus gattii isolates. Antimicrobial Agents Chemother. 2010;54(12):5139–5145. doi: 10.1128/AAC.00746-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Kidd SE, Hagen F, Tscharke RL, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc Natl Acad Sci USA. 2004;101(49):17258–17263. doi: 10.1073/pnas.0402981101. The first comprehensive study of the cryptococcosis outbreak and isolates recovered, demonstrating that the isolates comprise a unique outbreak strain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.MacDougall L, Kidd SE, Galanis E, et al. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg Infect Dis. 2007;13(1):42–50. doi: 10.3201/eid1301.060827. Examined environmental and clinical isolates recovered from Vancouver Island and other sites, including those from mainland British Columbia and several feline and human clinical isolates from Washington and Oregon. This was an important demonstration of spread of C. gattii both ‘off island’ and beyond, from the Pacific northwest USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Fraser JA, Giles SS, Wenink EC, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437(7063):1360–1364. doi: 10.1038/nature04220. A large study that raised multiple important observations and hypotheses regarding the unique genetics and virulence of C. gattii/VGII isolates. [DOI] [PubMed] [Google Scholar]
- 14••.Byrnes EJ, 3rd, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J Infect Dis. 2009;199(7):1081–1086. doi: 10.1086/597306. In this large and detailed evaluation of isolates using multilocus sequence typing, the new VGIIIc isolate was demonstrated from Oregon. Results provide clear documentation of a wider dispersal of C. gattii in the Pacific Northwest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser JA, Lim SM, Diezmann S, et al. Yeast diversity sampling on the San Juan Islands reveals no evidence for the spread of the Vancouver Island Cryptococcus gattii outbreak to this locale. FEMS Yeast Res. 2006;6(4):620–624. doi: 10.1111/j.1567-1364.2006.00075.x. [DOI] [PubMed] [Google Scholar]
- 16.Upton A, Fraser JA, Kidd SE, et al. First contemporary case of human infection with Cryptococcus gattii in puget sound: evidence for spread of the Vancouver island outbreak. J Clin Microbiol. 2007;45(9):3086–3088. doi: 10.1128/JCM.00593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Datta K, Bartlett KH, Baer R, et al. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg Infect Dis. 2009;15(8):1185–1191. doi: 10.3201/eid1508.081384. A comprehensive review of the emerging outbreak, produced by a multidisciplinary team of investigators in Canada and the USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Centers for Disease Control and Prevention (CDC) Emergence of Cryptococcus gattii – Pacific Northwest, 2004–2010. MMWR. 2010;59(28):865–868. This is a brief but important review of the cases recognized from the USA. [PubMed] [Google Scholar]
- 19•.Gillece JD, Schupp JM, Balajee SA, et al. Whole genome sequence analysis of Cryptococcus gattii from the Pacific Northwest reveals unexpected diversity. PLoS One. 2011;6(12):E28550. doi: 10.1371/journal.pone.0028550. A study in which whole genomes were evaluated, providing additional insight into the similarities and differences between C. gattii isolates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrnes EJ, 3rd, Li W, Lewit Y, et al. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6(4):E1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walraven CJ, Gerstein W, Hardison SE, et al. Fatal disseminated Cryptococcus gattii infection in New Mexico. PLoS One. 2011;6(12):E28625. doi: 10.1371/journal.pone.0028625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Harris JR, Lockhart SR, Debess E, et al. Cryptococcus gattii in the United States: clinical aspects of infection with an emerging pathogen. Clin Infect Dis. 2011;53(12):1188–1195. doi: 10.1093/cid/cir723. This is an important study that details more clinical characteristics of the cases recognized in the USA. [DOI] [PubMed] [Google Scholar]
- 23.McCulloh RJ, Phillips R, Perfect JR, Byrnes EJ, 3rd, Heitman J, Dufort E. Cryptococcus gattii genotype VGI infection in New England. Pediatr Infect Dis J. 2011;30(12):1111–1114. doi: 10.1097/INF.0b013e31822d14fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrnes EJ, 3rd, Li W, Lewit Y, et al. First reported case of Cryptococcus gattii in the Southeastern USA: implications for travel-associated acquisition of an emerging pathogen. PLoS One. 2009;4(6):E5851. doi: 10.1371/journal.pone.0005851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrnes EJ, 3rd, Li W, Ren P, et al. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog. 2011;7(9):E1002205. doi: 10.1371/journal.ppat.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Springer DJ, Chaturvedi V. Projecting global occurrence of Cryptococcus gattii. Emerg Infect Dis. 2010;16(1):14–20. doi: 10.3201/eid1601.090369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duncan C, Schwantje H, Stephen C, Campbell J, Bartlett K. Cryptococcus gattii in wildlife of Vancouver Island, British Columbia, Canada. J Wildl Dis. 2006;42(1):175–178. doi: 10.7589/0090-3558-42.1.175. [DOI] [PubMed] [Google Scholar]
- 28.Byrnes EJ, 3rd, Bildfell RJ, Dearing PL, Valentine BA, Heitman J. Cryptococcus gattii with bimorphic colony types in a dog in western Oregon: additional evidence for expansion of the Vancouver Island outbreak. J Vet Diagn Invest. 2009;21(1):133–136. doi: 10.1177/104063870902100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rotstein DS, West K, Levine G, et al. Cryptococcus gattiivgi in a spinner dolphin (Stenella longirostris) from Hawaii. J Zoo Wildl Med. 2010;41(1):181–183. doi: 10.1638/2009-0145.1. [DOI] [PubMed] [Google Scholar]
- 30.Miller WG, Padhye AA, van Bonn W, Jensen E, Brandt ME, Ridgway SH. Cryptococcosis in a bottlenose dolphin (Tursiops truncatus) caused by Cryptococcus neoformans var. gattii. J Clin Microbiol. 2002;40(2):721–724. doi: 10.1128/JCM.40.2.721-724.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norman SA, Raverty S, Zabek E, et al. Maternal–fetal transmission of Cryptococcus gattii in harbor porpoise. Emerg Infect Dis. 2011;17(2):304–305. doi: 10.3201/eid1702.101232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamoto K, Hatakeyama S, Itoyama S, et al. Cryptococcus gattii genotype VGIIa infection in man, Japan, 2007. Emerg Infect Dis. 2010;16(7):1155–1157. doi: 10.3201/eid1607.100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chowdhary A, Randhawa HS, Boekhout T, Hagen F, Klaassen CH, Meis JF. Temperate climate niche for Cryptococcus gattii in Northern Europe. Emerg Infect Dis. 2012;18(1):172–174. doi: 10.3201/eid1801.111190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iatta R, Hagen F, Fico C, Lopatriello N, Boekhout T, Montagna MT. Cryptococcus gattii infection in an immunocompetent patient from southern Italy. Mycopathologia. 2011 doi: 10.1007/s11046-011-9493-8. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 35•.MacDougall L, Fyfe M, Romney M, Starr M, Galanis E. Risk factors for Cryptococcus gattii infection, British Columbia, Canada. Emerg Infect Dis. 2011;17(2):193–199. doi: 10.3201/eid1702.101020. This study provides more insight into the clinical presentation and risks for C. gattii disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrett L, Marr KA, West S, Allada G. 74-year-old man from the Pacific Northwest with fever and a lung mass. Chest. 2011;140(3):814–817. doi: 10.1378/chest.10-2964. [DOI] [PubMed] [Google Scholar]
- 37.Dewar GJ, Kelly JK. Cryptococcus gattii: an emerging cause of pulmonary nodules. Can Respir J. 2008;15(3):153–157. doi: 10.1155/2008/392350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Iqbal N, DeBess EE, Wohrle R, et al. Correlation of genotype and in vitro susceptibilities of Cryptococcus gattii strains from the Pacific Northwest of the United States. J Clin Microbiol. 2010;48(2):539–544. doi: 10.1128/JCM.01505-09. This is the first study that clearly demonstrates a difference between geometric MICs to fluconazole according to specific C. gattii subtype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trilles L, Meyer W, Wanke B, Guarro J, Lazera M. Correlation of antifungal susceptibility and molecular type within the Cryptococcus neoformans/C. gattii species complex. Med Mycol. 2012;50(3):328–332. doi: 10.3109/13693786.2011.602126. [DOI] [PubMed] [Google Scholar]
- 40.Khan ZU, Randhawa HS, Chehadeh W, Chowdhary A, Kowshik T, Chandy R. Cryptococcus neoformans serotype A and Cryptococcus gattii serotype B isolates differ in their susceptibilities to fluconazole and voriconazole. Int J Antimicrobial Agents. 2009;33(6):559–563. doi: 10.1016/j.ijantimicag.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Chong HS, Dagg R, Malik R, Chen S, Carter D. In vitro susceptibility of the yeast pathogen cryptococcus to fluconazole and other azoles varies with molecular genotype. J Clin Microbiol. 2010;48(11):4115–4120. doi: 10.1128/JCM.01271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bromilow J, Corcoran T. Cryptococcus gattii infection causing fulminant intracranial hypertension. Br J Anaesthesia. 2007;99(4):528–531. doi: 10.1093/bja/aem208. [DOI] [PubMed] [Google Scholar]
- 43.Phillips P, Chapman K, Sharp M, et al. Dexamethasone in Cryptococcus gattii central nervous system infection. Clin Infect Dis. 2009;49(4):591–595. doi: 10.1086/603554. [DOI] [PubMed] [Google Scholar]
- 44.Lane M, McBride J, Archer J. Steroid responsive late deterioration in Cryptococcus neoformans variety gattii meningitis. Neurology. 2004;63(4):713–714. doi: 10.1212/01.wnl.0000134677.29120.62. [DOI] [PubMed] [Google Scholar]
- 45.Marr KA, Datta K, Pirofski LA, Barnes R. Cryptococcus gattii infection in healthy hosts: a sentinel for subclinical immunodeficiency? Clin Infect Dis. 2012;54(1):153–154. doi: 10.1093/cid/cir756. [DOI] [PubMed] [Google Scholar]