Abstract
Induced pluripotent stem cells (iPSCs) hold great promise for autologous cell therapies, but significant roadblocks remain to translating iPSCs to the bedside. For example, concerns about the presumed autologous transplantation potential of iPSCs have been raised by a recent paper demonstrating that iPSC-derived teratomas were rejected by syngeneic hosts. Additionally, the reprogramming process can alter genomic and epigenomic states, so a key goal at this point is to determine the clinical relevance of these changes and minimize those that prove to be deleterious. Finally, thus far few studies have examined the efficacy and tumorigenicity of iPSCs in clinically relevant transplantation scenarios, an essential requirement for the FDA. We discuss potential solutions to these hurdles to provide a roadmap for iPSCs to “jump the dish” and become useful therapies.
The goal of stem cell-based regenerative medicine is to treat disease states using cells, including the differentiated progeny of pluripotent stem cells (PSCs), as the therapeutic modality. In this way, regenerative medicine has the potential to transform conventional medicine, which has been dominated by surgery and drugs for centuries. The pluripotent nature of human embryonic stem cells (hESCs), which allows their potential use to repair almost any tissue, is only beginning to be harnessed for human therapies. Goldring et al. (2011) have recently reviewed safety issues pertaining to a range of promising stem cell-based therapeutics, including three clinical trials using ESCs to repair nerve cells and retinal pigment cells, which are not amenable to replacement by adult stem cells. However, three key issues have slowed the potential clinical use of hESCs: ethical issues, because a human blastocyst must be used to create the lines; immunological issues, because hESCs would be used for allotransplants; and safety issues, because hESCs can form teratomas and sometimes other, more malignant tumors.
When human induced pluripotent stem cells (hiPSCs) were first reported (Takahashi et al., 2007), part of the tremendous excitement surrounding them was their high level of similarity to hESCs, but at the same time, iPSCs had key potential advantages over hESCs. They seemed poised to avoid two out of the three central challenges facing the clinical use of hESCs: ethical and immune rejection issues. By using iPSCs for potential future regenerative medicine therapies, patients could, at least in theory, be given autologous transplants of iPSC-derived cells without using a human blastocyst and without immunosuppressive therapy. Not surprisingly, in the almost 5 years since the initial publication on murine iPSCs (miPSCs) (Takahashi and Yamanaka, 2006), as we have learned a great deal more about iPSCs, clinical expectations have become more realistic. While iPSCs are undoubtedly remarkably similar to hESCs, some laboratories report a number of differences that cast doubt upon the complete equivalence of the two cell types. In addition, iPSCs have their own unique issues that present different kinds of roadblocks to their future use in regenerative medicine therapies. These include the use of oncogenes for reprogramming and the time required to produce and characterize a new iPSC line, which may render autologous hiPSCs inherently unsuitable to treat acute conditions such as myocardial infarction and spinal cord injury. Even the immune tolerance of autologous iPSCs has recently been called into question (Zhao et al., 2011). At the same time, the tremendous potential of iPSCs for disease modeling has generated a great deal of excitement about iPSC-based “disease models in a dish” (Saha and Jaenisch, 2009). The crucial question facing the iPSC field at this time is whether iPSCs can escape the confines of the dish and go beyond disease modeling to get to the clinic to more directly help patients, as was originally hoped. Here we outline the main hurdles facing translation of iPSCs to the bedside and discuss the most promising solutions.
Immunity Issues
One of the most exciting aspects of the development of iPSCs was their potential use for patient-specific autologous transplants. While this remains an important potential attribute of iPSCs and their derivatives, enthusiasm was tempered a bit recently by the report of Zhao et al. (2011) who found that while murine ESC (mESC)-derived teratomas were accepted by syngeneic recipients, teratomas derived from miPSCs were rejected with massive CD4+ T cell infiltration. What might be the cause of this rejection in what should be a syngeneic context? It was not a result of MYC-based reprogramming or transgene integration, as miPSCs generated without MYC and with nonintegrating episomal vectors also encountered a significant immunologic response. Rather, the immunogenicity was apparently caused by overexpression of a few specific genes in miPSC-derived teratomas, suggesting that subtle epigenetic changes could have important therapeutic consequences. However, for many reasons the jury is still out on the immunity issue. We would argue that the focus of the Zhao study only on teratomas might very well have greatly overestimated the likelihood of autologous iPSCs to elicit an immune response. Because some tumors can be highly immunogenic, the teratoma context may confer an enhanced immunogenicity upon iPSC derivatives that does not manifest in iPSC-derived normal tissues. At least one of the overexpressed genes, HORMAD1, is expressed in developing germ cells and has been characterized as a tumor-specific antigen (Chen et al., 2005). Its expression could therefore be a result of germ cell differentiation within the teratoma, or a result of the tumor formation process itself, rather than an inherent characteristic of the iPSC lines studied. Teratoma assays require injecting large numbers of undifferentiated cells, which is very different from the way the cells will be used clinically. Indeed, there are hints that iPSCs that have been predifferentiated in vitro do not share the immune-activating properties of teratomas. A study from the Jaenisch group in which iPSCs were used successfully to treat sickle cell anemia without immune rejection seems to suggest that in some circumstances, iPSC derivatives are not immunogenic (Hanna et al., 2007). However, in this study the recipient mice were subjected to both radiation and immunosuppression, making it more difficult to draw conclusions. iPSC immunogenicity is a new, critical open question, but one that can be readily addressed by transplantation of normal cells or tissues derived from miPSCs into nonimmunodeficient, nonimmunosuppressed mice.
Because the Zhao study was only conducted in mice, another important open question is whether similar findings would be observed in a human context with hiPSCs. We predict that different iPSC lines will exhibit a range of immunotolerance in autologous hosts, so it may be fruitful to generate a panel of hiPSC lines from each patient and test them for autologous T cell reactivity in vitro. One potential way to begin addressing the immune tolerance of hiPSCs and their derivatives in vivo would be to study transplantation into mice with humanized immune systems capable of rejecting human allografts. Human peripheral blood mononuclear cells (PBMCs) can be used to reconstitute the immune system of immunodeficient mice, resulting in effective rejection of allogeneic human pancreatic islets (Vlad et al., 2008) and skin grafts (Issa et al., 2010). A similar experiment could be performed using hiPSCs autologous to the PBMCs in order to detect rejection of immune-matched iPSC grafts. It is currently unclear whether the immune capacity of these chimeric mice is sufficiently complex to mediate rejection of autologous iPSC derivatives that may differ only slightly from native human tissue, but if so, the results would begin to bridge the gap between immunologic experiments involving miPSCs in the murine immune context and clinical trials in human patients. We also predict that the specific tissue into which the stem cells are transplanted may greatly influence the extent of immune response in the recipient. Ultimately, if necessary, iPSC derivatives could be given as a transplant to patients with some degree of immunosuppression, such as the short-term leukocyte costimulatory blockade reported by Pearl et al. (2011) to enhance stem cell engraftment, but that would in some ways defeat the purpose of using iPSCs versus ESCs.
Genome Issues
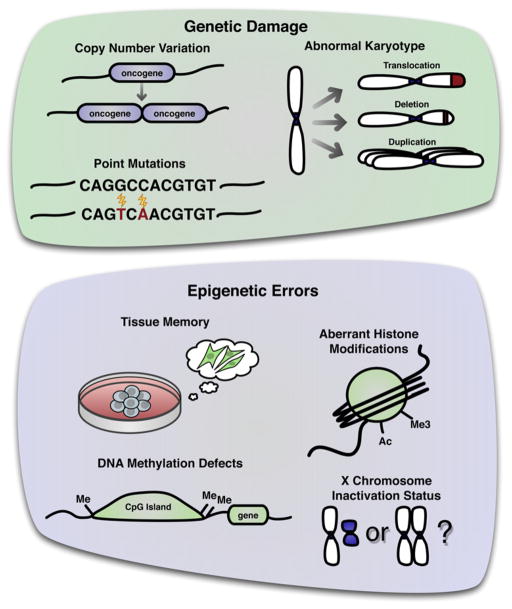
If iPSCs are to be used for therapies as we hope, we must understand the functional meaning, if any, of the different kinds of mutations that occur in iPSC lines to define a clinically acceptable level of genomic integrity. While some changes may be an inevitable result of extensively cultivating imperfect somatic cells, it is critical to determine their functional impact on the iPSCs, including any effect of mutational load on tumorigenicity, and how any risk of deleterious mutations can be minimized. Multiple kinds of genomic changes have been observed in hiPSCs, which may ultimately affect the therapeutic readiness of the cells (Figure 1). Chromosomal aneuploidy and translocations, megabase-scale duplications/deletions, and point mutations have all been described (Gore et al., 2011; Hussein et al., 2011; Laurent et al., 2011; Mayshar et al., 2010). As much as these mutations are cause for concern, nonetheless at this point there is no evidence proving or disproving that these mutations actually matter in a functional sense.
Figure 1. Genetic and Epigenetic Alterations Observed in hiPSCs.
Reprogramming can cause cells to have an abnormal karyotype (particularly gains of chromosome 12 and 17), copy number variation, and point mutations, all tending toward amplification/overexpression of oncogenes and deletion/inactivation of tumor suppressors. At the epigenetic level, reprogrammed cells can retain a memory of the starting tissue from which they were derived. The cells can exhibit DNA methylation defects, particularly at CpG island shores, and aberrant histone modifications. They can also vary in X chromosome inactivation status.
At the level of gross chromosomal abnormalities, karyotyping is routinely used to characterize genomic problems in hiPSCs as well as hESCs. Alternately, when gene expression profile data are available, these can also be used to identify chromosomal regions of overexpression or underexpression (Mayshar et al., 2010). These analyses point to chromosome 12 as a common duplication in both hiPSCs and hESCs after extended culturing (Baker et al., 2007; Mayshar et al., 2010). This chromosome contains the pluripotency genes Nanog and GDF3 as well as many cell cycle-related genes that may contribute to the selection of cells with these changes during culture. Duplication of chromosome 17 was previously reported to be an aberration specific to hESCs (Mayshar et al., 2010), but this duplication has also recently been observed in hiPSCs (Ben-David et al., 2011). These chromosomal anomalies are not a result of the reprogramming method used, because gains of whole or partial chromosomes have been identified in hiPSCs produced using a variety of techniques including nonintegrating methods such as synthetic mRNAs (Ben-David et al., 2011).
Karyotypes produced by G-banding can be used to detect large-scale chromosomal abnormalities such as aneuploidy and translocations. However, hiPSCs can also contain genomic changes at a smaller scale, undetectable by standard karyotyping, which nonetheless could have outsized consequences for cell biology. These smaller genetic alterations can be more labor-intensive to identify, requiring array- or sequencing-based high-throughput techniques. Extensive copy number variation (CNV) has been detected in early-passage hiPSCs using a high-resolution single nucleotide polymorphism (SNP) array. These CNVs tend to occur in common fragile sites, indicating that they are likely a result of replication stress (Hussein et al., 2011). It has been reported that in some cases, high-passage hiPSCs contain fewer CNVs than their early-passage precursors (Hussein et al., 2011). This suggests that most reprogramming-associated CNVs are detrimental to survival of the cells and are selected against during culture, but further study is required. The remaining CNVs that survive this selective pressure tend toward deletion of tumor suppressor genes and amplification of oncogenes (Laurent et al., 2011), highlighting the importance of monitoring these changes in cells that are intended for therapeutic use.
Still smaller genomic lesions have also been identified, including cancer-related point mutations in karyotypically normal hiPSCs (Gore et al., 2011). Some of these point mutations exist in the starting cell population, while the other mutations have a less clear origin and may occur during the reprogramming process and/or during expansion of the cells. Resolving when these mutations occur is an important priority because this data may shed light on not only their functional meaning, but also on potential methods to prevent their occurrence. All of the iPSC lines analyzed by Gore et al. were derived from fibroblasts, so it is quite likely that utilizing a more genomically protected cell source may minimize preexisting DNA mutations in the starting cell population. Dermal fibroblasts are predicted to be a relatively mutation-prone cell type given their high degree of exposure to mutagenic UV light. It is currently unclear whether any human somatic cell populations have significantly less mutational load than others, although there are some hints in the literature that suggest that this is likely the case. Somatic mutation rates in the mouse differ between organs, with higher rates of chromosomal aberrations in peripheral blood lymphocytes than in bone marrow (Tucker et al., 1999), and higher rates of point mutations in small intestine than in heart (Dollé et al., 2000). Mutation rates in accessible human tissues for reprogramming remain to be determined, but these data from the mouse suggest that cells from highly proliferative tissues such as blood and small intestine may contain a higher mutational load and therefore be less desirable as a cell source. In addition, a tissue’s relative protection from external factors may also play a role in the degree to which cells accumulate genetic lesions. For example, bone marrow cells may have a lower exposure to environmental toxins than blood or the gastrointestinal tract. As with CNVs, point mutations in iPSCs tend to cluster in cancer-associated genes, possibly pointing to connections between tumorigenic and pluripotency programming (Knoepfler, 2009). There have been no tumorigenicity studies comparing iPSCs with a relatively large number of mutations to less mutated iPSCs in a clinically relevant setting, so it is currently unclear what an acceptable mutation rate for a PSC line may be from a safety perspective. In addition, the potential functional importance of specific genomic alterations observed in iPSCs remains a key open question; it will be important to test whether the mutational load of iPSCs affects therapeutically relevant parameters such as tumorigenicity, immunogenicity, and impaired differentiation capacity. It also remains unknown if mutational load decreases with culture time as has been observed in at least some cases with CNVs (Hussein et al., 2011).
The difficulty of characterizing mutations and their effects, if any, on cellular functions illustrates the critical importance of developing reprogramming techniques that preserve genomic integrity. Introduction of reprogramming factors leads to increased DNA damage in the form of 8-oxoguanine, which is generally caused by oxidative stress, and histone γH2AX, a marker of double-strand breaks. DNA damage response elements including TP53/p53, CDKN2A/p16INK4a, and CDKN1A/p21CIP1 are also induced (Banito et al., 2009). Cells containing preexisting DNA damage, including irradiated cells and cells with short telomeres, tend to undergo p53-mediated growth arrest and apoptosis when put into reprogramming conditions (Marión et al., 2009). This may be one natural antitumorigenic mechanism to limit plasticity of cells containing DNA damage. Overriding these mechanisms enhances reprogramming efficiency, but potentially at the cost of allowing genetically damaged cells to be reprogrammed. The result of such strategies may be a higher proportion of unacceptably mutated iPSCs; indeed, Marión et al. (2009) observed that while knocking down p53 increases reprogramming efficiency, iPSCs produced from p53−/− fibroblasts contain more chromosomal breaks and fusions than iPSCs produced from wild-type fibroblasts. Conversely, reprogramming technologies that enhance innate genomic protection could conceivably produce fewer hiPSCs, but ones with fewer genomic modifications. If this speculation can be proven, it may be preferable from a clinical perspective to use less permissive reprogramming techniques that are designed to upregulate DNA repair processes and/or select for cells with intact DNA. Few studies reporting enhancement of reprogramming have examined whether their techniques allow cells with genomic changes to be reprogrammed, especially since some smaller genomic alterations have only been characterized in hiPSCs within the past year (Gore et al., 2011; Laurent et al., 2011). We would argue that focusing on developing methods to boost iPSC production efficiency is not enough. Instead, the goal should be to produce iPSCs with the fewest genomic alterations even if it is at low efficiency; for clinical purposes, theoretically all that is needed is a single bona fide iPSC line from a given patient.
One key way to minimize genomic damage is to exert control over oxidative stress during reprogramming and stem cell propagation. Interestingly, hiPSCs and hESCs share a similar ability to protect against genetic damage by limiting production of reactive oxygen species (ROS) and effectively clearing ROS from the cell (Armstrong et al., 2010). Compared to differentiated cells, partially reprogrammed cells also share genome-protective mechanisms with fully pluripotent cells, including maintenance of low intracellular superoxide levels and relatively few mitochondria (Armstrong et al., 2010). However, signs of oxidative damage appear even earlier than these genome-protective cellular changes, within a few days after reprogramming factor introduction (Banito et al., 2009), suggesting that genome protection may be amenable to enhancement especially during the first few days of reprogramming. Culture conditions can impact the prevalence of karyotypic abnormalities; for example, culture at physiological (5%) oxygen tension protects cardiac stem cells and hESCs from karyotypic changes (Li and Marbán, 2010). Physiological oxygen tension also enhances production of iPSCs compared with either normoxic (21%) or hypoxic (1%) conditions, increasing the efficiency and rate of reprogramming murine and human fibroblasts (Yoshida et al., 2009). However, iPSCs produced in 5% oxygen conditions have just as many point mutations as those produced at atmospheric oxygen (Gore et al., 2011), so it is still unclear whether hypoxic culture actually imparts any genomic protection during reprogramming. To mimic the effect of hypoxia, simply adding more antioxidants to the culture medium would seem like a plausible way to protect the genome. However, while addition of the anti-oxidant Vitamin C to culture medium has been reported to enhance reprogramming efficiency (Esteban et al., 2010), excessive antioxidant concentrations could actually increase the prevalence of genomic damage in stem cells by inhibiting DNA repair (Li and Marbán, 2010). A comprehensive study of genomic integrity of iPSCs produced at varied oxygen tension and antioxidant concentrations would help identify optimal conditions to reduce ROS damage while maintaining endogenous DNA repair at a high level.
Other methods of minimizing reprogramming-induced oxidative stress remain to be explored. These include ROS-limiting culture conditions known to enhance hESC pluripotency, such as reduced glucose levels in media (Crespo et al., 2010) or the addition of small molecule inhibitors of oxidative phosphorylation (Varum et al., 2009). Additionally, two hiPSC lines were reported to have lower expression levels of the antioxidant genes SOD2 and GSR compared with hESCs (Armstrong et al., 2010), suggesting the possibility that exogenously expressing these antioxidant genes during reprogramming may provide more ESC-like protection from oxidative damage. While the molecular mechanisms underlying reprogramming-associated DNA damage and repair are still being investigated, protecting the genome is clearly a critical and promising element of emerging cellular reprogramming strategies intended for clinical use. The importance of preserving genomic integrity has been a consideration from the very beginning of the iPSC field, as exemplified by the focus on removing MYC, a factor known to induce genomic instability (Felsher and Bishop, 1999). The relatively new data on mutations in iPSCs, including those produced in the absence of MYC, reinforce the potential importance of exploring innovative new approaches to genome preservation.
Epigenome Issues
Cellular reprogramming to pluripotency represents a herculean feat of epigenomic reorganization from a fully differentiated cell into an embryonic stem-like cell. The reprogrammed chromatin state is characterized at least in part by bivalent domains containing both transcriptionally activating (3meK4-H3) and repressive (3meK27) histone marks creating a poised gene state (Guenther et al., 2010), a state also observed in ESCs (Bernstein et al., 2006). DNA methylation also has to be reprogrammed across the genome, from a somatic cell state in which essentially all methylation occurs at CpG dinucleotides to a pluripotent state in which non-CpG sites account for 20%–30% of global DNA methylation (Lister et al., 2011). Not surprisingly, sometimes epigenomic reprogramming appears to be incomplete in iPSCs, especially at early passages soon after derivation (Lister et al., 2011; Pick et al., 2009; Polo et al., 2010; Stadtfeld et al., 2010). Several epigenetic differences between hiPSCs and hESCs have been described (Figure 1); however, it is unknown what effect these differences may have on differentiation or tumorigenicity of the cells. For example, differences in X chromosome inactivation (XCI) status have been described between different female hiPSC lines, usually with the implication that the best hiPSCs would have two active X chromosomes like their murine counterparts (Tchieu et al., 2010). Both hESC and hiPSC lines show heterogeneous XCI (Bruck and Benvenisty, 2011), which can be a dynamic process that changes with time in culture (Kiedrowski et al., 2011). In particular, derivation of hESCs in physiological (5%) oxygen conditions allows the production of cells with two active X chromosomes, while standard normoxic culture can induce irreversible XCI in these cells (Lengner et al., 2010). It is currently unclear whether these differences in XCI have any relation to the clinical safety and efficacy of the cells. It is hypothetically possible that hiPSCs that retain the XCI status of the parental fibroblasts may actually be safer because they avoid the possibility of aberrant X chromosome activation in their differentiated progeny, which is commonly seen in malignancies.
In addition to differences in XCI status, variation in imprinted gene expression has also attracted attention. Aberrant silencing of imprinted genes in miPSCs has been reported, which hinders the cells’ ability to contribute to chimeric mice (Stadtfeld et al., 2010). These differences may not ultimately impact the clinical utility of iPSCs if the imprinted gene products are not critical to the function of the cells’ differentiated progeny; however, proper expression of imprinted genes is critical during development of clinically relevant tissues such as the nervous system, suggesting that these genes may also be important during in vitro directed differentiation prior to transplantation. Also worrisome is the fact that some hiPSC lines overexpress cancer-associated imprinted genes (Pick et al., 2009). Based on the very limited data available, imprinting errors may turn out to be relatively rare events in hiPSCs, so screening a few cell lines for imprinted gene expression may be sufficient to identify correctly imprinted cells suitable for transplantation.
During reprogramming, DNA methylation patterns are massively altered to be remarkably similar, but not identical, to that of ESCs. Differential CpG methylation between iPSCs and ESCs falls roughly equally into two categories: (1) methylation patterns found in the iPSC parental cells, indicating epigenetic memory, and (2) methylation patterns specific to iPSCs that are found neither in ESCs nor the starting cells, many of which are shared among several independent iPSC lines (Lister et al., 2011). At least one common incompletely reprogrammed gene, C9orf64, appears to play a functional role in reprogramming, as RNAi ablation of this gene reduces reprogramming efficiency (Ohi et al., 2011). Differential methylation of CpG island shores appears to be a major way in which reprogramming alters the epigenetic landscape of cells (Doi et al., 2009). DNA methylation patterns in low-passage miPSCs retain a memory of the starting tissue, resulting in impaired differentiation toward unrelated lineages; differences in overall gene expression, methylation, and differentiation capacity between miPSCs derived from different tissues are subsequently eliminated by passage 16 (Polo et al., 2010). Incompletely reprogrammed genes tend to be isolated from other genes that are silenced during reprogramming, indicating that these early-passage differences may occur because isolated genes recruit silencing machinery less effectively (Ohi et al., 2011). However, some aberrant CG and non-CG methylation persists in hiPSCs even up to passage 65 and is retained after differentiation (Lister et al., 2011), suggesting that any abnormal gene expression resulting from imperfect reprogramming could persist even in the differentiated cell product. While epigenetic memory could be a hazard, it also has the potential to be useful. If epigenetic memory could be harnessed and maintained during long-term culture rather than obliterated, iPSCs could potentially be used to generate differentiated cell populations with greater ease and possibly greater purity than could be derived from ESCs. For example, hiPSCs derived from pancreatic islet beta cells show enhanced differentiation into insulin-producing cells compared with hESCs or hiPSCs derived from other cell types, even after moderate passaging (passage 10–20) in culture (Nur et al., 2011).
Rewriting histone modifications is a critical element of cellular reprogramming, as indicated by the plethora of small molecule reprogramming enhancers that act on chromatin-modifying enzymes that target histones. The reprogramming oncogene MYC also regulates global chromatin structure through its interaction with histone-modifying complexes including histone acetyltransferases (Knoepfler et al., 2006). This global effect of MYC may, in fact, be just as critical for enhancing reprogramming as its role as a classical transcription factor through which MYC contributes to maintenance and induction of pluripotency by repressing differentiation-associated gene expression (Smith et al., 2010; Varlakhanova et al., 2010). Reprogramming of human cells is enhanced by small-molecule histone deacetylase (HDAC) inhibitors such as valproic acid (Huangfu et al., 2008) and sodium butyrate (Mali et al., 2010), which facilitate chromatin remodeling events such as histone H3 lysine 9 acetylation. Inhibition of the G9a histone methyltransferase by BIX-01294 synergizes with sodium butyrate to reprogram human cells (Mali et al., 2010), likely by promoting an active chromatin state characterized by decreased histone methylation and increased acetylation. However, HDAC inhibitors have also been reported to induce double-strand breaks in DNA (Lee et al., 2010), so it remains to be determined whether these molecules themselves may induce karyotype abnormalities or other DNA sequence changes. More generally, it is assumed from the perspective of iPSC formation that small molecules such as HDAC inhibitors are either helpful or neutral, when the reality may be far more complex and could include deleterious effects.
How might a pluripotent epigenome be induced and preserved? Some of the epigenetic differences between hESCs and hiPSCs may reflect memory of the hiPSC parental tissue. However, some differences are almost certainly a result of the reprogramming process, since the use of isolated transcription factors is inherently quite different from generation of hESCs, which are derived from pluripotent ESCs that have yet to narrow their differentiation potential. It is possible that some of the epigenomic drugs such as those already used in iPSC production may have beneficial effects by preserving genomic integrity. The use of chromatin-modifying enzyme genes as reprogramming factors may lower the efficiency of iPSC production but give the bonus of producing iPSCs with fewer changes in their epigenomes. This important concept remains largely unaddressed in the field. Another possibility for producing iPSCs with more completely reprogrammed epigenomes is the use of miRNAs for reprogramming. Because of their pleiotropic function in rapidly regulating hundreds of mRNAs, reprogramming with miRNAs could potentially establish an ESC-like phenotype and epigenome more rapidly and completely than reprogramming with transcription factors. For example, the miRNA cluster miR302/367, which is strongly upregulated in hESCs compared with nonpluripotent cells (Laurent et al., 2008), is capable of rapidly reprogramming human fibroblasts to pluripotency in the absence of any exogenous transcription factors (Anokye-Danso et al., 2011; Miyoshi et al., 2011). These studies of miRNA-based reprogramming did not report any epigenomic information about the iPSC lines, so we look forward to an analysis of the rate of epigenomic aberrations in these cells compared with cells reprogrammed via transcription factors.
Tumorigenicity
The current gold standard test of pluripotency for hiPSCs and hESCs is teratoma formation (Müller et al., 2010), which is inherently a tumorigenesis assay. However, large numbers of undifferentiated cells implanted into an immunodeficient mouse bears little relevance to the in vivo environment that the cells or their differentiated derivatives will encounter in clinical use. In addition to teratoma assays, it will be vital to test the tumorigenicity of hiPSCs in more clinically relevant transplantation scenarios. Teratoma assays as commonly conducted in the stem cell field at present unfortunately have very little relevance to the tumorigenic potential of iPSCs in the context of human regenerative medicine therapies. The ideal preclinical tumorigenicity assay would be quite different from teratoma assays in that it would involve direct injection of iPSCs or their derivatives into the actual tissue of interest (e.g., injection into brain rather than subcutaneous or kidney capsule injection), the use of immunocompetent recipient mice—perhaps with the kind of transient immunosuppressive drug regimen used in human recipients, and rigorous assays for the presence of human cells (e.g., by qPCR for Alu repeats) at off-target organ sites. These three study components are all of high importance to the FDA, which is by comparison relatively uninterested in the ability of potential stem cell-based drugs to form teratomas in classical teratoma assays. The importance of such studies is illustrated by the fact that biotechnology companies currently in Phase I or Phase I/II trials for hESC-based therapies, as well as those conducting preclinical studies leading up to future Phase I trials, currently conduct such clinically relevant studies, often at the request of the FDA.
Preclinical testing of hiPSCs must therefore include clinically relevant transplantation scenarios that recapitulate the microenvironment cells will encounter in vivo, of which a few examples exist in the literature. Mesenchymal stem cells derived from hiPSCs engraft and induce functional improvement in a mouse hind limb ischemia model (Lian et al., 2010); the 21-day duration of transplantation follow-up is too short to assess tumorigenicity, but in separate assays, the cells did not exhibit teratoma-forming capacity after differentiation. Tsuji et al. (2010) classified miPSC lines as “safe” or “unsafe” based on residual teratoma forming capacity of neurospheres derived from the cells. This preselection step was sufficient to identify specific iPSC clones whose differentiated progeny engraft in injured murine spinal cord, participate in remyelination, and improve locomotor function without tumor formation. Of note, although one “unsafe” iPSC line did not produce teratomas in mouse spinal cord, it did produce clusters of Nanog+ cells; this highlights the importance of analyzing mice for other signs of tumorigenesis in addition to teratoma formation. It is not widely appreciated that hESCs also have been shown to have the potential, albeit somewhat limited, to form tumors beyond teratomas, including malignant tumors in SCID mice bearing engrafted human fetal tissue (Shih et al., 2007). Hepatic progenitors differentiated from retrovirus-derived human iPSCs were shown to engraft and regenerate cirrhotic mouse liver, with no evidence of tumor formation after a relatively lengthy 7 month follow-up (Liu et al., 2011). This lack of tumorigenicity may be partly due to efficient (>90%) differentiation to hepatic lineages; however, the study used intravenous injection (a method that lead to substantial cell loss in the lung) of an already relatively low number of cells (0.1–2 × 106 per mouse), so the lack of tumors may also be due to the delocalized route of administration and minimal effective cell dose. Swistowski et al. (2010) found that hiPSC-derived dopaminergic neurons engraft and improve function in a rat model of Parkinson’s disease with no evidence of teratoma formation at 12 weeks. A similar study of hiPSCs using a different differentiation protocol found proliferative nestin+ precursor cells in the rat brain (Cai et al., 2010), suggesting that the degree of differentiation achieved before transplantation may be a critical variable and that partially differentiated iPSC-derived progenitor cells could still form nonteratoma tumors if their proliferation is uncontrolled.
Much research has focused on removing or replacing the potent oncogene MYC in reprogramming in an effort to reduce tumorigenicity. MYC can be omitted or replaced by small molecules that target chromatin-modifying proteins and/or signaling pathways, yielding satisfactory levels of reprogramming. Substituting the MYCL1/L-Myc isoform is reported to reduce tumor formation in miPSC-derived chimeric mice (Nakagawa et al., 2010). Complicating the matter, endogenous MYC clearly also plays a role in iPSCs, repressing differentiation toward endodermal lineages in miPSCs at least in part by repressing expression of Gata6 (Smith et al., 2010). However, MYC is just the tip of the oncogenic iceberg. All known reprogramming-inducing genetic factors also have links to cancer, many of which are still being elucidated. KLF4 can function as either an oncogene (Wei et al., 2010) or a tumor suppressor (Zhao et al., 2004) in human cancers, depending on the cellular context. LIN28 contributes to a variety of human cancers by repressing expression of the let-7 family of miRNAs (Viswanathan et al., 2009). SOX2 functions as a potent oncogene in breast (Chen et al., 2008), lung, and esophageal cancers (Bass et al., 2009), while aberrant POU5F1 expression has been observed in osteosarcoma (Gibbs et al., 2005) and pancreatic cancer (Wen et al., 2010). Nanog is overexpressed in germ cell tumors (Hoei-Hansen et al., 2005), and its expression correlates with pathological grade in ovarian cancer (Pan et al., 2010). The miRNA cluster miR302/367 is overexpressed in germ cell tumors (Murray et al., 2010) and increases the growth of hESC-derived teratomas (Barroso-delJesus et al., 2011), suggesting an oncogenic role.
Concern has been raised about using integrated viral vectors to generate hiPSCs destined for the clinic, due to the possibility of insertional mutagenesis and reactivation of silenced reprogramming factors upon differentiation. To address this issue, several nonintegrating reprogramming techniques have been reported, including the use of episomal vectors (Yu et al., 2009), proteins (Kim et al., 2009), mRNAs (Warren et al., 2010), and miRNAs (Anokye-Danso et al., 2011). However, even transient overexpression of these oncogenes may produce lasting tumorigenic changes in the cells if they cause genomic instability. These potential problems may not be analogous to any process that occurs during hESC derivation. Consequently, rigorous preclinical testing is vital to the future success of iPSC-based therapies.
Conclusions
Thus far, much of the focus in the iPSC field has been on developing the most efficient methods for making iPSCs from a variety of parental cells, including those from patients who exhibit specific disease states. We argue for a shift in priorities. Future studies of hiPSCs should increase focus on issues most relevant to eventual clinical use of the cells, such as understanding the potential immunogenicity of autologous transplants, preserving genomic and epigenomic integrity during cellular reprogramming, and addressing tumorigenicity using clinically relevant transplantation protocols and not just teratoma assays. Key to this process will be two major goals: (1) studying the functional meaning of the genomic and epigenomic alterations described herein to define acceptable levels of changes, and (2) developing more rapid, accurate methods to screen iPSC lines for potentially unacceptable abnormalities. High-throughput techniques including microarray analysis to detect aberrant gene expression, SNP genotyping and comparative genomic hybridization to detect copy number changes associated with tumorigenicity, and resequencing of cancer-related genes to detect point mutations may be necessary to characterize iPSC lines for clinical use. Functional assays such as transplantation in an animal model, whether teratoma assays or, preferably, a more clinically relevant transplantation scenario, are other, more direct possible approaches to characterize the tumorigenic potential of a stem cell line. While molecular diagnostics alone do not have sufficient predictive power to be used as stand-alone tools for evaluation of tumorigenicity or metastatic potential of stem cell lines, they are rapidly evolving and may have substantial benefit when combined with other, more functional assays. More information could also be extracted from existing assays; for example, putative teratomas could be stained for markers of proliferation and pluripotency to quantify remaining levels of undifferentiated, highly proliferative cells possibly indicative of higher tumorigenic risk. However, all of these assays must be validated using clinically meaningful endpoints; for therapeutic purposes, a “healthy” iPSC will be defined by its capacity to generate functional differentiated cells with minimal risk of tumorigenesis or immunogenicity.
A broad study of the rate and nature of genomic abnormalities in hiPSCs produced by various reprogramming techniques (including the suggestions herein for preserving genomic and epigenomic integrity) would resolve questions related to the ability of these methods to preserve genomic integrity and/or select for cells with intact genomes. Taking these approaches may give iPSCs a boost in their trajectory, which has plateaued of late, to “jump the dish” and get into the clinic.
Acknowledgments
Funding for this work was provided by the California Institute for Regenerative Medicine via Grant RN2-00922-1 to P.K.
References
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, Przyborski S, Lako M. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 2010;28:661–673. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- Baker DEC, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-delJesus A, Lucena-Aguilar G, Sanchez L, Ligero G, Gutierrez-Aranda I, Menendez P. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J. 2011;25:1497–1508. doi: 10.1096/fj.10-172221. [DOI] [PubMed] [Google Scholar]
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Mayshar Y, Benvenisty N. Large-scale analysis reveals acquisition of lineage-specific chromosomal aberrations in human adult stem cells. Cell Stem Cell. 2011;9:97–102. doi: 10.1016/j.stem.2011.06.013. this issue. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bruck T, Benvenisty N. Meta-analysis of the heterogeneity of X chromosome inactivation in human pluripotent stem cells. Stem Cell Res. 2011;6:187–193. doi: 10.1016/j.scr.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Cai J, Yang M, Poremsky E, Kidd S, Schneider JS, Iacovitti L. Dopaminergic neurons derived from human induced pluripotent stem cells survive and integrate into 6-OHDA-lesioned rats. Stem Cells Dev. 2010;19:1017–1023. doi: 10.1089/scd.2009.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-T, Venditti CA, Theiler G, Stevenson BJ, Iseli C, Gure AO, Jongeneel CV, Old LJ, Simpson AJG. Identification of CT46/HORMAD1, an immunogenic cancer/testis antigen encoding a putative meiosis-related protein. Cancer Immun. 2005;5:9. [PubMed] [Google Scholar]
- Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, Sun L, Yang X, Wang Y, Zhang Y, Shang Y. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 2008;283:17969–17978. doi: 10.1074/jbc.M802917200. [DOI] [PubMed] [Google Scholar]
- Crespo FL, Sobrado VR, Gomez L, Cervera AM, McCreath KJ. Mitochondrial reactive oxygen species mediate cardiomyocyte formation from embryonic stem cells in high glucose. Stem Cells. 2010;28:1132–1142. doi: 10.1002/stem.441. [DOI] [PubMed] [Google Scholar]
- Doi A, Park I-H, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollé ME, Snyder WK, Gossen JA, Lohman PH, Vijg J. Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc Natl Acad Sci USA. 2000;97:8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs CP, Kukekov VG, Reith JD, Tchigrinova O, Suslov ON, Scott EW, Ghivizzani SC, Ignatova TN, Steindler DA. Stem-like cells in bone sarcomas: implications for tumorigenesis. Neoplasia. 2005;7:967–976. doi: 10.1593/neo.05394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring CEP, Duffy PA, Benvenisty N, Andrews PW, Ben-David U, Eakins R, French N, Hanley NA, Kelly L, Kitteringham NR, et al. Assessing the safety of stem cell therapeutics. Cell Stem Cell. 2011;8:618–628. doi: 10.1016/j.stem.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, Jaenisch R. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Hoei-Hansen CE, Almstrup K, Nielsen JE, Brask Sonne S, Graem N, Skakkebaek NE, Leffers H, Rajpert-De Meyts E. Stem cell pluripotency factor NANOG is expressed in human fetal gonocytes, testicular carcinoma in situ and germ cell tumours. Histopathology. 2005;47:48–56. doi: 10.1111/j.1365-2559.2005.02182.x. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Issa F, Hester J, Goto R, Nadig SN, Goodacre TE, Wood K. Ex vivo-expanded human regulatory T cells prevent the rejection of skin allografts in a humanized mouse model. Transplantation. 2010;90:1321–1327. doi: 10.1097/TP.0b013e3181ff8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski LA, Raca G, Laffin JJ, Nisler BS, Leonhard K, McIntire E, Mongomery KD. DNA methylation assay for X-chromosome inactivation in female human iPS cells. Stem Cell Rev. 2011 doi: 10.1007/s12015-011-9238-6. in press. Published online March 4, 2011. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS. Deconstructing stem cell tumorigenicity: a roadmap to safe regenerative medicine. Stem Cells. 2009;27:1050–1056. doi: 10.1002/stem.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, Fan JB, Loring JF. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Choy ML, Ngo L, Foster SS, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci USA. 2010;107:14639–14644. doi: 10.1073/pnas.1008522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Li TS, Marbán E. Physiological levels of reactive oxygen species are required to maintain genomic stability in stem cells. Stem Cells. 2010;28:1178–1185. doi: 10.1002/stem.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Q, Zhang Y, Zhang J, Zhang HK, Wu X, Zhang Y, Lam FFY, Kang S, Xia JC, Lai WH, et al. Functional mesenchymal stem cells derived from human induced pluripotent stem cells attenuate limb ischemia in mice. Circulation. 2010;121:1113–1123. doi: 10.1161/CIRCULATIONAHA.109.898312. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Anto-siewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kim Y, Sharkis S, Marchionni L, Jang YY. In vivo liver regeneration potential of human induced pluripotent stem cells from diverse origins. Sci Transl Med. 2011;3:82ra39. doi: 10.1126/scitranslmed.3002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Müller FJ, Goldmann J, Löser P, Loring JF. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell. 2010;6:412–414. doi: 10.1016/j.stem.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Murray MJ, Saini HK, van Dongen S, Palmer RD, Muralidhar B, Pett MR, Piipari M, Thornton CM, Nicholson JC, Enright AJ, Coleman N. The two most common histological subtypes of malignant germ cell tumour are distinguished by global microRNA profiles, associated with differential transcription factor expression. Mol Cancer. 2010;9:290. doi: 10.1186/1476-4598-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Takizawa N, Narita M, Ichisaka T, Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci USA. 2010;107:14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nur OB, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Jiao J, Zhou C, Cheng Q, Hu Y, Chen H. Nanog is highly expressed in ovarian serous cystadenocarcinoma and correlated with clinical stage and pathological grade. Pathobiology. 2010;77:283–288. doi: 10.1159/000320866. [DOI] [PubMed] [Google Scholar]
- Pearl JI, Lee AS, Leveson-Gower DB, Sun N, Ghosh Z, Lan F, Ransohoff J, Negrin RS, Davis MM, Wu JC. Short-term immunosuppression promotes engraftment of embryonic and induced pluripotent stem cells. Cell Stem Cell. 2011;8:309–317. doi: 10.1016/j.stem.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K, Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CC, Forman SJ, Chu P, Slovak M. Human embryonic stem cells are prone to generate primitive, undifferentiated tumors in engrafted human fetal tissues in severe combined immunodeficient mice. Stem Cells Dev. 2007;16:893–902. doi: 10.1089/scd.2007.0070. [DOI] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swistowski A, Peng J, Liu Q, Mali P, Rao MS, Cheng L, Zeng X. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells. 2010;28:1893–1904. doi: 10.1002/stem.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, Zack JA, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji O, Miura K, Okada Y, Fujiyoshi K, Mukaino M, Nagoshi N, Kitamura K, Kumagai G, Nishino M, Tomisato S, et al. Therapeutic potential of appropriately evaluated safe-induced pluripotent stem cells for spinal cord injury. Proc Natl Acad Sci USA. 2010;107:12704–12709. doi: 10.1073/pnas.0910106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JD, Spruill MD, Ramsey MJ, Director AD, Nath J. Frequency of spontaneous chromosome aberrations in mice: effects of age. Mutat Res. 1999;425:135–141. doi: 10.1016/s0027-5107(99)00036-6. [DOI] [PubMed] [Google Scholar]
- Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010;80:9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varum S, Momcilovic O, Castro C, Ben-Yehudah A, Ramalho-Santos J, Navara CS. Enhancement of human embryonic stem cell pluripotency through inhibition of the mitochondrial respiratory chain. Stem Cell Res. 2009;3:142–156. doi: 10.1016/j.scr.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad G, D’Agati VD, Zhang QY, Liu Z, Ho EK, Mohanakumar T, Hardy MA, Cortesini R, Suciu-Foca N. Immunoglobulin-like transcript 3-Fc suppresses T-cell responses to allogeneic human islet transplants in hu-NOD/SCID mice. Diabetes. 2008;57:1878–1886. doi: 10.2337/db08-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Wang L, Kanai M, Jia Z, Le X, Li Q, Wang H, Xie K. KLF4α up-regulation promotes cell cycle progression and reduces survival time of patients with pancreatic cancer. Gastroenterology. 2010;139:2135–2145. doi: 10.1053/j.gastro.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Park JY, Park KH, Chung HW, Bang S, Park SW, Song SY. Oct4 and Nanog expression is associated with early stages of pancreatic carcinogenesis. Pancreas. 2010;39:622–626. doi: 10.1097/MPA.0b013e3181c75f5e. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Krüppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]