Abstract
Despite the fact that ribosomal proteins are the constituents of an organelle that is present in every cell, they show a surprising level of regulation, and several of them have also been shown to have other extra-ribosomal functions, such in replication, transcription, splicing or even ageing. This review provides a comprehensive summary of these important aspects.
Keywords: protein synthesis, ribosome, ribosomal proteins, transcription, regulation, life span
Introduction
Protein synthesis requires accurate translation of the nucleotide sequence of messenger RNA (mRNA) to the amino acid sequence of a protein. This translation of mRNA to protein is carried out by the ribosome and transfer RNA (tRNA), along with other protein factors. In past years, studies on the structure of the ribosome have led us to understand this complex process of protein synthesis. The ribosome consists of two subunits, each of which is made up of ribosomal RNA (rRNA) and many ribosomal proteins. Structurally, ribosomes of prokaryotes and eukaryotes vary by the types of rRNA and protein molecules found in them. The prokaryotic 70S ribosome has a small 30S and a large 50S subunit. The 30S subunit consists of one 16S molecule of rRNA and about 21 proteins, while the 50S subunit consists of two rRNAs (5S and 23S) and 31 proteins. The eukaryotic 80S ribosome has a small 40S and a large 60S subunit. The 40S subunit consists of one 18S molecule of rRNA and about 33 proteins, whereas the 60S consists of three rRNAs (5S, 28S and 5.8S) and about 50 proteins [1].
During protein synthesis, the small ribosomal subunit plays a role in accurate codon-anticodon recognition between the mRNA and tRNA molecules, while the large subunit is mainly involved in the peptide bond formation of the growing amino acid chain. In addition, structural studies of the ribosome have now revealed that they are also involved in functions such as the translocation of tRNA and mRNA on the ribosome [2].
Apart from protein synthesis, many of the ribosomal proteins are shown to be involved in other cellular functions, independent of the ribosome [3]. Their first extra-ribosomal activity was observed for S1, as a replicase in the RNA phages, and numerous extra-ribosomal functions of these proteins have subsequently been discovered. This bifunctional tendency of ribosomal proteins can be explained by theories postulating the pre-existence of the ribosomal proteins as independent molecules before forming the components of the ribosome [3]. Another interesting functional aspect of the ribosomal proteins is their regulation. These proteins are shown to affect the mechanisms of development, apoptosis and ageing during their altered expression levels. In this review, information on the extra-ribosomal roles of these proteins is provided, along with information about their specific regulation in different cellular functions. Detailed lists of all functions and regulation are presented as Tables S1 (Table 4) and S2 (Table 5).
Table S1.
Function and regulation of eukaryotic small subunit ribosomal proteins
| Protein Name | Organism | Function | Reference | Find online at: |
|---|---|---|---|---|
| RPSA | Porcine | Candidate for binding and internalisation of externally added cellular prion protein in the gut | Knorr, C., Beuermann, C., Beck, J. and Brenig, B. (2007), 'Characterization of the porcine multicopy ribosomal protein SA/37-kDa laminin receptor gene family', Gene Vol. 395(1-2), pp. 135-143. | http://www.ncbi.nlm.nih.gov/pubmed/17434268 |
| RPS3A | Human | Cell apoptosis regulation | Naora, H. (1999), 'Involvement of ribosomal proteins in regulating cell growth and apoptosis: Translational modulation or recruitment for extraribosomal activity?', Immunol. Cell Biol. Vol. 77, pp. 197-205. | http://www.ncbi.nlm.nih.gov/pubmed/10361251 |
| RPS6 | Drosophila homologue of human S6 | Tumour suppressor in the haematopoietic system | Watson, K.L., Konrad, K.D., Woods, D.F. and Bryant, P.J. (1992), 'Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc. Natl. Acad. Sci. USA Vol. 89, pp. 11302-11306. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=50538 |
| RPS7 | Zebrafish | Mutations result in malignant peripheral nerve sheath tumour (zMPNST); RP genes may be 'haploinsufficient tumour suppressors' in zebrafish and cancer genes in humans | Amsterdam, A., Sadler, K.C., Lai, K., Farrington, S. et al. (2004), 'Many ribosomal protein genes are cancer genes in zebrafish', PLoS Biol. Vol. 2, p. E139. | http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371%2Fjournal.pbio.0020139&ct=1 |
| RPS8 | Zebrafish | Mutations result in malignant peripheral nerve sheath tumour (zMPNST); RP genes may be 'haploinsufficient tumour suppressors' in zebrafish and cancer genes in humans | Amsterdam, A., Sadler, K.C., Lai, K., Farrington, S. et al. (2004), 'Many ribosomal protein genes are cancer genes in zebrafish', PLoS Biol. Vol. 2, p. E139. | http://biology.plosjournals.org/perlserv/?request=get-document &doi=10.1371%2Fjournal.pbio.0020139&ct=1 |
| RPS9 | Human | Involved in retinal formation | Uechi, T., Tanaka, T. and Kenmochi, N. (2001), 'Complete map of the human ribosomal protein genes: Assignment of 80 genes to the cytogenetic map and implications for human disorders', Genomics Vol. 72, pp. 223-230. | http://www.ncbi.nlm.nih.gov/pubmed/11401437 |
| RPS10 | Arabidopsis thaliana | Developmental regulation | Majewski, P., Wołoszyńska, M. and Janńska, H. (2009), 'Developmentally early and late onset of Rps10 silencing in Arabidopsis thaliana: Genetic and environmental regulation', J. Exp. Bot. Vol. 60, pp. 1163-1178. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2657537&tool=pmcentrez |
| RPS13 | Human | Cell growth or proliferation regulation | Lai, M.D. and Xu, J. (2007), 'Ribosomal proteins and colorectal cancer', Curr. Genomics Vol. 8, pp. 43-49. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2474683 |
| RPS15 | Drosophila | Overexpression of S15a suppresses a utation in the Saccharomyces cerevisiae cdc33 gene, which encodes the cap-binding subunit of eukaryotic initiation factor 4F (eIF-4F); mutations of cdc33 lead to arrest in the cell cycle at the G1 to S transition. | Saeboe-Larssen, S. and Lambertsson, A. (1996), 'A novel Drosophila Minute locus encodes ribosomal protein S13', Genetics Vol. 143, pp. 877-885. | http://www.genetics.org/cgi/reprint/143/2/877 |
| Human | Role in nuclear export of 40S subunit precursors | Gazda, H., Sheen, M.R., Vlachos, A., Choesmel, V. et al. (2008), 'Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients', Am. J. Hum. Genet. Vol. 83, pp. 769-780. | http://www.ncbi.nlm.nih.gov/pubmed/19061985 | |
| RPS15A | Zebrafish | Mutations result in malignant peripheral nerve sheath tumour (zMPNST); RP genes may be 'haploinsufficient tumour suppressors' in zebrafish and cancer genes in humans | Amsterdam, A., Sadler, K.C., Lai, K., Farrington, S. et al. (2004), 'Many ribosomal protein genes are cancer genes in zebrafish', PLoS Biol. Vol. 2, p. E139. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=406397 |
| RPS18 | Arabidopsis thaliana | Developmental regulation | Lai, M.D. and Xu, J. (2007), 'Ribosomal proteins and colorectal cancer', Curr. Genomics Vol. 8, pp. 43-49. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2474683 |
| Arabidopsis thaliana | Mutation in S18 associated with growth retardation and abnormal leaf development | Naora, H. (1999), 'Involvement of ribosomal proteins in regulating cell growth and apoptosis: Translational modulation or recruitment for extraribosomal activity?', Immunol. Cell Biol. Vol. 77, pp. 197-205. | http://www.ncbi.nlm.nih.gov/pubmed/10361251 | |
| Zebrafish | Mutations result in malignant peripheral nerve sheath tumour (zMPNST); RP genes may be 'haploinsufficient tumour suppressors' in zebrafish and cancer genes in humans | Amsterdam, A., Sadler, K.C., Lai, K., Farrington, S. et al. (2004), 'Many ribosomal protein genes are cancer genes in zebrafish', PLoS Biol. Vol. 2, p. E139. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=406397 | |
| RPS19 | Ascaris lumbricoides | Developmental regulation | Lai, M.D. and Xu, J. (2007), 'Ribosomal proteins and colorectal cancer', Curr. Genomics Vol. 8, pp. 43-49. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2474683 |
| Human | Tumour progression, invasion, metastasis, differentiation' | Lai, M.D. and Xu, J. (2007), 'Ribosomal proteins and colorectal cancer', Curr. Genomics Vol. 8, pp. 43-49. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2474683 | |
| Human | Degeneration of retina | Uechi, T., Tanaka, T. and Kenmochi, N. (2001), 'Complete map of the human ribosomal protein genes: Assignment of 80 genes to the cytogenetic map and implications for human disorders', Genomics Vol. 72, pp. 223-230. | http://www.ncbi.nlm.nih.gov/pubmed/11401437 | |
| Human | Dimer acts as a monocyte chemotactic factor in phagocytic clearance of apoptotic cells | Naora, H. (1999), 'Involvement of ribosomal proteins in regulating cell growth and apoptosis: Translational modulation or recruitment for extraribosomal activity?', Immunol. Cell Biol. Vol. 77, pp. 197-205. | http://www.ncbi.nlm.nih.gov/pubmed/10361251 | |
| Zebrafish | Haematopoietic and developmental abnormalities | Danilova, N., Sakamoto, K.M. and Lin, S. et al. (2008), 'Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family', Blood Vol. 112, pp. 5228-5537. | http://www.ncbi.nlm.nih.gov/pubmed/18515656 | |
| RPS20 | Yeast | Overexpression of S20 suppresses temperature-sensitive RNA pol III (but no specificity?) | Hermann-Le Denmat, S., Sipiczki, M. and Thuriaux, P. (1994), 'Suppression of yeast RNA polymerase III mutations by the URP2 gene encoding a protein homologous to the mammalian ribosomal protein S20', J. Mol. Biol. Vol. 240, pp. 1-7. | http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WK7-45PV6 2P-1S&_user=4887109&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000062864&_version=1&_urlVersion=0&_userid=4887109&md5=88a77e1986 f7765e9374d649cc9b23a8 |
| Human | mRNA downregulated in onset of apoptosis in leukaemic cells | Naora, H. (1999), 'Involvement of ribosomal proteins in regulating cell growth and apoptosis: Translational modulation or recruitment for extraribosomal activity?', Immunol. Cell Biol. Vol. 77, pp. 197-205. | http://www.ncbi.nlm.nih.gov/pubmed/10361251 | |
| RPS21 | Drosophila | Acts as a translation initiation factor rather than as a core ribosomal protein | Török, I., Herrmann-Horle, D., Kiss, I., Tick, G. et al. (1999), 'Down-regulation of RpS21, a putative translation initiation factor interacting with P40, produces viable minute imagos and larval lethality with overgrown hematopoietic organs and imaginal discs', Mol. Cell Biol. Vol. 19, pp. 2308-2321. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=84023&tool=pmcentrez |
| RPS27A | Human | Cell growth or proliferation regulation | Ye, J.L. and Zhang, Y.Z. (2007), 'The connection between tumor and ubiquitin-ribosomal protein S27a, ubiquitin and ribosomal protein', Sheng Wu Gong Cheng Xue Bao Vol. 23, pp. 982-988. [Article in Chinese] | http://www.ncbi.nlm.nih.gov/pubmed/18257223?ordinalpos=5&itool=EntrezSystem2.PEntrez. Pubmed.Pubmed_Results Panel.Pubmed_DefaultReport Panel.Pubmed_RVDocSum |
| Human | Cell growth or proliferation regulation | Lai, M.D. and Xu, J. (2007), 'Ribosomal proteins and colorectal cancer', Curr. Genomics Vol. 8, pp. 43-49. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2474683 | |
| Human | Cell malignant transformation | Ye, J.L. and Zhang, Y.Z. (2007), 'The connection between tumor and ubiquitin-ribosomal protein S27a, ubiquitin and ribosomal protein', Sheng Wu Gong Cheng Xue Bao Vol. 23, pp. 982-988. [Article in Chinese] | http://www.ncbi.nlm.nih.gov/pubmed/18257223?ordinalpos=5&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum | |
| RPS28 | Yeast | Binds to the 3' UTR of its mRNA to stimulate its deadenylation and degradation | Badis, G., Saveanua, C., Fromont-Racinea, M. and Jacquie, A. (2004), 'Targeted mRNA degradation by deadenylation-independent decapping', Mol. Cell Vol. 15, pp. 5-15. | http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WSR-4CRXKG3-3&_user=4887109&_rdoc=1&_fmt=&_orig=search &_sort=d&view=c&_acct=C000062864&_version=1&_urlVersion=0&_userid=4887109&md5=9b5ba025da819e725850644ba547d47c |
| RPS29 | Human | Tumour suppression gene regulation | Lai, M.D. and Xu, J. (2007), 'Ribosomal proteins and colorectal cancer', Curr. Genomics Vol. 8, pp. 43-49. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2474683 |
| Human | Increases tumour supporessor activity of Krev-1' | Naora, H. (1999), 'Involvement of ribosomal proteins in regulating cell growth and apoptosis: Translational modulation or recruitment for extraribosomal activity?', Immunol. Cell Biol. Vol. 77, pp. 197-205. | http://www.ncbi.nlm.nih.gov/pubmed/10361251 | |
| Zebrafish | Mutations result in malignant peripheral nerve sheath tumour (zMPNST); RP genes may be 'haploinsufficient tumour suppressors' in zebrafish and cancer genes in humans | Amsterdam, A., Sadler, K.C., Lai, K., Farrington, S. et al. (2004), 'Many ribosomal protein genes are cancer genes in zebrafish', PLoS Biol. Vol. 2, p. E139. | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=406397 | |
Date last accessed for all websites is 17th June, 2010
Table S2.
Function and regulation of eukaryotic large subunit ribosomal proteins
| Protein Name | Organism | Function | Reference | Find online at |
|---|---|---|---|---|
| RPL4 | Rat | Required for rapid neurite regeneration | Twiss, J.L., Smith, D.S., Chang, B. and Shooter, E.M. (2000), 'Translational control of ribosomal protein L4 mRNA is required for rapid neurite regeneration', Neurobiol. Dis. Vol. 7, pp. 416-428. | http://www.ncbi.nlm.nih.gov/pubmed/10964612 |
| S. cerevisiae | Binds to single-stranded RNA/DNA | Cusick, M.E. (1994), 'Purification and identification of two major single-stranded binding proteins of yeast Saccharomyces cerevisiae as ribosomal protein L4 and histone H2B', Biochim. Biophys. Acta. Vol. 1217, pp. 31-40. | http://www.ncbi.nlm.nih.gov/pubmed/8286414 | |
| RPL7A | Human | Part of chimeric protein encoded by trk-2h oncogene | Ziemiecki, A., Müller, R.G., Fu, X.C., Hynes, N.E. et al. (1990), 'Oncogenic activation of the human trk proto-oncogene by recombination with the ribosomal large subunit protein L7a', EMBO J. Vol. 9, pp. 191-196. | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC551645/ |
| Zebrafish | Categorised as an ocular gene; downregulated in eyeless masterblind zebrafish. | Wang, H., Kesinger, J.W., Zhou, Q., Wren, J.D. et al. (2008), 'Identification and characterization of zebrafish ocular formation genes', Genome Vol. 51, pp. 222-235. | http://www.ncbi.nlm.nih.gov/pubmed/18356958 | |
| RPL7 | Human | Coregulator of vitamin D receptor-retinoid X receptor-mediated transactivation of genes | Berghöfer-Hochheimer, Y., Zurek, C., Wölfl, S., Hemmerich, P. et al. (1998), 'L7 protein is a coregulator of vitamin D receptor-retinoid X receptor-mediated transactivation', J. Cell. Biochem. Vol. 69, pp. 1-12. | http://www.ncbi.nlm.nih.gov/pubmed/9513041 |
| Rana sylvatica | Upregulated under freezing conditions | Wu, S., De Croos, J.N. and Storey, K.B. (2008), 'Cold acclimation-induced up-regulation of the ribosomal protein L7 gene in the freeze tolerant wood frog, Rana sylvatica', Gene Vol. 424, pp. 48-55. | http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6T39-4T3DCV0-1&_user=4887109&_coverDate=11%2F15%2F2008&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_searchStrId=1246451135&_rerunOrigin=google&_acct=C000062864&_version=1&_urlVersion=0&_userid=4887109&md5=4e11f74a6e6a29fe16aa172087195d0d | |
| RPL10 | Arabidopsis | A component of the NIK-mediated antiviral signaling | Rocha, C.S., Santos, A.A., Machado, J.P. and Fontes, E.P. (2008), 'The ribosomal protein L10/QM-like protein is a component of the NIK-mediated antiviral signaling', Virology Vol. 380, pp. 165-169. | http://www.ncbi.nlm.nih.gov/pubmed/18789471 |
| RPL13 | Hamster cells | Upregulated in response to DNA damage | Kobayashi, T., Sasaki, Y., Oshima, Y., Yamamoto, H. et al. (2006), 'Activation of the ribosomal protein L13 gene in human gastrointestinal cancer', Int. J. Mol. Med. Vol. 18, pp. 161-170. | http://www.ncbi.nlm.nih.gov/pubmed/16786168 |
| RPL22 | Mammals | Identical to heparin-binding protein, HBp15 | Fujita, Y., Okamoto, T., Noshiro, M., McKeehan, W.L. et al. (1994), 'A novel heparin-binding protein, HBp15, is identified as mammalian ribosomal protein L22', Biochem. Biophys. Res. Commun. Vol. 199, pp. 706-713. | http://www.ncbi.nlm.nih.gov/pubmed/8135813 |
| Drosophila | Interacts with casein kinase II | Zhao, W., Bidwai, A.P. and Glover, C.V. (2002), 'Interaction of casein kinase II with ribosomal protein L22 of Drosophila melanogaster', Biochem. Biophys. Res. Commun. Vol. 298, pp. 60-66. | http://www.ncbi.nlm.nih.gov/pubmed/12379220 | |
| Human | Binds Epstein-Barr virus (EBV)-encoded RNA (EBER) in EBV-infected cells | Le, S., Sternglanz, R. and Greider, C.W. (2000), 'Identification of two RNA-binding proteins associated with human telomerase RNA', Mol. Biol. Cell Vol. 11, pp. 999-1010. | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC14826/ | |
| Human | Binds human telomerase RNA | Le, S., Sternglanz, R. and Greider, C.W. (2000), 'Identification of two RNA-binding proteins associated with human telomerase RNA', Mol. Biol. Cell Vol. 11, pp. 999-1010. | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC14826/ | |
| RPL23A | Human | May play a role in growth inhibition | Jiang, H., Lin, J.J., Tao, J. and Fisher, P.B. (1997), 'Suppression of human ribosomal protein L23A expression during cell growth inhibition by interferon-beta', Oncogene Vol. 14, pp. 473-480. | http://www.nature.com/onc/journal/v14/n4/abs/1200858a.html |
| RPL24 | Arabidopsis | Gynoecium development | Nishimura, T., Wada, T. and Okada, K. (2004), 'A key factor of translation reinitiation, ribosomal protein L24, is involved in gynoecium development in Arabidopsis', Biochem. Soc. Trans. Vol. 32, pp. 611-613. | http://www.ncbi.nlm.nih.gov/pubmed/15270688?dopt=Abstract |
| Marine shrimp | Differential expression in gonads | Zhang, Z., Wang, Y., Jiang, Y., Lin, P. et al. (2007), 'Ribosomal protein L24 is differentially expressed in ovary and testis of the marine shrimp Marsupenaeus japonicus', Comp. Biochem. Physiol. B Biochem. Mol. Biol. Vol. 147, pp. 466-474. | http://www.ncbi.nlm.nih.gov/pubmed/17462931 | |
| RPL35A | Human | Cell death inhibition | Lopez, C.D., Martinovsky, G. and Naumovski, L. (2002), 'Inhibition of cell death by ribosomal protein L35a', Cancer Lett. Vol. 180, pp. 195-202. | http://www.ncbi.nlm.nih.gov/pubmed/12175552 |
| RPP0 | Human | Interacts with GCIP, and over-expression in breast and liver cancer results in cell proliferation | Chang, T.W., Chen, C.C., Chen, K.Y., Su, J.H. et al. (2008), 'Ribosomal phosphoprotein P0 interacts with GCIP and overexpression of P0 is associated with cellular proliferation in breast and liver carcinoma cells', Oncogene Vol. 27, pp. 332-338. | http://www.ncbi.nlm.nih.gov/pubmed/17621266 |
| RPLP1 | Mouse | Over-expression leads to cell proliferation of mouse embryonic fibroblasts | Artero-Castro, A., Kondoh, H., Fernández-Marcos, P.J., Serrano, M. et al. (2009), 'Rplp1 bypasses replicative senescence and contributes to transformation', Exp. Cell Res. Vol. 315, pp. 1372-1383. | http://www.ncbi.nlm.nih.gov/pubmed/19233166 |
| MRPL41 | Human and mice | Suppresses cell growth | Yoo, Y.A., Kim, M.J., Park, J.K., Chung, Y.M. et al. (2005), 'Mitochondrial ribosomal protein L41 suppresses cell growth in association with p53 and p27Kip1', Mol. Cell. Biol. Vol. 25, pp. 6603-6616. | http://www.ncbi.nlm.nih.gov/pubmed/16024796 |
Date last accessed for all websites is 17th June, 2010
Extra-ribosomal properties of the ribosomal proteins
Ribosomal proteins and gene expression
Temporal regulation of gene expression is critical for cell survival and function. Chromatin modification, transcription, translation, RNA processing and post-translational modification are the major checkpoints for a cell to regulate gene expression. Many of the prokaryotic and eukaryotic ribosomal proteins are involved in the regulation of their own expression or expression of other genes at different levels of gene regulation (Table 1).
Table 1.
Ribosomal proteins involved in gene regulation mechanisms
| Gene regulation level | Ribosomal protein (RP) | Organism | Function | Reference |
|---|---|---|---|---|
| Chromatin | S2 | Escherichia coli | Negative regulator of rpsB and tsf expression | 4 |
| S3 | Homo sapiens | Becomes a part of nuclear factor-κB complex that interacts with specific sites in the genome, on tumour necrosis factor stimulation | 6 | |
| S4 | Bacillus subtilis | Autoregulates rpsD gene expression | 5 | |
| L13a | H. sapiens | Inflammatory gene expression | 7 | |
| Transcription | S1 | E. coli | Transcription anti-termination and stimulates transcriptional activity of RNA polymerase | 8,9 |
| S4 | E. coli | Transcription anti-termination | 10 | |
| S10 | E. coli | Transcription anti-termination | 11 | |
| L3 | E. coli | Transcription anti-termination | 10 | |
| L4 | E. coli | Inhibits transcription of S10 operon mRNA and transcription anti-termination | 3,10 | |
| S14 | H. sapiens | Self-regulation at both transcriptional and translational levels | 3,12 | |
| S20 | Saccharomyces cerevisiae | Transcription anti-termination | 3 | |
| S0 and S21 (in association with each other) | S. cerevisiae | Promote maturation of 3' end of 18S rRNA | 13 | |
| L11 | Rattus rattus | Inhibits the transcriptional activity of peroxisome proliferator-activated receptor-alpha, a nuclear receptor | 14 | |
| L13 | E. coli | Transcription anti-termination | 10 | |
| Post-transcription | S14 | S. cerevisiae | Post-transcriptional repression of RPS14B [CRY2] expression | 15 |
| RNA processing and splicing | S12 | E. coli | Acts as RNA chaperone in the folding process of T4 phage intron RNA | 16 |
| S12 | H. sapiens | RNA splicing and modification | 12 | |
| S13 | S. cerevisiae and H. sapiens | Binds to the first intron of its transcript to inhibit splicing. Overproduction of RPS13 interferes with splicing of its own pre-mRNA by a feedback mechanism. Negatively controls splicing of its own pre-mRNA |
17,18 | |
| S14 | H. sapiens | Required for 18S pre-RNA processing and 40S subunit formation | 19 | |
| L4 | Mus musculus | Interacts with Gu(alpha) which is involved in rRNA processing | 20 | |
| Translation | S4 | E. coli | Translational repressor of α operon (operon genes; S13, S11, S4, L17) | 21 |
| S8 | E. coli | Translational repressor of spc operon | 22 | |
| S15 | E. coli | Self-translation regulation | 23 | |
| L1 | E. coli | Self-translation regulation | 12 | |
| L4 | E. coli | Suppresses translation of S10 operon mRNA. Self-translation regulation |
3,12 | |
| L10 | E. coli | Self-translation regulation | 12 | |
| S26 | H. sapiens | Self-translation regulation | 12 | |
| S30 | S. cerevisiae | Self-translation regulation | 12 | |
| L13a | H. sapiens | Silence translation of ceruloplasmin (Cp) mRNA | 24 | |
| Post-translation | S20 | E. coli | Post-translational inhibition of ornithine and arginine decarboxylase enzymes | 25 |
Ribosomal proteins and nucleic acid replication
During viral infection, viruses recruit some of the host machinery in order to produce new viral particles. The synthesis of new viral particles requires the replication of the viral genome, and in most of the DNA viruses the duplication of their genome is carried out by the host replication system. Ribosomal proteins are shown to take part in the genome replication in both DNA and RNA viruses. The ribosomal protein L14 helps Rep helicase to unwind the DNA during replication of the bacteriophage genome[12], and S1 is a subunit of Qβ replicase that replicates the genome of RNA coliphage Qβ [3]. In yeast, L3 helps in replication or maintenance of the double-stranded RNA genome [26].
Ribosomal proteins and DNA repair
Any damage to DNA disrupts the genome's integrity and thus proves fatal to the cell. The causes of such DNA damage are either metabolic processes within the cell or environmental factors like radiation/mutagens. Several DNA repair mechanisms exist within the cell to correct DNA damage. The type of mechanism employed is determined, in turn, by the type of damage. Ribosomal proteins are shown to function in DNA repair mechanisms in both prokaryotes and eukaryotes (Table 2).
Table 2.
Ribosomal proteins in DNA repair mechanisms
| Ribosomal protein | Organism | Function | Reference |
|---|---|---|---|
| S9 | E. coli | Involved in SOS repair mechanism by participating with polymerase UmuC | 3 |
| S3 | Drosophila spp. | DNA repair endonuclease. Corrects damage resulting from oxidative and ionising radiation | 27 |
| H. sapiens | Knockdown of S3 protects human cells from genotoxic stress. This is the converse of the situation in Drosophila S3 |
28 | |
| P0/LP0 (constituent of ribosomal stalk structure) | Drosophila, H. sapiens | Apurinic/apyrimidinic endonuclease activity | 29 |
Regulation of ribosomal proteins
Ribosomal proteins and the cell cycle
The cell undergoes different phases of growth and division during the cell cycle. The progression of a cell through these phases is controlled by cyclin/cyclin-dependent kinases (Cdk) and regulatory molecules of cell cycle checkpoints. Ribosomal proteins have been shown to alter the cell cycle fate by interacting with these molecules as an extra-ribosomal function. Human L34 inhibits the cell cycling proteins Cdk4 and Cdk5 [30]. L26 binds to the 5' untranslated region (UTR) of p53 mRNA upon DNA damage and increases translation of p53, a key player in cell cycle regulation and apoptosis [31].
Many of the other ribosomal proteins function to control the cell cycle and apoptosis through their expression levels. Abnormal expression levels of L7[32] and L13a[33] in humans interfere with cell cycle progression by arresting the cell cycle and inducing apoptosis. The involvement of ribosomal proteins in apoptosis is further evidenced by their interaction with Mdm2, a ubiquitin ligase that keeps a check on P53 levels under normal cellular conditions. The mammalian ribosomal protein L26 interacts with Mdm2 and thus regulates p53 levels [34]. Many more eukaryotic ribosomal proteins (S7, S19, S20, S27L, L5, L22 and L23) function in p53-mediated apoptosis [35-38]. In humans, the ribosomal protein S3 is shown to induce caspase-dependent apoptosis [12]. Also, some of the ribosomal proteins involved in apoptosis are over-expressed in cancers (Table 3).
Table 3.
Expression pattern of ribosomal proteins in cancers
| Ribosomal protein | Expression pattern | Cancer type | Reference |
|---|---|---|---|
| S2 | Over-expressed | Prostate cancer, head and neck carcinomas | 39,40 |
| S3, S6, S8, S12 | Over-expressed | Colon cancer | 40 |
| S3A, S4, S17 | Over-expressed | Feline leukaemia virus-induced lymphomas | 40 |
| S11 | Over-expressed | Colorectal cancer | 41 |
| L7A | Over-expressed | Colorectal cancer | 42 |
| Under-expressed | Osteosarcoma | 43 | |
| L13 | Over-expressed | Gastrointestinal cancer | 44 |
| L15 | Over-expressed | Oesophageal cancer | 45 |
| Over-expressed | Gastric cancer | 46 | |
| L19 | Over-expressed | Human breast cancer Used as marker for human prostate cancer | 47,48 |
| L23A, L27, L30 | Over-expressed | Hepatocellular carcinoma | 49 |
| L30 | Over-expressed | Medulloblastoma | 50 |
Ribosomal proteins and disease
Any defects in ribosomal proteins affect the synthesis of proteins that are required by a cell for carrying out vital cellular functions. Apart from protein synthesis, some of the ribosomal proteins are implicated in disease conditions owing to abnormal expression levels or expression of mutated genes. A mutation in ribosomal protein S19 was initially characterised as the cause of Diamond-Blackfan anaemia (DBA), a congenital erythroid aplasia [51]. Subsequently, ribosomal proteins S17, S15, S24, S7, L5 and L11 were also found to be involved in DBA [52]. It also has been shown that ribosomal proteins S3A (mouse) and S19 (zebrafish) function in erythropoiesis [18,53]. The function of these ribosomal proteins in erythropoiesis and DBA might give some clues as to how defects in the ribosomal proteins lead to the low red blood cell count in DBA patients.
In some disease conditions, the expression levels of the ribosomal proteins play an important role, as in Turner syndrome and human cataracts. Turner syndrome has been linked to a deficiency in human ribosomal proteins 4X and 4Y (isoforms of rps4)[54], and expression of L7A, L15 and L21 is downregulated in human cataracts [55]. A similar syndrome, named Noonan's syndrome, has been linked to ribosomal protein gene rpl6. This gene was found to be located in the same chromosome locus as Noonan's syndrome [56]. Other ribosomal proteins, such as S14, L24 and S26, are associated with 5q syndrome, mouse Bst and diabetes, respectively [19,57,58].
Ribosomal proteins and developmental regulation
During the development of an organism, the cells undergo growth and differentiation to give rise to tissues and organs. These processes are regulated by spatial and temporal control of gene expression. The ribosomal proteins that are involved in protein synthesis are also found to regulate development in many species. In Arabidopsis, some of the ribosomal protein genes are termed embryo defective, as mutated forms of these genes are lethal to embryo development [59]. A similar study in zebrafish has shown that ribosomal protein L11 affects embryological development in this species [60]. In animals, ribosomal proteins are involved in processes such as oogenesis and gonad development. The ribosomal protein S2 in Drosophila melanogaster and S15A in sea urchins play a role in oogenesis, while S4 in human is involved in gonad development [3]. Developmental defects in genes such as Drosophila minutes, mouse Bst (belly spot and tail), which encodes rpL24, and Dsk (dark skin mutants), which encodes rpS19, are also the result of defective ribosomal proteins. Organisms with these conditions exhibit various growth defects and have reduced adult size.
Since protein synthesis is the essential process that needs to be regulated during development, expression levels of ribosomal proteins are also regulated during the different developmental stages (Figure 1). Any change in this expression profile thus affects the protein machinery that is necessary for the normal development of an organism.
Figure 1.
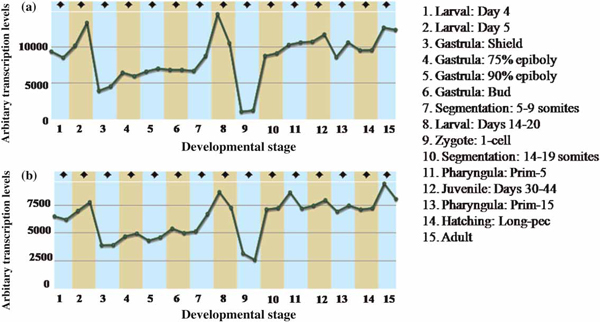
(a) rps4x transcription profile during zebrafish development. (b) rp///transcription profile during zebrafish development. See the Array Express Archive from the European Bioinformatics Institute: http://www.ebi.ac.uk/;http://www.ebi.ac.uk/microarray-as/ae/(accessed 23rd March, 2010).
Ribosomal proteins and lifespan regulation
Many recent studies have come up with different mechanisms by which an organism regulates its life span. The insulin/insulin-like growth factor 1 signalling (IIS) pathway and caloric restriction (CR) has been the major players of lifespan regulation in many species [61]. In the insulin signalling pathway, the components of this pathway, such as abnormal DAuer Formation (DAF)-2 or the downstream factor DAF-16, regulate the expression of various genes involved in metabolism, the stress response and other processes that shorten life span [61,62]. In CR, the life span of an organism is increased by decreasing the caloric intake. There is not much evidence of the mechanism by which CR affects the life span but some genes have been identified in Caenorhabditis elegans that influence life span regulation through CR [61]. It is further observed that the genes involved in CR mechanism are also linked to the IIS pathway [63,64]. Another player of longevity is the nutrient-responsive pathway mammalian target of rapamycin (mTOR) [65]. Both IIS and mTOR have a common downstream factor, ribosomal protein S6 kinase 1, which functions in regulating the mammalian life span [66]. Thus, these different pathways interact with each other to regulate longevity.
Also, many of the genes essential for growth and development are shown to extend the life span of a wide range of organisms. Among these genes are those involved in protein synthesis. The inactivation of translation initiation factors and ribosomal proteins S3, S8 and S11 was observed to increase the mean life span in Caenorhabditis elegans [61]. This indicates that the cell conserves its energy by keeping a check on protein synthesis.
Clearly, ribosomal proteins have additional functions outside the ribosome which are also regulated. One would expect that this would not be the case for such 'housekeeping' factors (indeed, several ribosomal protein genes are used as controls to normalise for gene regulation). Why does such regulation exist, and is it important? One answer could be that differential regulation might slow or speed up the process of protein synthesis. In the case of life span extension, it seems that downregulation of protein synthesis is involved. Another interesting aspect is how these extra-ribosomal functions have evolved; one possibility is via gene duplication, something that has been suggested for plant development and also in yeast [38,59]. These properties provide an important evolutionary paradigm in which nature uses existing genes for diversification.
Table S3.
Function and regulation of prokaryotic small subunit ribosomal proteins
| Protein Name | Organism | Function | Reference | Find online at |
|---|---|---|---|---|
| RPS1 | E. coli | Stimulates the T4 endo-ribonuclease Reg B | Aliprandi et al., S1 Ribosomal Protein Functions in Translation Initiation and Ribonuclease RegB Activation are mediated by similar RNA-Protein Interactions. (2008). The Journal of Biological Chemistry 283(19):13289-13301. | http://www.ncbi.nlm.nih.gov/pubmed/18211890 |
| Poly (A) binding protein in E. coli | Kalapos MP, Paulus H, Sarkar N. (1997). Identification of ribosomal protein S1 as a poly(A) binding protein in Escherichia coli. Biochimie 79(8):493-502. | http://www.ncbi.nlm.nih.gov/pubmed/9451450 | ||
| Interact with non-coding RNA DsrA and with rpoS mRNA and has a small role in altering the structures of these RNAs | Rositsa I. Koleva, Christina A. Austin, Jeffrey M. Kowaleski, Daniel S. Neems, Leyi Wang, Calvin P.H. Vary, Paula Jean Schlax. (2006). Interactions of ribosomal protein S1 with DsrA and rpoS mRNA. Biochemical and Biophysical Research Communications 348: 662-668. | http://www.ncbi.nlm.nih.gov/pubmed/16890206 | ||
| Binds to tmRNA, which tags truncated/trans-translated proteins for degradation | Matthieu Saguy, Reynald Gillet, Patricia Skorski, Sylvie Hermann-Le Denmat and Brice Felden. (2007). Ribosomal protein S1 influences trans-translation in vitro and in vivo. Nucleic Acids Research 35(7): 2368-2376. | http://nar.oxfordjournals.org/cgi/content/abstract/gkm100v1 | ||
| Over expression results in protection of mRNA degradation by PNPase | Briani et al.; (2008). Polynucleotide phosphorylase hinders mRNA degradation upon ribosomal protein S1 overexpression in Escherichia coli. RNA 4(11):2417-2429. | http://rnajournal.cshlp.org/content/14/11/2417.abstract | ||
| RPS3 | E. coli | Identical to H-protein in E. coli (Binds DNA and is associated with E. coli nucleoid) | Robert C.Bruckner and Michael M.Cox. (1989). The histone-like H protein of Escherichia coli is ribosomal protein S3. Nucleic Acids Research 17(8). | http://nar.oxfordjournals.org/cgi/content/abstract/17/8/3145 |
| RPS4 | E. coli | Overproduction of S4 stimulate rRNA synthesis | Takabe, Y., Miura, A., Bedwell, D., Tam, M. and Nomura, M. (1985). Increased expression of ribosomal genes during inhibition of ribosome assembly in Escherichia coli. Journal of Molecular Biology 184: 23-30. | http://www.ncbi.nlm.nih.gov/pubmed/3897554 |
| RPS6 | Myxococcus xanthus | Heat inducible protein | Maria De Angelis, Raffaella Di Cagno, Claude Huet, Carmine Crecchio, Patrick F. Fox, and Marco Gobbetti. (2004). Heat Shock Response in Lactobacillus plantarum. Applied and Environmental Microbiology 70 (3): 1336-1346. | http://aem.asm.org/cgi/content/abstract/70/3/1336 |
| RPS16 | E. coli | Acts as an endonuclease | Jacques Oberto, Eliette Elisabeth Mouray, Olivier Pellegrini, P. Mikael Wikstrom and Josette Rouviere-Yaniv. (1996). The Escherichia coli ribosomal protein S16 is an Endonuclease. Molecular Microbiology 19(6): 1319-1330. | http://www3.interscience. wiley.com/journal/119219619/abstract |
Table S4.
Function and regulation of prokaryotic large subunit ribosomal proteins
| Protein Name | Organism | Function | Reference | Find online at |
|---|---|---|---|---|
| RPL2 | E. coli | Zinc-binding protein | Katayama A, Tsujii A, Wada A, Nishino T, Ishihama A. Systematic search for zinc-binding proteins in Escherichia coli Eur. J. Biochem. 269(9):2403-2413. | http://www.ncbi.nlm.nih.gov/pubmed/11985624?ordinalpos=2&itool=EntrezSystem2. PEntrez.Pubmed.Pubmed_ResultsPanel. Pubmed_DefaultReportPanel.Pubmed_RVDocSum |
| RPL4 | E. coli | Allosterically regulates RNase E-dependent RNA degradation 'inhibiting RNase E-specific cleavage in vitro, stabilising mRNAs targeted by RNase E in vivo, and controlling plasmid DNA replication by stabilizing an antisense regulatory RNA normally attacked by RNase E' also upregulated in stress, which accompanies inactivation of RNase E and increased half-life of stress-responsive transcripts | Singh D, Chang SJ, Lin PH, Averina OV, Kaberdin VR, Lin-Chao S. (2009), Regulation of ribonuclease E activity by the L4 ribosomal protein of Escherichia coli. Proc. Natl. Acad. Sci. USA. 106(3):864-869. Epub 2009 Jan 14. | http://www.ncbi.nlm.nih.gov/pubmed/19144914?ordinalpos=1&itool=EntrezSystem2. PEntrez.Pubmed.Pubmed_ResultsPanel. Pubmed_DefaultReportPanel.Pubmed_RVDocSum |
| RPL11 | E. coli | Involved in regulating the activity of (p)ppGpp synthetase I | Yang X, Ishiguro EE. (2001), Involvement of the N terminus of ribosomal protein L11 in regulation of the RelA protein of Escherichia coli. J. Bacteriol. 183(22):6532-6537. | http://www.ncbi.nlm.nih.gov/pubmed/11673421?ordinalpos=7&itool=EntrezSystem2. PEntrez.Pubmed.Pubmed_ResultsPanel. Pubmed_DefaultReportPanel.Pubmed_RVDocSum |
| RPL13 | E. coli | Zinc binding protein | Katayama A, Tsujii A, Wada A, Nishino T, Ishihama A. Systematic search for zinc-binding proteins in Escherichia coli. Eur. J. Biochem. 269(9):2403-2413. | http://www.ncbi.nlm.nih.gov/pubmed/11985624?ordinalpos=2&itool=EntrezSystem2. PEntrez.Pubmed.Pubmed_ResultsPanel. Pubmed_DefaultReportPanel.Pubmed_RVDocSum |
| RPL25 | E. coli/Bacillus subtilis | General stress protein Ctc: might be required for accurate translation under stress conditions | Schmalisch M, Langbein I, Stülke J. (2002), The general stress protein Ctc of Bacillus subtilis is a ribosomal protein. J. Mol. Microbiol. Biotechnol. 4(5):495-501. | http://www.ncbi.nlm.nih.gov/pubmed/12432960?ordinalpos=1&itool=EntrezSystem2. PEntrez.Pubmed.Pubmed_ResultsPanel. Pubmed_DiscoveryPanel. Pubmed_Discovery_RA&linkpos=2& log$=relatedarticles&logdbfrom=pubmed |
References
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Molecular cell biology. 4. W.H. Freeman and Co., New York, NY; 2000. [Google Scholar]
- Zimmermann RA. The double life of ribosomal proteins. Cell. 2003;115:130–132. doi: 10.1016/S0092-8674(03)00804-3. [DOI] [PubMed] [Google Scholar]
- Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21:164–165. [PubMed] [Google Scholar]
- Aseev LV, Levandovskaya AA, Tchufistova LS, Scaptsova NV. et al. A new regulatory circuit in ribosomal protein operons: S2-mediated control of the rpsB-tsf expression in vivo. RNA. 2008;14:1882–1894. doi: 10.1261/rna.1099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. The rpsd gene, encoding ribosomal-protein S4, is autogenously regulated in Bacillus subtilis. J Bacteriol. 1991;173:4595–4602. doi: 10.1128/jb.173.15.4595-4602.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F, Anderson DE, Barnitz RA, Snow A. et al. Ribosomal protein S3: A KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Ray PS, Arif A, Brady AK. et al. DAPK-ZIPK-L13a axis constitutes a negative-feedback module regulating inflammatory gene expression. Mol Cell. 2008;32:371–382. doi: 10.1016/j.molcel.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogridge J, Greenblatt J. Specific binding of Escherichia coli ribosomal protein S1 to boxA transcriptional antiterminator RNA. J Bacteriol. 1998;180:2248–2252. doi: 10.1128/jb.180.8.2248-2252.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhodolets MV, Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- Torres M, Condon C, Balada JM, Squires C. et al. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 2001;20:3811–3820. doi: 10.1093/emboj/20.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren F, Das A. Formation of termination-resistant transcription complex at phage-lambda nut locus: Effects of altered translation and a ribosomal mutation. Proc Natl Acad Sci USA. 1984;81:3612–3616. doi: 10.1073/pnas.81.12.3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MD, Xu J. Ribosomal proteins and colorectal cancer. Curr Genomics. 2007;8:43–49. doi: 10.2174/138920207780076938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb-Massey A, Caffrey JM, Logsden P, Taylor S. et al. Ribosomal proteins Rps0 and Rps21 of Saccharomyces cerevisiae have overlapping functions in the maturation of the 3' end of 18S rRNA. Nucleic Acids Res. 2003;31:6798–6805. doi: 10.1093/nar/gkg899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JP, Davis JW, Gopinathan L, Leas TL. et al. The ribosomal protein rpL11 associates with and inhibits the transcriptional activity of peroxisome proliferator-activated receptor-alpha. Toxicol Sci. 2006;89:535–546. doi: 10.1093/toxsci/kfj040. [DOI] [PubMed] [Google Scholar]
- Li Z, Paulovich AG, Woolford JL. Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre-Messenger-RNA. Mol Cell Biol. 1995;15:6454–6464. doi: 10.1128/mcb.15.11.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clodi E, Semrad K, Schroeder R. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 1999;18:3776–3782. doi: 10.1093/emboj/18.13.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malygin AA, Parakhnevitch NM, Ivanov AV, Eperon IC. et al. Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res. 2007;35:6414–6423. doi: 10.1093/nar/gkm701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem Biophys Res Commun. 2009;379:167–170. doi: 10.1016/j.bbrc.2008.12.083. [DOI] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY. et al. Identification of RPS14 as a 5q(-) syndrome gene by RNA interference screen. Nature. 2008;451:252–253. doi: 10.1038/451252a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Henning D, Valdez BC. Functional interaction between RNA helicase II Gu alpha and ribosomal protein L4. FEBS J. 2005;272:3788–3802. doi: 10.1111/j.1742-4658.2005.04811.x. [DOI] [PubMed] [Google Scholar]
- Jinksrobertson S, Nomura M. Ribosomal protein-S4 acts in trans as a translational repressor to regulate expression of the alpha-operon in Escherichia coli. J Bacteriol. 1982;151:193–202. doi: 10.1128/jb.151.1.193-202.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheakis LC, Nomura M. Feedback regulation of the spc operon in Escherichia coli: Translational coupling and messenger RNA processing. J Bacteriol. 1988;170:4484–4492. doi: 10.1128/jb.170.10.4484-4492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard L, Mathy N, Grunberg-Manago M, Ehresmann B. et al. Identification in a pseudoknot of a U.G motif essential for the regulation of the expression of ribosomal protein S15. Proc Natl Acad Sci USA. 1998;95:2564–2567. doi: 10.1073/pnas.95.5.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK. et al. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/S0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Panagiotidis CA, Huang SC, Canellakis ES. Relationship of the expression of the S20 and L34 ribosomal proteins to polyamine biosynthesis in Escherichia coli. Int J Biochem Cell Biol. 1995;27:157–168. doi: 10.1016/1357-2725(94)00068-M. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Ridley SP, Fried HM, Ball SG. Ribosomal protein L3 is involved in replication or maintenance of the killer double-stranded RNA genome of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1982;79:4706–4708. doi: 10.1073/pnas.79.15.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoub A, Augeri L, Kelley MR, Doetsch PW. et al. A Drosophila ribosomal protein contains 8-oxoguanine and abasic site DNA repair activities. EMBO J. 1996;15:2306–2312. [PMC free article] [PubMed] [Google Scholar]
- Hegde V, Yadavilli S, Deutsch WA. Knockdown of ribosomal protein S3 protects human cells from genotoxic stress. DNA Repair. 2007;6:94–99. doi: 10.1016/j.dnarep.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Yacoub A, Kelley MR, Deutsch WA. Drosophila ribosomal protein PO contains apurinic/apyrimidinic endonuclease activity. Nucleic Acids Res. 1996;24:4298–4303. doi: 10.1093/nar/24.21.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthamer M, Chaudhuri B. Identification of ribosomal protein L34 as a novel Cdk5 inhibitor. Biochem Biophys Res Commun. 1999;255:631–638. doi: 10.1006/bbrc.1999.0145. [DOI] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Neumann F, Krawinkel U. Constitutive expression of human ribosomal protein L7 arrests the cell cycle in G(1) and induces apoptosis in Jurkat T-lymphoma cells. Exp Cell Res. 1997;230:252–261. doi: 10.1006/excr.1996.3417. [DOI] [PubMed] [Google Scholar]
- Chen FW, Ioannou YA. Ribosomal proteins in cell proliferation and apoptosis. Int Rev Immunol. 1999;18:429–448. doi: 10.3109/08830189909088492. [DOI] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB. et al. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhang Z, Li M, Wang W. et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: Binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V. et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Sun Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 2007;26:2707–2716. doi: 10.1038/sj.onc.1210073. [DOI] [PubMed] [Google Scholar]
- Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Hu YJ, Stearns ME. RPS2: A novel therapeutic target in prostate cancer. J Exp Clin Cancer Res. 2009;28:6. doi: 10.1186/1756-9966-28-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naora H. Involvement of ribosomal proteins in regulating cell growth and apoptosis: Translational modulation or recruitment for extra-ribosomal activity? Immunol Cell Biol. 1999;77:197–205. doi: 10.1046/j.1440-1711.1999.00816.x. [DOI] [PubMed] [Google Scholar]
- Kasai H, Nadano D, Hidaka E, Higuchi K. et al. Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J Histochem Cytochem. 2003;51:567–573. doi: 10.1177/002215540305100502. [DOI] [PubMed] [Google Scholar]
- Wang YX, Cheong D, Chan S, Hooi SC. Ribosomal protein L7a gene is up-regulated but not fused to the tyrosine kinase receptor as chimeric trk oncogene in human colorectal carcinoma. Int J Oncol. 2000;16:757–762. doi: 10.3892/ijo.16.4.757. [DOI] [PubMed] [Google Scholar]
- Zheng SE, Yao Y, Dong Y, Lin F. et al. Down-regulation of ribosomal protein L7A in human osteosarcoma. J Cancer Res Clin Oncol. 2009;135:1025–1031. doi: 10.1007/s00432-008-0538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Sasaki Y, Oshima Y, Yamamoto H. et al. Activation of the ribosomal protein L13 gene in human gastrointestinal cancer. Int J Mol Med. 2006;18:161–170. [PubMed] [Google Scholar]
- Wang Q, Yang CH, Zhou J, Wang XQ. et al. Cloning and characterization of full-length human ribosomal protein L15 cDNA which was overexpressed in esophageal cancer. Gene. 2001;263:205–209. doi: 10.1016/S0378-1119(00)00570-9. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhao LN, Li KZ, Ling R. et al. Overexpression of ribosomal protein L15 is associated with cell proliferation in gastric cancer. BMC Cancer. 2006;6:91. doi: 10.1186/1471-2407-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JL, Coggin DL, King CR. High-level expression of the ribosomal protein L19 in human breast tumors that overexpress Erbb-2. Cancer Res. 1993;53:1403–1408. [PubMed] [Google Scholar]
- Bee A, Ke YQ, Forootan S, Lin K. et al. Ribosomal protein L19 is a prognostic marker for human prostate cancer. Clin Cancer Res. 2006;12:2061–2065. doi: 10.1158/1078-0432.CCR-05-2445. [DOI] [PubMed] [Google Scholar]
- Kondoh N, Shuda M, Tanaka K, Wakatsuki T. et al. Enhanced expression of S8, L12, L23a, L27 and L30 ribosomal protein mRNAs in human hepatocellular carcinoma. Anticancer Res. 2001;21:2429–2433. [PubMed] [Google Scholar]
- De Bortoli M, Castellino RC, Lu XY, Deyo J. et al. Medulloblastoma outcome is adversely associated with overexpression of EEF1D, RPL30, and RPS20 on the long arm of chromosome 8. BMC Cancer. 2006;6:223. doi: 10.1186/1471-2407-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M. et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–75. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Gazda HT, Sheen MR, Vlachos A, Choesmel V. et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Zinn AR, Page DC, Nishimoto T. Functional equivalence of human X-encoded and Y-encoded isoforms of ribosomal protein-S4 consistent with a role in Turner syndrome. Nat Genet. 1993;4:268–271. doi: 10.1038/ng0793-268. [DOI] [PubMed] [Google Scholar]
- Zhang WY, Hawse J, Huang QL, Sheetz N. et al. Decreased expression of ribosomal proteins in human age-related cataract. Invest Ophthalmol Vis Sci. 2002;43:198–204. [PMC free article] [PubMed] [Google Scholar]
- Kenmochi N, Yoshihama M, Higa S, Tanaka T. The human ribosomal protein L6 gene in a critical region for Noonan syndrome. J Hum Genet. 2000;45:290–293. doi: 10.1007/s100380070018. [DOI] [PubMed] [Google Scholar]
- Oliver ER, Saunders TL, Tarle SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K. et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:1020–1032. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME. A role for the ribosome in development. Trends Plant Sci. 2009;14:512–519. doi: 10.1016/j.tplants.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Uechi T, Higa S, Torihara H. et al. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS One. 2009;4:e4152. doi: 10.1371/journal.pone.0004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. Plos Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A. et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–284. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Canto C, Auwerx J. Caloric restriction, SIRT1 and longevity. Trends Endocrinol Metab. 2009;20:325–331. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL. et al. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Tullet JMA, Wieser D, Irvine E. et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]


