Abstract
Fatty acid-binding proteins (FABPs) are members of the intracellular lipid-binding protein (iLBP) family and are involved in reversibly binding intracellular hydrophobic ligands and trafficking them throughout cellular compartments, including the peroxisomes, mitochondria, endoplasmic reticulum and nucleus. FABPs are small, structurally conserved cytosolic proteins consisting of a water-filled, interior-binding pocket surrounded by ten anti-parallel beta sheets, forming a beta barrel. At the superior surface, two alpha-helices cap the pocket and are thought to regulate binding. FABPs have broad specificity, including the ability to bind long-chain (C16-C20) fatty acids, eicosanoids, bile salts and peroxisome proliferators. FABPs demonstrate strong evolutionary conservation and are present in a spectrum of species including Drosophila melanogaster, Caenorhabditis elegans, mouse and human. The human genome consists of nine putatively functional protein-coding FABP genes. The most recently identified family member, FABP12, has been less studied.
Keywords: FABP, intracellular lipid-binding proteins, uptake and trafficking of lipids, metabolic diseases, human genome
Introduction
Hydrophobic ligands, such as fatty acids (FAs) and their acyl-CoA derivatives (FA-CoA), serve many biological functions within the cell. They serve as metabolic energy sources, substrates for membranes and signalling molecules for metabolic regulation [1,2]. The insoluble properties of FAs make for the requirement for chaperones to bind and transfer them throughout various cellular compartments, including the peroxisomes, mitochondria, endoplasmic reticulum, lipid droplets and nucleus. A family of highly expressed intracellular lipid-binding proteins (iLBPs)--the fatty acid-binding proteins (FABPs)--serves to bind these free ligands with high affinity. FABPs are ubiquitously expressed throughout tissues that are highly active in FA metabolism and comprise several isoforms. To date, nine FABP protein-coding genes have been identified in the human genome. These include liver(L-FABP), intestine- (I-FABP), heart- (H-FABP), adipocyte- (A-FABP), epidermal- (E-FABP), ileal-(Il-FABP), brain- (B-FABP), myelin- (M-FABP) and testis-FABP (T-FABP). These different isoforms were first named for the organ in which they were first identified or mostly predominate, but their expression profiles are not exclusive to that specific organ. For example, L-FABP is not only expressed in the liver, but also in the intestine, pancreas, kidney, lung and stomach. In addition to the nine known FABPs that have been widely studied, a newer member of the family, FABP12, has recently been discovered. The gene has been identified, but published reports on the protein encoded by this gene are not yet available. Therefore, in this reviews only the identification of the gene will be addressed.
Domain structure of FABP proteins
Although members of the FABP family share moderate sequence homologies of 20-70 per cent, their tertiary structures are virtually superimposable [3-5]. Sequences of human FABPs are aligned in Figure 1. These ~15-kilodalton proteins comprise ten anti-parallel β-barrel (βA-βJ) structures containing a solvent-accessible ligand-binding pocket, capped by an N-terminal helix-turn-helix motif (αI-αII) which is thought to act as the regulatory portal for binding. The helical N-terminus is involved in the regulation of FA transfer from membranes via electrostatic interactions [6-8]. Detailed superimposed images of the FABP family have been demonstrated previously [5]. All FABPs are capable of binding long-chain FAs (LCFAs; C12-20), which differ in their selectivity, affinity and binding mechanism [1,6]. Generally, these lipid chaperones bind only one ligand per molecule of protein, with the exception of L-FABP, which is capable of binding two ligands simultaneously. The ligand-binding process has been hypothesised, suggesting that FA enters a solvent-accessible area of the protein through a dynamic region made of α-helix II and the turns between βC-βD and βE-βF loops before binding in the cavity of the protein [9]. This has been termed the 'portal hypothesis', and is supported by a number of groups. Affinities toward ligands are generally increased with more hydrophobic molecules, and decreased with molecules of decreasing chain length and increasing sites of unsaturation. Dissociation constants have been calculated to be in the nanomolar to micromolar range [1,10,11]. Published reports have also demonstrated that proteins from different tissues reveal large differences in binding thermodynamics. ΔH° values differ by about 15 kcal/mol for different FABPs and by about 21 kcal/mol when compared with cellular retinoic acid-binding protein (CRABP) 1. Heat capacity changes generated by FA binding (ΔCp) range from 0 to-1.3 kcal/mol/K [11]. X-ray crystallography and nuclear magnetic resonance (NMR) studies have revealed FA-binding characteristics within the water-filled cavity, where bound FAs occupy approximately one-third the total pocket volume and share the cavity with many ordered and disordered water molecules [12]. Generally, the FA is orientated with the carboxylate group facing inwards. In this binding coordination, the carboxylate group of the FA typically involves tyrosine and arginine residues [12]. The binding conformations for an individual FABP vary slightly for different FAs, further distinguishing the differences between FABP family members.
Figure 1.
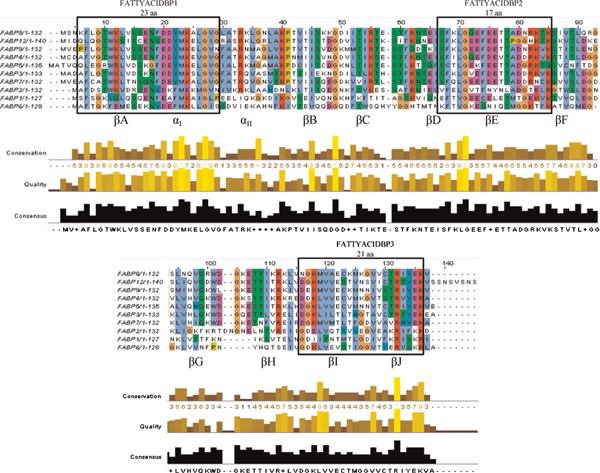
Amino acid sequences of the human FABP family members 1-9 and 12 are demonstrated using the ClustalW2 multiple sequences alignment software program,[176]demonstrated in Jalview [177]. The colour of amino acids is determined by the default annotation created by the analysis of multiply aligned sequences (AMAS), where it allows the identification of functional residues by comparing subgroups of sequences arranged on a tree (http://www.compbio.dundee.ac.uk/Software/Amas/amas.html). Conservation between amino acids is demonstrated below the alignment, where yellow (value of 10) is 100 per cent conserved, and absence of bars (value of 0) represents zero conservation among the sequences at that point. The consensus sequence for the family is shown below the conservation annotation. Common among FAPBs is a three-element fingerprint domain, separated by motifs named FATTYACIDBP1-3 (Kyoto Encyclopedia of Genes and Genomes, PRINTS: PR00178). Each motif is identified by a black box outline. Characteristic structural elements of all iLBP family members, alpha helices and beta-sheets, are all shown in grey boxes. Amino acid sequences were obtained from the National Center for Biology Information (NCBI) website (www.ncbi.nlm.nih.gov/): FABP1 (GenBank: CAG46887.1), FABP2 (GenBank: AAH69617.1), FABP3 (GenBank: CAG33148.1), FABP4 (GenBank: CAG33184.1), FABP5 (NCBI Reference Sequence: NP_001435.1), FABP6 (GenBank: AAH22489.1), FABP7 (GenBank: CAG33338.1), FABP8 (NCBI Reference Sequence: NP_002668.1), FABP9 (NCBI Reference Sequence: NP_001073995.1) and FABP12 (NCBI Reference Sequence: NP_001098751.1).
The overall gene structure of FABPs is highly conserved, consisting of four exons separated by three introns [13-16]. The position of exons and introns are similar for all FABPs, but the intron length is variable between isoforms. Typically, the lengths of exons are relatively short, with 23-24 amino acids encoded by exon 1, 57-58 amino acids by exon 2, 34-36 amino acids by exon 3 and 16-17 amino acids by exon 4 [17,18]. The length of intron 1 in human FABP3, FABP8 and FABP2 differs considerably, at 3.4, 2.3 and 1.2 kilobases (kb), respectively [13,15,19]. The conservation of gene structure is also maintained in other members of the iLBP family, where the genes encoding CRABPs and cellular retinol-binding proteins (CRBPs) all contain four exons and three introns, although the average lengths of introns are significantly longer than those of FABP genes [20,21]. Additionally, FABP genes contain the canonical TATA box upstream of the transcription start site [6]. For example, the TATA box sequence of FABP4 (5'-TTTAAAA-3') is located 32 nucleotides upstream of the TCA transcription start site [22].
All FABPs share a conserved three-element fingerprint domain, separated by motifs termed FATTYACIDBP1-3 (Kyoto Encyclopedia of Genes and Genomes, PRINTS: PR00178). Motif 1 contains a G-x-W triplet, which is involved in the initial formation of β-strand A (βA) and is homologous with a similar motif in another small hydrophobic ligand-binding protein family, the lipocalins [1,23]. Motif 2 contains strands 4 and 5 (βD-βE) and motif 3 spans strands 9 and 10 (βI-βJ). Each motif is outlined in Figure 1. Potential functional domains of FABPs include those found in the FABP4 gene, which has a nuclear localisation signal, nuclear export site and hormone-sensitive lipase (HSL)-binding site [24-26].
Chromosomal mapping of FABP family members has shown both dispersion and synteny, as demonstrated in Table 1. FABP1-3, 6 and 7 all exist on separate chromosomes in the human, whereas FABP4, 5, 8 and 9 all co-localise at chromosome (Chr) 8q21. A more detailed analysis within the chromosome containing these genes reveals clustering within a 300,000-base pair (bp) region [18]. In this same region, prediction programs identified an additional gene homologous to FABP8. It has since been cloned and named as the tenth member of the human FABP family, FABP12 [18]. In addition to protein-coding FABPs, pseudogenes for FABP3 and FABP5 have been identified and do not code for functional proteins in humans [27,28]. A list of known FABP pseudogenes to date is provided in the Human Gene Nomenclature Committee (HGNC) database (http://www.genenames.org).
Table 1.
Human FABP genes, as listed in the Human Gene Nomenclature Committee (HGNC) and Online Mendelian Inheritance in Man (OMIM) databases
| Gene | Common name |
Aliases for proteins | Previous symbols |
Localisation | Chromosomal location |
OMIM ID/ HGNC ID |
Number of amino acids |
|---|---|---|---|---|---|---|---|
| FABP1 | Liver FABP | L-FABP, hepatic FABP, Z protein, heme-binding protein |
Liver, intestine, pancreas, kidney, lung, stomach |
2p11 | 134650/ 3555 |
127 | |
| FABP2 | Intestinal FABP |
I-FABP, gut FABP (gFABP) | Intestine, liver | 4q28-q31 | 134640/ 3556 |
132 | |
| FABP3 | Heart FABP |
H-FABP, O-FABP, mammary-derived growth inhibitor (MDGI) |
FABP11 | Cardiac and skeletal muscle, brain, kidney, lung, stomach, testis, adrenal gland, mammary gland, placenta, ovary, brown adipose tissue |
1p33-p31 | 134650/ 3557 |
133 |
| FABP4 | Adipocyte FABP |
A-FABP, aP2 | Adipocytes, macrophages, dendritic cells, skeletal muscle fibres |
8q21 | 600434/ 3559 |
132 | |
| FABP5 | Epidermal FABP |
E-FABP, keratinocyte-type FABP (KFABP), psoriasis-associated-FABP (PA-FABP) |
Skin, tongue, adipocyte, macrophage, dendritic cells, mammary gland, brain, stomach, intestine, kidney, liver, lung, heart, skeletal muscle, testis, retina, lens, spleen, placenta |
8q21.13 | 605168/ 3560 |
135 | |
| FABP6 | Ileal FABP | Il-FABP, Ileal lipid-binding protein (ILLBP), intestinal bile acid-binding protein (I-BABP), gastrophin |
Ileum, ovary, adrenal gland, stomach |
5q23-q35 | 600422/ 3561 |
128 | |
| FABP7 | Brain FABP | B-FABP, brain lipid-binding protein (BLBP), MRG |
Brain, central nervous system (CNS), glial cell, retina, mammary gland |
6q22-q23 | 602965/ 3562 |
132 | |
| FABP8 | Myelin FABP |
M-FABP, peripheral myelin protein 2 (PMP2) |
Peripheral nervous system, Schwann cells |
8q21.3-q22.1 | 170715/ 9117 |
132 | |
| FABP9 | Testis FABP | T-FABP, testis lipid-binding protein (TLBP), PERF, PERF 15 |
Testis, salivary gland, mammary gland |
8q21.13 | --/3563 | 132 | |
| FABP12 | --- | --- | Retinoblastoma cell,a retina (ganglion and inner nuclear layer cells),b testicular germ cells,b cerebral cortex,b kidney,b epididymisb |
8q21.13 | --/34524 | 140 |
aExpression found in humans, versus brodents
Evolution of FABPs
It has been well established that significant structural homology between the members of family FABP exist, not only between human isoforms but between other species as well. This is highly suggestive of duplication and divergence from a single ancestral lipid-binding gene. FABPs belong to a larger family of iLBPs that also contain the CRABPs and CRBPs [29]. The iLPB family is divided into four subfamilies based on the types of ligands to which they bind [5]. The first subfamily binds vitamin A derivatives and contains the CRABPs and CRBPs. Subfamily II includes FABP1 and FABP6, binding bulkier hydrophobic groups such as bile acids (BAs), heme and acyl-CoA. The third subfamily includes FABP2, which exclusively binds LCFA. The fourth, and largest, subfamily includes FABP3, FABP4, FABP5, FABP7, FABP8 and FABP9, which bind a variety of ligands including LCFA, eiscosanoids and retinoids. The iLBP family is also part of a superfamily of lipophilic ligand-binding proteins which are widely expressed throughout the animal kingdom, the calycins [30]. Other members of the calycin superfamily include the avidins and the lipocalins [23,30,31]. Interestingly, the homology between these family members is relatively low (≤ 10 per cent amino acid identity), yet they still share the characteristic β-barrel tertiary structure [30,32]. iLBPs are found in both vertebrates and invertebrates throughout the animal kingdom [32].
The phylogenesis of human FABPs is summarised as a dendrogram in Figure 2, demonstrating the divergence of the primary sequences over time. It has been estimated that the iLBP family evolved approximately 1,000 million years ago (MYA) by subsequent duplications of a single ancestral gene, further propagating the increasing number of tissue-specific homologues that we know today [33]. Homology searches between either ancestral iLBP genes or present-day iLBP sequences have demonstrated the absence of this family in yeast (Saccharomyces cerevisiae) and plant (Arabidopsis thaliana) genomes, thus confining this family to the animal kingdom. This further suggests evidence for emergence of the ancestral iLBP gene post-divergence of animals from plants and fungi (1,200-1,000 MYA) [32]. The evolutionary rates of iLBP members have been estimated using the average distance between mammalian orthologues (rodents, man, rabbit, cow and pig), indicating that the amino acid substitution rate within the family differs widely (0.016-1.24 amino acid substitutions per amino acid site per billion years) [32]. The amino acid substitution rate for the FABP family ranges from 0.60-1.24. Earlier analyses of the evolutionary relatedness of proteins within the FABP family showed similarity, while identifying the positions of at least 14 gene duplications [20,34]. In addition, an index of evolutionary conservation between the family averages 1.16, with CRABP being the most highly conserved proteins in the iLBP family [32].
Figure 2.
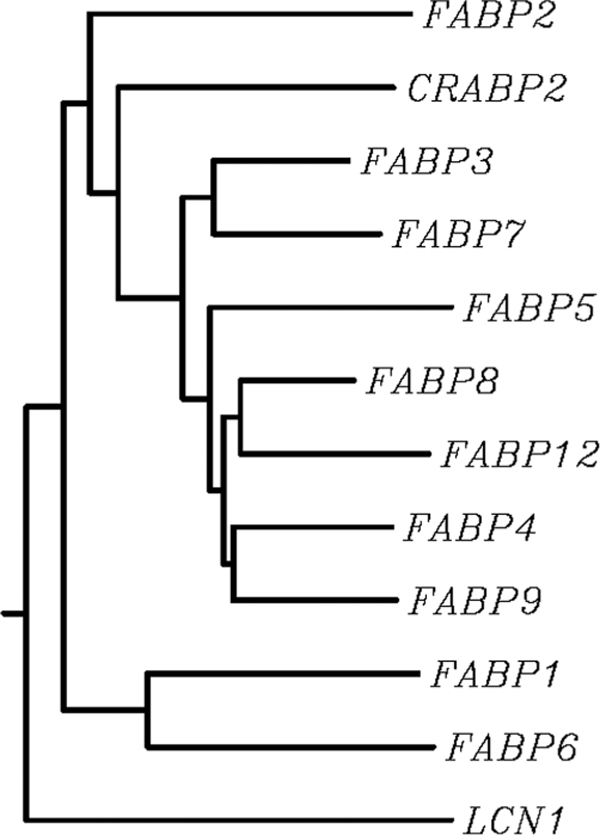
Phylogenetic analysis of human FABP family members. Using ClustalW alignment software of known peptide sequences for FABP1-9, FABP12, CRABP2 and LCN1, a dendogram with branch lengths was constructed. Clustering analysis showed divergence of iLBP family members by their known and established subfamilies: Subfamily I - FABP1, FABP6, Subfamily III - FABP2, Subfamily IV - FABP3, FABP4, FABP5, FABP7, FABP8, FABP9, and the newly established FABP family member FABP12. Subfamily II includes CRABP and CRBP: only CRABP2 was included in this analysis, to demonstrate similarities in the divergences of the iLBP family. Additionally, a member of the calcyin superfamily of hydrophobic ligand-binding proteins, lipocalin 1 (LCN1, GenBank accession no. NP_002288), was included in this analysis as an outgroup, as calcyins share ≤ 10 per cent homology with iLBP members.
The human FABP gene family
As mentioned previously, FABPs are ubiquitously expressed but differ in stoichiometry, affinity and specificity toward ligands. Expression of these proteins is mostly regulated at the transcriptional level, due to resulting synergistic interactions at conserved motifs within gene promoters [6,35]. Multiple cis-acting regulatory elements in the 5'-flanking region of these genes and trans-acting nuclear factors that either activate or repress transcription have been identified, and will be discussed separately for each FABP gene in the following sections of this review. Often, the expression of FABPs in a tissue directly reflects its lipid-metabolising capacity; that is, in hepatocytes, adipocytes and car-diomyocytes, FABPs make up 1-5 per cent of all soluble cytosolic proteins, due to the high concentration of FAs used for lipid biosynthesis, storage and metabolism [36]. In other tissues, less involved in active lipid metabolism, FABPs are not as highly expressed.
Numerous functions have been proposed for FABPs. Their presence in the cell is essential for the binding of hydrophobic molecules, reducing the detergent-like properties of high FA concentrations and making them more soluble. Additionally, FABPs traffic these ligands to various cellular compartments for storage, oxidation, membrane synthesis, signalling and activation of nuclear receptors. It has been demonstrated that FABPs can target FA to transcription factors such as the peroxisome proliferator-activated receptor (PPAR) family in the lumen of the nucleus, where the FABP1, FABP3, FABP4 and FABP5 genes are all regulated by PPARs via activation from FA or other ligands [37,38]. Many FABPs interact physically with phospholipid-rich membranes and other proteins for ligand transfer. For example, L-FABP interacts with membranes, the mitochondrial protein carnitine palmitoyl transferase 1 and transcription factors (PPARα) [7,39,40]. Additionally, FABPs also bind eicosanoid intermediates and protect these substrates against peroxidation, suggesting antioxidant-type behaviour [41,42]. This further suggests that, in addition to the general binding and trafficking functions of FABPs, they must have diverse and highly specified roles in regulating the metabolism and actions of the ligands they bind.
FABP1 (L-FABP)
The FABP1 human gene is located at Chr 2p12-q11 [15,43]. The gene has several regulatory elements, differing in specificity and function. For example, one TATA-box (5'-TATAAAA-3') is located within the promoter region and is recognised by the Ets superfamily of proteins [15,29]. The gene promoter of FABP1 also contains sterol response elements-1 and -2 (SRE1, SRE2), two hepatocyte nuclear factor (HNF) 1-binding sites, activator protein-1 (AP-1)-binding sites and a CCAAT/enhancer-binding protein (C/EBP) site [6,39,44]. In addition, the presence of a peroxisome proliferator response element (PPRE) is important for the regulation of FABP1, as well as other genes involved in metabolism [29,33,37,45]. Treatment with hypercholesterolaemic and hyperlipidaemic drugs (statins and fibrates) upregulates FABP1 expression in the liver [44,46]. This mechanism is important to consider in diseases in which metabolic homeostasis is altered, such as non-alcoholic fatty liver disease, metabolic syndrome (MetS), type 2 diabetes mellitus (T2DM) and alcoholic liver disease.
L-FABP is abundantly expressed in the liver, accounting for up to 5 per cent of the total cytosolic protein. The liver isoform of the FABP family of proteins is unique, attributed to its ability to bind multiple ligands at once. It has a larger solvent-accessible core (440 Å3) in comparison to M-FABP (330 Å3), H-FABP (323 Å3), A-FABP (310 Å3) or I-FABP (234 Å3);[47] this allows for more diverse binding to substrates, further indicating a unique functionality by comparison with other FABP family members. X-ray crystallography and NMR analysis of both apo-and holo-isoforms of L-FABP have revealed differences in the structural orientation of the protein in both states [47,48]. When oleate occupies both binding sites, site-specific perturbations are found near the linker between αII and βB, between strands βC and βD and βE and βF, as well as other areas within the binding cavity [48]. In other words, it appears as though the alpha-helix 'cap' and the β-turn of sheets E and F fold inwards to encapsulate the binding pocket. The orientation of the first FA/ligand, in the designated primary binding site, is in a bent or U-shaped conformation, with the carboxyl group interacting with Arg122, Ser39 and Ser124 [47]. Other amino acids in contact with the FA are Ile41, Phe63, Glu72, Thr73, Thr93 and Thr102 [47]. The second binding site is more heavily involved in electrostatic interactions, interacting with Leu28, Gly32, Ile35, Ile53, Tyr54, Gly55, Lys57, Met113 and Arg122 [47]. The FA extends with its carboxylate group at the opening of the binding pocket and is exposed to the aqueous environment. The molecular dynamics of ligand dissociation from L-FABP has been investigated using computer simulations but is still largely speculative. Three potential 'portals' for FA exit are proposed, located at: the αII and βC/βD loop (portal I); βG/βH and βE/βF loops and the C-terminal end of αI and N-terminal end of the αII/αI loop (portal II); and at the bottom of the cavity (portal III) [49]. The first binding site is considered the 'high affinity' site (with Kds ranging from 4-60 nM) and the second site is the 'low affinity' site (with Kds ranging from 0.3-12 μM for FA and FA-CoAs) [11,39,50-52]. L-FABP also binds intermediates of FA oxidation (fatty acyl-carnitines) and glyceride synthesis (1-oleoylglycerol), as well as lysophospholipids, cholesterol, BAs, prostaglandins, lipoxygenase products, retinoids, heme and biliru-bin (for extensive review refer to Atshaves et al.[39]). For larger ligands, such as BAs, the stoichiometry (1:1) and affinity are both significantly reduced [53]. In addition to possessing affinity for endogenous substrates, L-FABP also binds to a variety of xeno-biotic drugs, including beta-blockers (atenolol, nadolol), non-steroidal anti-inflammatory drugs (ibuprofen, ketorolac), fibrates (benzafibrate, clofibrate, gemfibrozil) and benzodiazepines (diazepam, lorazepam) with affinities in the nano- to micro-molar range [54].
The high level of expression, binding properties and function in regulating a variety of cellular processes (inflammation, immunity, metabolism and energy homeostasis) demonstrate the importance of L-FABP. Therefore, it would be detrimental if inactivation and/or loss of this protein occurred. Gene knockdown studies in mice have revealed a significant impact on metabolic regulation and weight gain when challenged with high-fat or high-cholesterol-containing diets [55-57]. To date, a highly conserved c.340A > G missense mutation in exon 3 of the human FABP1 gene has been identified, which results in a Thr → Ala substitution at position 94 (T94A) [58]. Carriers for this single nucleotide polymorphism (SNP) were identified in a subset of French Canadian subjects, and had higher baseline plasma-free FA levels, lower body mass index (BMI) and a smaller waist circumference than T94 homozygotes. When challenged with fenofibrate, subjects were at increased risk for exhibiting higher plasma trigly-ceride (TG) levels (≥ 2.00 mmol/L). The T94A mutant was later found to be associated with quantitative traits associated with MetS, cardiovascular disease and T2DM (elevated fasting TG and LDL-cholesterol content) [59]. The functional consequences of this SNP on L-FABP binding have not been investigated, but can be inferred--based on the location of the mutation--to affect binding in the cavity of the protein.
FABP2 (I-FABP)
The human FABP2 gene is located at Chr 4q28-q31 [60]. Three conserved domains of 14 bp are located in the 5'-flanking region of FABP2 which specifically bind to nuclear proteins [6,60]. In mice, one of these conserved 14-bp domains (5'-TGAACTTTGAACTT-3') was found to be necessary for establishing and maintaining its region-specific expression along the duodenal-to-colonic axis of perpetually renewing gut epithelium; this same domain is found in other genes transcribed in the intestine. Nucleotides between -1178 and -277 have been promote expression of FABP2 in the ileum and colon, whereas nucleotides between -277 and -185 suppress expression in these regions [61]. Two regulatory elements, the CCAAT and TATA box sequences, are also found in the promoter region of the FABP2 gene [15]. Several more elements are found within FABP2 and include cyclic adenosine monophosphate (cAMP)-responsive elements (24-bp region), which bind colonic nuclear factors; a C/EBP binding site; and HNF4 and apolipoprotein regulatory protein-1 (ARP-1) binding sites [29,6,62].
This intestinal isoform of the FABP family mediates fat absorption through binding and intracellular trafficking of free LCFAs. It is highly expressed throughout the intestine, with the highest levels in the distal portion of the organ. L-FABP and Il-FABP are also expressed in different segments of intestine; therefore, it is difficult to determine the individual contributions of each protein in the uptake, trafficking and metabolism of FA [1,63]. Comparison of their binding properties (stoichiometry, specificity and affinity) reveals strong differences between all three FABPs expressed in the intestine [63]. Nevertheless, it can be inferred that I-FABP actively participates in dietary lipid metabolism due to its high specificity and selectivity for LCFAs. Although L-FABP and I-FABP bind saturated LCFAs with relatively the same affinity (but at different stoichiometric ratios), I-FABP exhibits lower affinity for unsaturated LCFAs [64,65]. In addition, the binding cavity of I-FABP is much smaller than that of L-FABP, which contributes to the lower binding ratio (1:1). Similar to L-FABP, the orientation of bound FA is in a slightly bent conformation, with its hydrocarbon tail extending toward the alpha helices (αI-αII) and the carboxylate group of the FA buried in the cavity and interacting with Arg106, Gln115 and two ordered water molecules [9]. Through the use of amide 15N relaxation and 1H exchange NMR experiments, it has been determined that the majority of residues do not undergo conformational changes between apo-and holo-I-FABP [66]. Residues Val26-Asp35, Ser52-Arg56 and Ala73-Thr76 (αII and the βC-βD and βE-βF turns) undergo significant conformational change upon ligand binding, however, forming a 'cap' over the FA. In addition to binding LCFAs (Kd = 120 nM for palmitate), I-FABP binds lipophilic drugs with lower affinity (Kd = 62.5 μM for ibuprofen; Kd = 56.0 μM for benzafibrate) [67,68].
Extensive studies have been performed on FABP2 gene variants better to understand their possible contributions to metabolic diseases. Several SNPs in the FABP2 gene have been identified, but only one has been associated with the pathologies attributed to MetS [69-75]. Gene variants include seven alleles (including the wild-type allele and trinucleotide repeats of 10-15 consecutive ATT sequences) in a microsatellite region of intron 2 [76,77]. Three SNPs have been found--two being silent variants occurring in the coding regions of the gene (T → C at codon 71 and A → G at codon 118) [70]. The third polymorphism in human FABP2 is located at codon 54 (Ala → Thr) and is a missense variant associated with dyslipidaemia, insulin resistance and obesity [69-71]. Frequencies for the alanine-encoding (Ala54) and threonine-encoding (Thr54) FABP2 alleles of 0.71 and 0.29 have been found in a population of 760 Pima Indians (which has the highest reported prevalence of T2DM) [70]. Similar frequencies (0.69 and 0.31) were observed in 56 Caucasian DNA samples. In this study, Thr54 homozygotes had a higher mean two-hour plasma insulin concentration during an oral glucose tolerance test than did Ala54 homozygotes. Functional effects of the SNP were assessed with recombinant protein and were shown to reduce the affinity for oleate and arachidonate from 191 and 179 nM to 381 and 380 nM, respectively [70]. Based on these studies, it has been inferred that FABP2 is not a major 'diabetes gene'; however, it may be associated with the propensity for T2DM. Association studies have also been performed with the Ala54Thr SNP and parameters of obesity and cardiovascular risk factors in a group of 264 Spanish obese individuals [78]. Allelic frequencies for Ala54/Thr54 heterozygotes and Thr54/Thr54 homozygotes were 34.8 per cent and 7.2 per cent, respectively, and were significantly associated with elevated levels of C-reactive protein. In contrast to previously published reports, a newer study has failed to associate the Ala54Thr SNP with anthropometric and biochemical parameters of MetS in 760 obese subjects [79]. Another FABP2 variant was found to be located in the 3' non-coding region, where GCGCA → GCACA [80]. It is believed that this SNP does not affect the gene product, but little is understood about the sequences that flank coding regions of genes [72]. Of the evidence provided here, it is clear that more studies need to be conducted further to understand if gene variants of FABP2 significantly contribute to pathologies associated with metabolic disorders.
FABP3 (H-FABP)
Human FABP3 is located at Chr 1p33-p31, whereby distinct tissue distribution is regulated by concise promoter regions within the gene [81]. A 0.4 kb promoter region specifies expression of the protein in muscle, while elements further upstream in the 1.2 kb promoter are required for highly enriched expression of H-FABP in the heart. Multiple transcription factor binding sites are present within the promoter, including E box (CATGTG; consensus CANNTG) and TATA boxes. Full promoter activity is controlled via a CArG-like motif (5'-CTT CCT ATT TCG GGA GC-3') that functions similar to SREs. The promoter also contains a potential response element for ERRα, while a PPAR response element is present in conserved regions of the FABP3 gene [37,81,82]. Other motifs in the 5'-flanking region of porcine FABP3 have been identified, including AP-1, AP-2, C/EBP, mammary active factor and Stat-5 binding sites [83].
H-FABP constitutes 4-8 per cent of the cytosolic protein in mammalian heart, and is highly expressed in both cardiac and skeletal muscle and, to a lesser extent, in stomach, brain, lung and mammary gland [84]. An important function for H-FABP in skeletal muscle is to traffic FAs towards the mitochondria, where they undergo β-oxidation for energy expenditure. The presence of H-FABP in blood has been proposed as an early biochemical marker of acute myocardial infarction, as it is rapidly released from the myocardium into the circulation after cells are damaged [85-87]. It is also being considered as a cerebral spinal fluid marker for the fatal neurodegenerative disease, Creutzfeldt-Jakob disease [88]. H-FABP shares similar affinity for FAs with Kds in the nanomolar range, with affinities for palmitate, oleate and arachidonic acid being 0.96 μM, 0.43 μM and 0.37 μM, respectively [89]. Additionally, H-FABP binds non-prostanoid oxygenated FAs (epoxyeicosatrienoic acids, hydroxyeicosatetraenoic acids and dihy-droxyeiosatrienoic acids) with similar affinity (Kd' = 0.05-14.2 μM) [42]. The conformation of a bound FA differs slightly from that with I-FABP, being orientated in a more confined U-shape, with the carboxylate group buried within the core of H-FABP [90]. In addition, different FAs (palmitic, oleic, elaidac and stearic acids) exhibit similar orientations in the crystallised holo-form [90,91]. An ordered network of hydrogen bonds allows for ligand binding, and involves the side chains of five residues of H-FABP (Arg126, Tyr128, Thr40, Arg106 and Thr53), as well as two water molecules with the carboxylate group of oleate. The portal region of H-FABP is similar to that of I-FABP, and is defined by the side chain residues of Val25, Thr29, Phe57, Ala75 and Asp76. It also has been postulated that lysine residues on the edge of this 'binding portal' add positivity to this region and may serve to draw the carboxylate group of FA toward the portal via electrostatic interactions. Solvent mapping of H-FABP has also highlighted a secondary portal region (Glu72, His93 and Arg106), which may act as the conduit for solvent efflux as a ligand either enters or exits the primary binding portal. Phe16 is also involved in van der Waals interactions with the bound ligand, and is hypothesised to be a key determinant for ligand specificity and affinity in H-FABP [90]. Mutation of this residue proved the importance of Phe16 in ligand binding [92].
Initial characterisation of FABP3 using southern hybridisation revealed multiple hybridising fragments of cDNAs in both humans and mouse, highlighting the presence of pseudogenes. An intronless FABP3 gene (FABP3P2) has been identified in region 13q13-q14 and found to be 81 per cent homologous to FABP3; however, no cDNAs have been detected in human skeletal muscle or foetal brain libraries [27]. In addition to pseudogenes, several polymorphisms of FABP3 have been identified and are presumed to contribute to traits associated with obesity, T2DM and MetS. In an attempt to screen for polymorphic markers and candidate genes for T2DM, 14 polymorphisms of FABP3 have been identified [93]. These include two insertion/deletions, two short tandem repeats and ten SNPs (two in the promoter, nine within introns, two in the 3'-untranslated region and on the 3' end of the gene). Among the identified polymorphisms found in 24 Korean DNA samples, five polymorphic sites (c.-530_-532delCTC, c.-345T > C, c.348 + 429(CA)9-18, c.246 + 1806G > C and c.634 + 483delT) were chosen for larger-scale screening in 669 individuals and found to have allele frequencies of 0.184 (c.-530_-532delCTC), 0.495 (c.-345T > C), 0.177 (c.246 + 1806G > C) and 0.139 (c.634 + 483delT), respectively. Logistics analysis between patients and normal subjects were performed and revealed the association of one insertion/deletion polymorphism at the 3' end (c.634 + 483delT) of FABP3 with an increased risk of T2DM. FABP3 polymorphisms were also compared with five metabolic parameters (levels of cholesterol, TG, HDL, BMI and waist/hip ratio) and the only significant association, again, was with the 3' end ins/del (c.634 + 483delT) SNP. These studies have shown only moderate associations between FABP3 polymorphisms with the risk for T2DM and related phenotypes. Other association studies performed in Beijing-You chickens and pigs have related SNPs to intramuscular fat deposition and other fatness traits (back fat thickness and body weight) [94-96]. A newly published report analysing postmenopausal osteoporosis in Hungarian women found a significant correlation between the homozygous recessive genotype of rs10914367 in the promoter region of FABP3 and increased hip bone mineral density [97]. This polymorphism tags the first exon and intron of FABP3, which contain the transcription factor binding sites forkhead box protein P3 (GCCAAC), C/EBP-β (CCAA) and progesterone receptor isoforms A and B (AACACCA). These physical interactions between transcription factor binding sites are ablated in the case of the polymorphism, however. It is evident that only correlative studies have been performed with polymorphisms of the FABP3 gene, and more detailed analyses need to be performed to clarify the contribution of genetic variations in the pathomechanisms of various diseases.
FABP4 (A-FABP)
FABP4 is located on Chr 8q21 and is regulated by a number of cis-acting regulatory elements and trans-acting nuclear factors in the 5' flanking region of the genes that activates or represses transcription [29,98,99]. Two fat-specific elements (FSE) are found within the FABP4 gene, including four copies (three direct and one inverted) of the 13 bp FSE1 (5'-GGCTCTGGTCATG-3') and 15 bp FSE2 (5'-ACTCAGAGGAAAAG-3') [22]. The canonical TATA box is located 31 bp upstream of the transcription start site. Additionally, the gene has overlapping positive (AP-1) and negative (cAMP) regulatory elements, with a binding sequence for the trans-activating C/EBP upstream of these regulatory elements [29]. Further upstream of the 5'-flanking region of FABP4 are five cis-acting adipocyte regulatory elements (ARE1, ARE2, ARE4, ARE6 and ARE7) [100]. The ARE1 site targets a member of the nuclear factor (NF) 1 family and, when mutated, reduces the activity of the enhancer in adipocytes by 76 per cent. Both ARE2 and ARE4 are recognised by adipocyte regulatory factor 2 (ARF2), stimulating promoter activity in several cell types, while ARE6 and ARE7 stimulate promoter activity only in adipocytes and are bound by adipocyte regulatory factor 6 (ARF6) [29]. FABP4 also contains several functional PPREs approximately 5.5 kb upstream of the transcription start site, allowing for the transcriptional regulation of FABP4 by fatty acids, PPARγ, insulin and agonists of PPARγ, such as thiazolidinediones [37,101,102].
Adipocyte FABP (aP2) is highly homologous to myelin P2 protein, and is significantly expressed in white and brown adipose tissue, monocytes and macrophages [33]. Total expression in adipocytes is proposed to be 1-3 per cent of the total cytosolic protein and is a known gene marker for the maturation of adipocytes (50-fold upregulation) [103]. In addition to the chaperone-like activity responsible for integrating lipid signals and organelle responses, A-FABP interacts with HSL, potentially modulating its catalytic activity and integrating signalling networks that control inflammatory responses and lipid hormone production in adipocytes [1,26]. In macrophages, A-FABP also mediates inflammatory responses through the inhibitor of kappa kinase nuclear factor κB (IKK-NF-κB) and c-Jun N terminal kinase (JNK)-AP-1 pathways and attenuates cholesterol efflux through inhibition of the PPARγ-liver X receptor α(LXRα)-ATP binding cassette A1 (ABCA1) pathway [1,104,105]. Recently, A-FABP has been found to be expressed within the skeletal muscle fibres, but is much less abundant than H-FABP [106]. The significance of muscular expression is hypothesised to be for metabolic adaptation, based on the fact that mRNA expression of A-FABP is elevated in endurance-trained subjects. A-FABP appears specifically to bind LCFAs with high affinity (Kd = 80, 83, 57, 92 and 182 nM for stearate, palmitate, oleate, linoleate and arachidonate, respectively) [11]. A-FABP also binds retinoic acid, but with lower affinity (Kd = 50 μM) [107]. Crystal structures have been solved for the apo-form and holo-forms liganded with oleate, stearate, palmitate, hexadecane sulphonate and arachidonate and modified with the reactive aldehyde 4-hydroxynonenal [108-112]. Upon ligand binding, both the surface area and cavity volume increase and changes in the topology of the electrostatic potential and surface area are observed around the ligand portal [113]. The stoichiometry of binding is similar to that of I-FABP and H-FABP (1:1), while the orientation of stearate is slightly bent and U-shaped for arachidonic acid. The hydrocarbon tail of bound FA (stearic and oleic acid) is located in the cavity formed by side chains of hydrophobic amino acids, with the first segment of the FA (C-4) making van der Waals contact with Cys117, the middle segment (C5-C14) interacting with Tyr19 and a salt bridge formed by Asp76 and Arg78, and the last segment (C15-C18) protruding towards the solvent in close proximity to Val32, Phe57 and Lys58 [110]. Tyr19 is phosphorylated by the insulin receptor kinase, which has been shown significantly to reduce both the affinity for LCFA and the release of ligand, demonstrating the involvement of A-FABP in the lipogenic pathway [111,114,115]. The carboxylate group of the bound FA forms a network of hydrogen bonds and other electrostatic interactions with Arg126, Tyr128, and Arg106 through an intervening water molecule [112]. Structural differences induced from binding affect the portal region and involve the end of αII (Met35, Ala36, Lys 37 and Pro38) and Phe57. These changes are absent when arachidonic acid is the bound ligand because its orientation is entirely buried within the cavity, although additional amino acids are involved in binding (Ser53 and Pro38).
A-FABP is becoming a hot target in the area of drug development for T2DM, based on animal studies involving a mutated protein and the resulting observed protection from insulin resistance when fed a high-fat diet as compared with wild-type controls [116,117]. Additionally, significantly higher concentrations are secreted from adipocytes into the serum of overweight/obese patients than into serum of lean persons [118]. These serum concentrations of A-FABP also correlated positively (p < 0.005) with waist circumference, blood pressure, dyslipidaemia, fasting insulin and the homeostasis model assessment insulin resistance index. Components of MetS also were assessed (p < 0.05), suggesting that A-FABP is a central regulator of systemic insulin sensitivity. Polymorphisms of the FABP4 gene have also been associated with resistance to obesity and other associated pathologies that include dyslipidaemia, insulin resistance, T2DM and cardiovascular disease. A multitude of SNPS (119 via SNPHunter[119] and 130 from the National Center for Biotechnology Information [NCBI] SNP database) have been identified, but only a few representative studies will be addressed in this overview. Five SNPs have been identified in Caucasian men and women, and include sequence variants at positions T-87C, C2600T, T2613C and an insertion of G at position 4265, which are outside the coding regions of the gene [120]. A silent variant, G4356C, was found on exon 4. Allele frequencies for these SNPs in 96 primarily Caucasian men and women were 4.69 per cent, 16.67 per cent, 0.52 per cent, 16.67 per cent and 0.52 per cent, respectively. The T-87C variant is located at the C/EBP binding site, and it was found that this mutation exhibited significantly reduced competitive-binding capacity (≥ 50 per cent) for C/EBPα compared with the wild-type (WT) FABP4 promoter. This variant also significantly impaired transcriptional activation of the promoter by C/EBPα in cultured cells. In a population cohort of 7,899 subjects, carriers for the T-87C variant (2.2 per cent allele frequency) had significantly lower fasting TGs and were associated with reduced risk for coronary heart disease and T2DM. Additionally, 1,247 subjects from Costa Rica (non-fatal myocardial infarction case-control study) were screened, and carriers for T-87C were found to have a 63 per cent reduction in adipose tissue A-FABP mRNA expression, indicating that T-87C is a loss-of-function allele in vivo. Two additional SNPs within the 5' region of FABP4: A-376C and A-374C have been identified from 314 Hispanic and Caucasian males [121]. When A-376C and a SNP of PPARγ (Pro12Ala) were examined with measures of insulin sensitivity and body composition, the individual variants were not significantly associated; however, when both sites were considered as one model, they were significantly associated. FABP4 variants have also been assessed in cases of childhood obesity, where the frequency of rs1054135 G/A was higher in obese compared with non-obese individuals and found to contribute to higher FABP4 levels. Meanwhile, rs16909233 G/A was found to be associated with insulin resistance in the context of obesity [122]. In contrast to fat-related pathologies, SNPs of FABP4 have been analysed for contribution to the development of polycystic ovary syndrome (PCOS) [123]. Three SNPs were identified in a population of Chinese women, where the genotype frequencies of two alleles (T-911C and G-737C) did not differ between PCOS and case control subjects. An insertion/deletion CA SNP at -798 to -799 bp was found to have higher frequencies in PCOS subjects, but was not associated with the clinical/biochemical parameters associated with PCOS.
FABP5 (E-FABP)
FABP5 is located at Chr 8q21.13, where putative response elements within the promoter region may be responsible for the wide level of expression observed throughout tissues. A more detailed analysis involving the mouse FABP5 gene revealed promoter elements and transcription factor binding sites in the 5' region [124]. The TATA box is at nucleotide (nt) -40 (5' -ATATAA-3'); a CCAAT box at nt -80 (5'-ATTGG-3'); and GC-rich regions at nt -110 (5'-GGGGCGG-3'). Putative binding sites for myogenic differentiation factor (MyoD), HNF1, C/EBPα and β, GATA binding protein 1 (GATA1) and myeloid zinc finger 1 (MZF1) exist within the promoter of FABP5. Additionally, PPREs present in the gene allow for functional interactions between PPARβ/δ [38]. To date, 15 known pseudogenes have been identified in humans, and are classified as FABP5P1-15 (HGNC).
E-FABP is highly expressed in epidermal cells, but also in a plethora of other tissues, including mammary gland, brain, liver, kidney, lung, adipocytes, macrophages, tongue and testis [1,33,36]. It was first identified in human psoriatic keratinocytes (due to its significant upregulation) and then later in human epidermis [125,126]. It is difficult to decipher the exact functionality of the protein (in addition to its generic role of FA binding and trafficking) because all of these tissues also express additional FABPs. Gene ablation of A-FABP in adipocytes results in a significant upregulation of E-FABP expression, and ablation of both genes in mice suggests importance in systemic glucose and lipid homeostasis [33,116,127]. Similar to A-FABP, E-FABP physically interacts with HSL when in the holo-form, further characterising separate and distinct functions of FABPs [127]. It has also been hypothesised that E-FABP plays an important role in PPARβ/δ-mediated keratinocyte differentiation [38]. E-FABP differs from other members of the FABP/iLBP family in several ways. First, it displays a different electrophoretic mobility compared with FABPs and CRBPs of other tissues in the range pI = 6.2-6.4 for recombinant human E-FABP [128]. Secondly, it possesses six conserved cysteine residues, which is unusually high in FABPs, with Cys120 and Cys127 participating in a disulphide bond (the only reported case for the entire iLBP family) [128,129]. Additionally, it has been found to complex with psoriasin (S100A7), a calcium-binding protein expressed in keratinocytes in psoriasis and in malignant breast epithelial cells [130,131]. The physiological ligand(s) of E-FABP are not completely known; however, the preferred ligand for E-FABP in recombinant protein binding assays is stearic acid (Kd = 0.29 μM) at a stoichiometric ratio of 1:1, while sites of unsaturation appear to reduce affinity (Kd = 1.6 μM and 1.73 μM for oleic acid and arachidonic acid, respectively) [128]. X-ray crystallography has elucidated that interactions with bound ligands are similar to those of other FABP complexes, where the carboxylic head-group forms salt bridges with Arg109 and Arg129 and hydrogen bonds with Tyr131 [128]. Additionally, E-FABP exhibits the lowest conformational stability of FABPs in the presence of urea [129].
Various studies have been conducted utilising models that involve either the ablation or overexpression of E-FABP, stressing an important contribution in the regulation of insulin responses inflammation, the water permeability barrier in the epidermis and even inducing a metastatic phenotype in rat mammary epithelial cells [132-135]. It has also become a serum marker for the association with MetS (independent of A-FABP), as it was found in a population of 497 Chinese subjects and correlated positively with parameters of adiposity, adverse lipid profiles, serum insulin, A-FABP and C-reactive protein levels, and was higher in subjects with MetS (p < 0.001 versus no MetS) [136]. Polymorphisms in human FABP5 have been identified in several studies. A 340G > C (Gly114Arg) variant of FABP5 was found in a cohort of 285 Japanese autistic patients, but failed to make a genetic association with both autism and schizophrenia [137]. Phylogenetic analysis of the pig and human genomes shows a comparative map between these orthologues; seven polymorphisms of FABP5 (g.1165G > T, g.1441G > A, g.3000T > G, g.3478G > A, g.3813A > G, g.4106C > T and g.4218C > T) were identified in Iberian x Landrace crosses and found to be associated with fatness traits [138].
FABP6 (Il-FABP)
FABP6 was originally identified in both pig and mouse, and termed gastrophin and ileal LBP, respectively [139-141]. Localisation of the human orthologue on Chr 5q23-q35 has revealed, like other FAPBs, several conserved elements that transcriptionally regulate FABP6 [142]. The proximal promoters of human, rabbit and mouse FABP6 consist of a highly conserved but imperfect inverted repeat of the nuclear receptor half-site sequence (5'-(G/A)GGTGAATAACCT-3') separated by one nt (IR-1) at position -160/-148 bp [143]. The IR-1 allows for the farnesoid X receptor/retinoic acid receptor heterodimer to bind to the FABP6 DNA sequence, where binding is activated in the presence of BAs. Therefore, the IR-1 motif within the FABP6 promoter is a functional BA response element. A consensus element for the caudal-related homeobox factor 2 is present within FABP6 [144]. A putative PPAR-binding site is located at the -198 bp position and -186 bp upstream of the transcription initiation site, where gene induction via benzafibrate is PPARα- or PPARβ/δ-dependent [145]. Additionally, an SRE has been identified at position -72/-62 (5'-GTGGGGTGAC-3'), which is highly homologous with an SRE-binding protein (SREBP) binding site (SRE-1) found in the promoters of the LDL receptor, HMG-CoA synthase and glycerol 3-phosphate acyltransferase [146]. A putative Sp-1 binding site (GC box) (5'-GGGGC-3') has also been identified at positions -72/-62, proximal to the SRE, further demonstrating involvement in cholesterol homeostasis. This is confirmed when mice are fed a high cholesterol-containing diet, where Il-FABP is hypothesised to be upregulated though the SREBP1c/LXR pathway, although no LXR response element-like sequence has been found in the FABP6 promoter.
Il-FABP is highly expressed in the ileum, and to a lesser extent in the ovaries, placenta and adrenal gland [142]. The ileum plays a critical role in entero-hepatic circulation of BA, further demonstrating the importance for Il-FABP and as specificity for binding and trafficking BAs. Il-FABP also binds BA and FA (although with less affinity) and interacts with the ileal BA transporter protein at the cytoplasmic face of the ileocyte, to aid in uptake and trafficking of BAs [147]. The binding properties of Il-FABP are not yet well known and existing publications conflict as to whether one or two binding sites exist. Binding constants have not been calculated for mono-, di-, tri-hydroxylated, conjugated or unconjugated BAs, but Il-FABP exhibits significantly less affinity for palmitate and oleate than for L-FABP [63,142]. Comparisons of the binding of nine different conjugated and non-conjugated BAs to Il-FABP have been investigated by monitoring changes in intrinsic tyrosine fluorescence; affinities for ligands increased in the order of taurine-conjugated > glycine-conjugated > unconjugated BA [148]. The solution structure of human Il-FABP has been analysed using 2D- and 3D-homo and heteronuclear NMR techniques, revealing binding interactions and structural information [149]. Bound cholyltaurine is deeply buried within the core of Il-FABP and its flexible side chain is located near the FA portal as an entry region into the binding core. This study concluded that these interactions act at 1:1 protein to ligand binding ratios, and involve a large number of intermolecular nuclear overhauser effects (NOEs) between cholyltaurine and amino acids Trp49, Met59, Asn61, Phe63, Ile71, Phe79, Val83, Leu90, Val92, Phe94 and Tyr97. Interactions between photoaffinity-labelled rabbit Il-FABP and photo-reactive analogues of cholyltaurine indicated the contact point of the 3-position of the cholyltaurine derivative to be at residues His100, Thr101 and Ser102 of Il-FABP [150]. The contact point for the negatively charged side chain of cholyltaurine was identified at Arg122, where blocking studies with phenylglyoxal further reduced binding affinity for the ligand. Titration calorimetry demonstrated a lower affinity for oleate (Kd = 36 μM) [151]. X-ray crystallographic studies with zebrafish Il-FABP revealed conformational changes between apo-and holo-Il-FABP bound with cholate, where the hairpin connecting strands βG-βH obtains a more open conformation upon ligand binding [152]. This study also showed that several cholate molecules can bind to hydrophobic patches on the surface of the protein, and that two to three cholate molecules can bind within the core of a single Il-FABP molecule. Ligand selectivity has also been investigated using glycocholic (GCA) and glycocheno-deoxycholic (GCDA) acids; where each ligand occupies individual binding sites of human Il-FABP (when incubated individually with the protein), but when equimolar amounts of GCA and GCDA are mixed with protein, GCDA binds exclusively to site 1 and GCA to site 2 [153]. It was concluded that this ligand selectivity is energetically favoured, and is governed by the presence/absence of a hydroxyl group at the C-12 position of the steroid ring system.
It has been established that Il-FABP exhibits high affinity for BA and must participate in the regulation of bile homeostasis in the intestine; therefore, it seems that factors that affect the expression/activity of this protein may significantly contribute to the pathogenesis of many diseases. In a fat assimilation study, a variant of FABP6 (Thr79Met) was the most significant marker gene associated with T2DM [154]. The Met allele of the Thr79Met substitution showed decreased risk for T2DM in a nested case-control study of 192 incident T2DM subjects and 384 controls from the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam cohort. The associative relationship for risk of T2DM and the Thr79Met SNP was further investigated in a larger cohort of 543 incident T2DM cases (EPIC-Potsdam cohort) and a case control study including 939 T2DM cases from the Co-operative Health Research in the Region Augsburg (KORA) [155]. The overall frequency of the homozygous rare (Met79/Met79) allele was 16 per cent. This allele was significantly associated with a lower risk of T2DM (p = 0.04) when adjusted for age, sex, BMI and waist circumference in the EPIC-Potsdam cohort; however, this significance was not reached in the association studies with the KORA subject cohort (p = 0.06). When association studies were analysed in different BMI groups (BMI <25 kg/m2, BMI 25-30 kg/m2, BMI > 30 kg/m2), it was found that Met79/Met79 obese individuals (BMI >30 kg/m2) were at 53 per cent (EPIC-Potsdam; p = 0.02) and 31 per cent (KORA; p = 0.05) reduced relative risk for diabetes compared with Thr-allele carriers (Thr79/Thr79, Thr79/Met79). When the Thr79Met SNP was analysed for associations with anthropometric and metabolic traits related to T2DM and lipid metabolism (glucose, insulin, cholesterol, triglycerides and lipoproteins) in 2,122 non-diabetic subjects, no significant differences in age- and sex-adjusted mean values were established.
FABP7 (B-FABP)
Human FABP7 has been identified at Chr 6q22-q23, where expression of the transcript during foetal stages is thought to play an essential role in the development of the brain [156]. Regulatory elements of FABP7 control gene expression and regional expression within the brain and central nervous system (CNS). A canonical TATA box is located 30 bp upstream of the transcription start site, along with a Pax gene consensus-binding sequence (5'-AATATATTTTCACTCACACTTCGTATC-3') at -548 to -565 [157]. Temporal and spatial FABP7 expression in both radial and Bergman glial cells is regulated by a radial glial cell-specific element (RGE), and is located between 0.3 and 0.8 kb upstream of the transcription start site [6,157]. Within the same region, a Pbx/Pit-1/Oct/Unc-86 (POU) binding site for the CNS-specific Pbx proteins Pbx-1, Brn-1 and Brn-2 (5'-ATCAATCTC-3') has been found at positions -370 to -362 bp [158]. This Pbx/POU binding site has also been shown to be a critical element for the expression of B-FABP in the CNS. A hormone response element has also been identified at -286 to -275. It has been established that transcriptional regulation of FABP7 in vivo is complex; in addition to the RGE, another region controls expression in the dorsal root ganglion and notochord (-800 bp to -1,200 bp) and a silencer required for suppression in the dorsal spinal cord (-1,200 bp to -1,600 bp) also has been identified [157]. However, two NF1 binding sites have been found at positions -35 to -58 bp and -237 to -260 bp in malignant glioma cell lines [159].
Tissue expression of B-FABP is both spatially and temporally correlated with neuronal differentiation in many regions of the CNS, including the postnatal cerebellum, embryonic spinal cord and cerebral cortex [160]. In mice, high levels of expression have been demonstrated in developing and postnatal cerebellum (postnatal day 0-10), with lower levels in the young adult (day 20) and negligible expression in the adult [161]. B-FABP is also highly expressed in radial glial cells in the developing brain (preperinatal striatum) and at diminished levels in mature glia of the white matter. It is also noteworthy that neurones in the grey matter do not express B-FABP; however, both H-FABP and E-FABP are present [36]. The binding properties of B-FABP exhibit certain characteristics of other FABP family members. For example, affinities for oleic acid and arachidonic acid are ~0.4 μM and 0.25 μM, respectively in mouse B-FABP; by contrast, B-FABP does not bind palmitic acid or retinoic acid [162]. Additional binding studies revealed that B-FABP exhibits higher affinity towards FAs with longer chain lengths, with n-3 polyunsaturated FAs (docosahexaenoic acid [DHA], eicosapentaenoic acid [EPA] and α-einolenic acid) being the preferred ligands. Titration calorimetry binding studies of human B-FABP revealed affinities for monounsaturated n-9 FAs and long-chain polyunsaturated n-6 and n-3 FAs to be (Kd = 41-47 nM, 115-206 nM and 27-53 nM, respectively [163]. Polyunsaturated FAs (DHA, EPA and arachidonic acid) are highly concentrated within membrane phospholipids in the brain, by comparison with other tissues, and may further explain how B-FABP functions in the brain during the development of the CNS by ensuring the constant supply of FA during cellular maturation. Crystal structure studies have revealed the conformation of a bound oleic acid to be in the typical bent conformation within the binding pocket, at a 1:1 ratio [163]. The carboxylate group of bound oleate interacts with Try128, Thr53, Arg106 and Arg126. Additionally, Phe16 is thought to regulate ligand binding, and the side chains of residues around the outer surface (Try19, Met20, Leu23, Thr36, Pro38, Val40, Thr53, and Arg78) of the binding site appear to stabilise the folded conformation of the bound FA. By comparison with oleate, the conformation of the acyl chain of DHA adopts a more helical conformation, where the double bonds of DHA are in the cis-configuration and have rotational flexibility. The carboxylate moiety interacts with Try128 and Arg126, and the double bonds form extensive π- π interactions with Phe104, Met115 and Phe16.
Utilising gene knockdown studies in mice,[164] B-FABP has been shown to have effects on brain development, emotion, learning and memory. Although B-FABP -/- mice showed no gross morphological or histological alterations in the brain, distinct behavioural differences were found in these mice [33]. These differences included enhanced anxiety and increased fear memory, decreased levels of DHA during the neonatal period, altered FA content in the amygdala of the adult brain (increased arachidonic and palmitic acids) and decreased N-methyl-D-aspartate receptor-mediated current to DHA [164]. In mice, FABP7 was also traced using quantitative train loci analysis for impaired prepulse inhibition as an endophenotype for schizophrenia [165]. Additionally, a population-based genetic association study of 570 schizophrenics and 570 age/sex frequency-matched controls identified eight FABP7 SNPs (SNP1 -4058T > C and SNP2 -2323G < A in the 5'-flanking region; SNP3 IVS1-63T > G in intron 1; SNP4 182C > T (Thr61Met) in exon 2; SNP5 IVS3-1047G > T and SNP6 IVS3-555G > T in intron 3; and SNP7 Ex4 + 810T > C and SNP8 Ex4 + 3890T > C in the 3'-flanking region of the gene). Significant associations were detected with the Thr61Met missense SNP4 (p = 0.038), but only in male cohorts. Following modelling studies, it was proposed that this SNP causes decreased binding of DHA and alters protein stability. Another FABP7 SNP (376G > C, Val126Leu) was identified in 285 Japanese autistic individuals, but no genetic association of the SNP with autism and schizophrenia could be established [137]. A study assessing candidate genes for ethanol-preferring phenotypes in rodent models of human alcoholism uncovered that FABP7 is differentially regulated by alcohol, although the expression pattern failed to segregate phenotypes that either prefer or avoid behavioural alcohol consumption [166]. Additionally, it has been shown that B-FABP is overexpressed in the brains of foetuses with Down's syndrome (DS), and FABP7 is hypothesised to contribute to DS-associated neurological disorders [167].
FABP8 (M-FABP)
FABP8 is located at Chr 8q21.3-q22.1. The 5'-flanking region contains two AP-1 binding sites, a candidate TATA element and two CAAT sequences [13,29,168]. Along with P0 glycoprotein and myelin basic protein, M-FABP (peripheral myelin protein 2) is one of the major proteins comprising peripheral nervous system myelin (up to 15 per cent soluble protein) [29]. It is also expressed in small amounts in the CNS myelin (spinal cord and brain stem) [169]. Myelin is the multilamellar compacted membrane structure that surrounds and insulates axons and thus facilitates conduction of nerve impulses [13]. In addition to binding LCFA in Schwann cells, the function of M-FABP is largely unknown. It is hypothesised to stabilise myelin membranes by supplying LCFA and contributing to membrane biogenesis, which can be attributed to a high composition of lipid (75-80 per cent). In recombinant binding assays, it shared a similar affinity for LCFA as other FABP family members (Kd = 0.62, 0.31 and 0.37 μM for palmitate, oleate and arachidonate, respectively) [148]. X-ray crystallography has revealed that residues Arg106, Arg126 and Tyr128 interact with the carboxyl group of bound oleic acid [170]. Computational docking studies and fluorescence spectroscopy measurements have also demonstrated that cholesterol is a likely ligand for M-FABP in vivo [169]. More studies with this family member are required, as SNP associations with human diseases have not yet been established.
FABP9 (T-FABP)
FABP9 is another poorly understood member of the FABP family. It is located at Chr 8q21.13. T-FABP, or PERF15, is a major protein found in the inner acrosomal membrane and outer face of the nuclear envelope of mammalian sperm [171]. It shares the most homology with M-FABP (MP2), A-FABP and H-FABP; therefore, it is hypothesised that it shares similar binding affinities for LCFA. It is expressed during spermatogenesis and in mammalian testis [172,173]. It is known that the FA composition of sperm is related to fertility, so the proposed function of T-FABP has been hypothesised to protect sperm FA from oxidation, thus maintaining their ability to fertilise oocytes [174]. Other roles of T-FABP have been investigated with FABP9 -/- mice, where gene-ablated mice had significant increases in sperm head abnormalities (8 per cent greater than WT), but still remained fertile [175]. The total lipid profile of the FABP9 -/-mice did not change when compared with WT controls; however, it was proposed that FABP12 may share a compensatory role.
FABP12
The FABP12 gene is the newest member of the FABP family and, to date, little information is available on the functional properties of the protein. FABP12 is phylogenetically restricted; it has been identified in human, rat and mouse but no counterpart has been identified in chicken and zebrafish genomes [18]. It has been mapped to the same chromosomal region as FABP4, FABP5, FABP8 and FABP9 (8q21.13). Regulatory elements that control gene transcription have not been verified. FABP12 mRNA has been identified in the retina and testis at high levels, and to lesser extent in the cerebral cortex, kidney and epididymis of rat and mouse tissues. The transcript has also been identified in human retinoblastoma cells. Identification of the FABP12 gene provides further evidence of the evolutionary divergences of the FABP protein family from a single ancestral gene, as it is hypothesised to have formed from a tandem gene duplication.
Conclusions
The FABP gene family embodies a group of diverse proteins which participate in a spectrum of tissue-specific pathways involved in lipid homeostasis. To date, nine putatively functional protein-coding FABP genes have been identified in humans. A newer member of the family, FABP12, has been less studied. The evolutionary conservation of these genes suggests that FABPs have diverse and highly specified roles in regulating the metabolism and actions of the ligands they bind. As we understand these functions more clearly, FABPs will be highlighted as important targets for drug development and therapy for many metabolic diseases.
Acknowledgements
This work was supported in part by Grants NIH/NIAAA R37AA09300, NIH/NIAAA R01 DK074487-01 and NIH/NIAAA NRSA (RLS) 5 F31 AA018898-02.
References
- Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur MJ, Atshaves BP, Frolov A, Foxworth WD. et al. Cellular uptake and intracellular trafficking of long chain fatty acids. J Lipid Res. 1999;40:1371–1383. [PubMed] [Google Scholar]
- Storch J, Thumser AE. Tissue-specific functions in the fatty acid-binding protein family. J Biol Chem. 2010;285:32679–32683. doi: 10.1074/jbc.R110.135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszak L, Winter N, Xu Z, Bernlohr DA. et al. Lipid-binding proteins: A family of fatty acid and retinoid transport proteins. Adv Protein Chem. 1994;45:89–151. doi: 10.1016/s0065-3233(08)60639-7. [DOI] [PubMed] [Google Scholar]
- Marcelino AM, Smock RG, Gierasch LM. Evolutionary coupling of structural and functional sequence information in the intracellular lipid-binding protein family. Proteins. 2006;63:373–384. doi: 10.1002/prot.20860. [DOI] [PubMed] [Google Scholar]
- Chmurzynska A. The multigene family of fatty acid-binding proteins (FABPs): Function, structure and polymorphism. J Appl Genet. 2006;47:39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- Corsico B, Cistola DP, Frieden C, Storch J. The helical domain of intestinal fatty acid binding protein is critical for collisional transfer of fatty acids to phospholipid membranes. Proc Natl Acad Sci USA. 1998;95:12174–12178. doi: 10.1073/pnas.95.21.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou HL, Kahn PC, Storch J. Role of the helical domain in fatty acid transfer from adipocyte and heart fatty acid-binding proteins to membranes: analysis of chimeric proteins. J Biol Chem. 2002;277:1806–1815. doi: 10.1074/jbc.M107987200. [DOI] [PubMed] [Google Scholar]
- Sacchettini JC, Gordon JI, Banaszak LJ. Crystal structure of rat intestinal fatty-acid-binding protein. Refinement and analysis of the Escherichia coli-derived protein with bound palmitate. J Mol Biol. 1989;208:327–339. doi: 10.1016/0022-2836(89)90392-6. [DOI] [PubMed] [Google Scholar]
- Storch J, McDermott L. Structural and functional analysis of fatty acid-binding proteins. J Lipid Res. 2009;50(Suppl):S126–S131. doi: 10.1194/jlr.R800084-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richieri GV, Ogata RT, Zimmerman AW, Veerkamp JH. et al. Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry. 2000;39:7197–7204. doi: 10.1021/bi000314z. [DOI] [PubMed] [Google Scholar]
- Richieri GV, Ogata RT, Kleinfeld AM. Fatty acid interactions with native and mutant fatty acid binding proteins. Mol Cell Biochem. 1999;192:77–85. doi: 10.1023/A:1006826405151. [DOI] [PubMed] [Google Scholar]
- Hayasaka K, Himoro M, Takada G, Takahashi E. et al. Structure and localization of the gene encoding human peripheral myelin protein 2 (PMP2) Genomics. 1993;18:244–248. doi: 10.1006/geno.1993.1462. [DOI] [PubMed] [Google Scholar]
- Liu RZ, Denovan-Wright EM, Wright JM. Structure, linkage mapping and expression of the heart-type fatty acid-binding protein gene (fabp3) from zebrafish (Danio rerio) Eur J Biochem. 2003;270:3223–3234. doi: 10.1046/j.1432-1033.2003.03705.x. [DOI] [PubMed] [Google Scholar]
- Sweetser DA, Birkenmeier EH, Klisak IJ, Zollman S. et al. The human and rodent intestinal fatty acid binding protein genes. A comparative analysis of their structure, expression, and linkage relationships. J Biol Chem. 1987;262:16060–16071. [PubMed] [Google Scholar]
- Sweetser DA, Lowe JB, Gordon JI. The nucleotide sequence of the rat liver fatty acid-binding protein gene. Evidence that exon 1 encodes an oligopeptide domain shared by a family of proteins which bind hydrophobic ligands. J Biol Chem. 1986;261:5553–5561. [PubMed] [Google Scholar]
- Zimmerman AW, Veerkamp JH. New insights into the structure and function of fatty acid-binding proteins. Cell Mol Life Sci. 2002;59:1096–1116. doi: 10.1007/s00018-002-8490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RZ, Li X, Godbout R. A novel fatty acid-binding protein (FABP) gene resulting from tandem gene duplication in mammals: Transcription in rat retina and testis. Genomics. 2008;92:436–445. doi: 10.1016/j.ygeno.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Phelan CM, Larsson C, Baird S, Futreal PA. et al. The human mammary-derived growth inhibitor (MDGI) gene: Genomic structure and mutation analysis in human breast tumors. Genomics. 1996;34:63–68. doi: 10.1006/geno.1996.0241. [DOI] [PubMed] [Google Scholar]
- Bernlohr DA, Simpson MA, Hertzel AV, Banaszak LJ. Intracellular lipid-binding proteins and their genes. Annu Rev Nutr. 1997;17:277–303. doi: 10.1146/annurev.nutr.17.1.277. [DOI] [PubMed] [Google Scholar]
- Li E, Norris AW. Structure/function of cytoplasmic vitamin A-binding proteins. Annu Rev Nutr. 1996;16:205–234. doi: 10.1146/annurev.nu.16.070196.001225. [DOI] [PubMed] [Google Scholar]
- Hunt CR, Ro JH, Dobson DE, Min HY. et al. Adipocyte P2 gene: Developmental expression and homology of 5'-flanking sequences among fat cell-specific genes. Proc Natl Acad Sci USA. 1986;83:3786–3790. doi: 10.1073/pnas.83.11.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower DR, North AC, Attwood T. Structure and sequence relationships in the lipocalins and related proteins. Protein Sci. 1993;2:753–761. doi: 10.1002/pro.5560020507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers SD, Nedrow KL, Gillilan RE, Noy N. Continuous nucleocytoplasmic shuttling underlies transcriptional activation of PPAR gamma by FABP4. Biochemistry. 2007;46:6744–6752. doi: 10.1021/bi700047a. [DOI] [PubMed] [Google Scholar]
- Gillilan RE, Ayers SD, Noy N. Structural basis for activation of fatty acid-binding protein 4. J Mol Biol. 2007;372:1246–1260. doi: 10.1016/j.jmb.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, Thompson BR, Sanders MA, Bernlohr DA. Interaction of the adipocyte fatty acid-binding protein with the hormone-sensitive lipase: Regulation by fatty acids and phosphorylation. J Biol Chem. 2007;282:32424–32432. doi: 10.1074/jbc.M703730200. [DOI] [PubMed] [Google Scholar]
- Prinsen CF, Weghuis DO, Kessel AG, Veerkamp JH. Identification of a human heart FABP pseudogene located on chromosome 13. Gene. 1997;193:245–251. doi: 10.1016/S0378-1119(97)00129-7. [DOI] [PubMed] [Google Scholar]
- Treuner M, Kozak CA, Gallahan D, Grosse R. et al. Cloning and characterization of the mouse gene encoding mammary-derived growth inhibitor/heart-fatty acid-binding protein. Gene. 1994;147:237–242. doi: 10.1016/0378-1119(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Veerkamp JH, Maatman RG. Cytoplasmic fatty acid-binding proteins: Their structure and genes. Prog Lipid Res. 1995;34:17–52. doi: 10.1016/0163-7827(94)00005-7. [DOI] [PubMed] [Google Scholar]
- Flower DR. Structural relationship of streptavidin to the calycin protein superfamily. FEBS Lett. 1993;333:99–102. doi: 10.1016/0014-5793(93)80382-5. [DOI] [PubMed] [Google Scholar]
- Flower DR. The lipocalin protein family: Structure and function. Biochem J. 1996;318:1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap FG, van der Vusse GJ, Glatz JF. Evolution of the family of intracellular lipid binding proteins in vertebrates. Mol Cell Biochem. 2002;239:69–77. doi: 10.1023/A:1020519011939. [DOI] [PubMed] [Google Scholar]
- Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- Schleicher CH, Cordoba OL, Santome JA, Dell'Angelica EC. Molecular evolution of the multigene family of intracellular lipid-binding proteins. Biochem Mol Biol Int. 1995;36:1117–1125. [PubMed] [Google Scholar]
- Glatz JF, van der Vusse GJ. Cellular fatty acid-binding proteins: Their function and physiological significance. Prog Lipid Res. 1996;35:243–282. doi: 10.1016/S0163-7827(96)00006-9. [DOI] [PubMed] [Google Scholar]
- Haunerland NH, Spener F. Fatty acid-binding proteins: Insights from genetic manipulations. Prog Lipid Res. 2004;43:328–349. doi: 10.1016/j.plipres.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Schachtrup C, Emmler T, Bleck B, Sandqvist A. et al. Functional analysis of peroxisome-proliferator-responsive element motifs in genes of fatty acid-binding proteins. Biochem J. 2004;382:239–245. doi: 10.1042/BJ20031340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan NS, Shaw NS, Vinckenbosch N, Liu P, Yasmin R. et al. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atshaves BP, Martin GG, Hostetler HA, McIntosh AL. et al. Liver fatty acid-binding protein and obesity. J Nutr Biochem. 2010;21:1015–1032. doi: 10.1016/j.jnutbio.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetler HA, McIntosh AL, Atshaves BP, Storey SM. et al. L-FABP directly interacts with PPAR alpha in cultured primary hepatocytes. J Lipid Res. 2009;50:1663–1675. doi: 10.1194/jlr.M900058-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek BA, Cistola DP, Hamilton JA, Kaduce TL. et al. Fatty acid binding proteins reduce 15-lipoxygenase-induced oxygenation of linoleic acid and arachidonic acid. Biochim Biophys Acta. 1997;1346:75–85. doi: 10.1016/S0005-2760(97)00021-0. [DOI] [PubMed] [Google Scholar]
- Widstrom RL, Norris AW, Spector AA. Binding of cytochrome P450 mono oxygenase and lipoxygenase pathway products by heart fatty acid-binding protein. Biochemistry. 2001;40:1070–1076. doi: 10.1021/bi001602y. [DOI] [PubMed] [Google Scholar]
- Chen SH, Van Tuinen P, Ledbetter DH, Smith LC. et al. Human liver fatty acid binding protein gene is located on chromosome 2. Somat Cell Mol Genet. 1986;12:303–306. doi: 10.1007/BF01570790. [DOI] [PubMed] [Google Scholar]
- Landrier JF, Thomas C, Grober J, Duez H. et al. Statin induction of liver fatty acid-binding protein (L-FABP) gene expression is peroxisome proliferator-activated receptor-alpha-dependent. J Biol Chem. 2004;279:45512–45518. doi: 10.1074/jbc.M407461200. [DOI] [PubMed] [Google Scholar]
- Issemann I, Prince R, Tugwood J, Green S. A role for fatty acids and liver fatty acid binding protein in peroxisome proliferation? Biochem Soc Trans. 1992;20:824–827. doi: 10.1042/bst0200824. [DOI] [PubMed] [Google Scholar]
- Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010. [DOI] [PMC free article] [PubMed]
- Thompson J, Winter N, Terwey D, Bratt J. et al. The crystal structure of the liver fatty acid-binding protein. A complex with two bound oleates. J Biol Chem. 1997;272:7140–7150. doi: 10.1074/jbc.272.11.7140. [DOI] [PubMed] [Google Scholar]
- He Y, Yang X, Wang H, Estephan R. et al. Solution-state molecular structure of apo and oleate-liganded liver fatty acid-binding protein. Biochemistry. 2007;46:12543–12556. doi: 10.1021/bi701092r. [DOI] [PubMed] [Google Scholar]
- Long D, Mu Y, Yang D. Molecular dynamics simulation of ligand dissociation from liver fatty acid binding protein. PLoS One. 2009;4:e6081. doi: 10.1371/journal.pone.0006081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AW, Spector AA. Very long chain n-3 and n-6 polyunsaturated fatty acids bind strongly to liver fatty acid-binding protein. J Lipid Res. 2002;43:646–653. [PubMed] [Google Scholar]
- Wolfrum C, Borchers T, Sacchettini JC, Spener F. Binding of fatty acids and peroxisome proliferators to orthologous fatty acid binding proteins from human, murine, and bovine liver. Biochemistry. 2000;39:1469–1474. doi: 10.1021/bi991638u. [DOI] [PubMed] [Google Scholar]
- Frolov A, Cho TH, Murphy EJ, Schroeder F. Isoforms of rat liver fatty acid binding protein differ in structure and affinity for fatty acids and fatty acyl CoAs. Biochemistry. 1997;36:6545–6555. doi: 10.1021/bi970205t. [DOI] [PubMed] [Google Scholar]
- Thumser AE, Wilton DC. The binding of cholesterol and bile salts to recombinant rat liver fatty acid-binding protein. Biochem J. 1996;320:729–733. doi: 10.1042/bj3200729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang S, Velkov T, Horne J, Porter CJ. Characterization of the drug binding specificity of rat liver fatty acid binding protein. J Med Chem. 2008;51:3755–3764. doi: 10.1021/jm701192w. [DOI] [PubMed] [Google Scholar]
- Martin GG, Atshaves BP, McIntosh AL, Mackie JT. et al. Liver fatty acid binding protein gene ablation potentiates hepatic cholesterol accumulation in cholesterol-fed female mice. Am J Physiol Gastrointest Liver Physiol. 2006;290:G36–G48. doi: 10.1152/ajpgi.00510.2004. [DOI] [PubMed] [Google Scholar]
- Martin GG, Huang H, Atshaves BP, Binas B. et al. Ablation of the liver fatty acid binding protein gene decreases fatty acyl CoA binding capacity and alters fatty acyl CoA pool distribution in mouse liver. Biochemistry. 2003;42:11520–11532. doi: 10.1021/bi0346749. [DOI] [PubMed] [Google Scholar]
- Newberry EP, Xie Y, Kennedy S, Han X. et al. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid-binding protein gene. J Biol Chem. 2003;278:51664–51672. doi: 10.1074/jbc.M309377200. [DOI] [PubMed] [Google Scholar]
- Brouillette C, Bosse Y, Perusse L, Gaudet D. et al. Effect of liver fatty acid binding protein (FABP) T94A missense mutation on plasma lipoprotein responsiveness to treatment with feno fibrate. J Hum Genet. 2004;49:424–432. doi: 10.1007/s10038-004-0171-2. [DOI] [PubMed] [Google Scholar]
- Fisher E, Weikert C, Klapper M, Lindner I. et al. L-FABP T94A is associated with fasting triglycerides and LDL-cholesterol in women. Mol Genet Metab. 2007;91:278–284. doi: 10.1016/j.ymgme.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Green RP, Cohn SM, Sacchettini JC, Jackson KE. et al. The mouse intestinal fatty acid binding protein gene: nucleotide sequence, pattern of developmental and regional expression, and proposed structure of its protein product. DNA Cell Biol. 1992;11:31–41. doi: 10.1089/dna.1992.11.31. [DOI] [PubMed] [Google Scholar]
- Cohn SM, Simon TC, Roth KA, Birkenmeier EH. et al. Use of transgenic mice to map cis-acting elements in the intestinal fatty acid binding protein gene (Fabpi) that control its cell lineage-specific and regional patterns of expression along the duodenal-colonic and crypt-villus axes of the gut epithelium. J Cell Biol. 1992;119:27–44. doi: 10.1083/jcb.119.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottman JN, Gordon JI. Comparison of the patterns of expression of rat intestinal fatty acid binding protein/human growth hormone fusion genes in cultured intestinal epithelial cell lines and in the gut epithelium of transgenic mice. J Biol Chem. 1993;268:11994–12002. [PubMed] [Google Scholar]
- Besnard P, Niot I, Poirier H, Clement L. et al. New insights into the fatty acid-binding protein (FABP) family in the small intestine. Mol Cell Biochem. 2002;239:139–147. doi: 10.1023/A:1020505512364. [DOI] [PubMed] [Google Scholar]
- Lowe JB, Sacchettini JC, Laposata M, McQuillan JJ. et al. Expression of rat intestinal fatty acid-binding protein in Escherichia coli. Purification and comparison of ligand binding characteristics with that of Escherichia coli-derived rat liver fatty acid-binding protein. J Biol Chem. 1987;262:5931–5937. [PubMed] [Google Scholar]
- Nemecz G, Hubbell T, Jefferson JR, Lowe JB. et al. Interaction of fatty acids with recombinant rat intestinal and liver fatty acid-binding proteins. Arch Biochem Biophys. 1991;286:300–309. doi: 10.1016/0003-9861(91)90044-J. [DOI] [PubMed] [Google Scholar]
- Hodsdon ME, Cistola DP. Ligand binding alters the backbone mobility of intestinal fatty acid-binding protein as monitored by 15N NMR relaxation and 1H exchange. Biochemistry. 1997;36:2278–2290. doi: 10.1021/bi962018l. [DOI] [PubMed] [Google Scholar]
- Velkov T, Chuang S, Wielens J, Sakellaris H. et al. The interaction of lipophilic drugs with intestinal fatty acid-binding protein. J Biol Chem. 2005;280:17769–17776. doi: 10.1074/jbc.M410193200. [DOI] [PubMed] [Google Scholar]
- Velkov T, Lim ML, Horne J, Simpson JS. et al. Characterization of lipophilic drug binding to rat intestinal fatty acid binding protein. Mol Cell Biochem. 2009;326:87–95. doi: 10.1007/s11010-008-0009-x. [DOI] [PubMed] [Google Scholar]
- Ishii T, Hirose H, Kawai T, Hayashi K. et al. Effects of intestinal fatty acid-binding protein gene Ala54Thr polymorphism and beta3-adrenergic receptor gene Trp64Arg polymorphism on insulin resistance and fasting plasma glucose in young to older Japanese men. Metabolism. 2001;50:1301–1307. doi: 10.1053/meta.2001.27228. [DOI] [PubMed] [Google Scholar]
- Baier LJ, Sacchettini JC, Knowler WC, Eads J. et al. An amino acid substitution in the human intestinal fatty acid binding protein is associated with increased fatty acid binding, increased fat oxidation, and insulin resistance. J Clin Invest. 1995;95:1281–1287. doi: 10.1172/JCI117778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agren JJ, Valve R, Vidgren H, Laakso M. et al. Postprandial lipemic response is modified by the polymorphism at codon 54 of the fatty acid-binding protein 2 gene. Arterioscler Thromb Vasc Biol. 1998;18:1606–1610. doi: 10.1161/01.ATV.18.10.1606. [DOI] [PubMed] [Google Scholar]
- Weiss EP, Brown MD, Shuldiner AR, Hagberg JM. Fatty acid binding protein-2 gene variants and insulin resistance: Gene and gene-environment interaction effects. Physiol Genomics. 2002;10:145–157. doi: 10.1152/physiolgenomics.00070.2001. [DOI] [PubMed] [Google Scholar]
- Hegele RA, Harris SB, Hanley AJ, Sadikian S. et al. Genetic variation of intestinal fatty acid-binding protein associated with variation in body mass in aboriginal Canadians. J Clin Endocrinol Metab. 1996;81:4334–4337. doi: 10.1210/jc.81.12.4334. [DOI] [PubMed] [Google Scholar]
- Kim CH, Yun SK, Byun DW, Yoo MH. et al. Codon 54 polymorphism of the fatty acid binding protein 2 gene is associated with increased fat oxidation and hyperinsulinemia, but not with intestinal fatty acid absorption in Korean men. Metabolism. 2001;50:473–476. doi: 10.1053/meta.2001.21022. [DOI] [PubMed] [Google Scholar]
- Lei HH, Coresh J, Shuldiner AR, Boerwinkle E. et al. Variants of the insulin receptor substrate-1 and fatty acid binding protein 2 genes and the risk of type 2 diabetes, obesity, and hyperinsulinemia in African-Americans: The Atherosclerosis Risk in Communities Study. Diabetes. 1999;48:1868–1872. doi: 10.2337/diabetes.48.9.1868. [DOI] [PubMed] [Google Scholar]
- Humphreys P, McCarthy M, Tuomilehto J, Tuomilehto-Wolf E. et al. Chromosome 4q locus associated with insulin resistance in Pima Indians. Studies in three European NIDDM populations. Diabetes. 1994;43:800–804. doi: 10.2337/diabetes.43.6.800. [DOI] [PubMed] [Google Scholar]
- Prochazka M, Lillioja S, Tait JF, Knowler WC. et al. Linkage of chromosomal markers on 4q with a putative gene determining maximal insulin action in Pima Indians. Diabetes. 1993;42:514–519. doi: 10.2337/diabetes.42.4.514. [DOI] [PubMed] [Google Scholar]
- de Luis DA, Ruiz E, Cordero M, Munoz C. et al. Polymorphism Ala54Thr of fatty acid-binding protein 2: Allelic frequencies and influence on cardiovascular risk factors in a multicenter study of Castilla y Leon. Ann Nutr Metab. 2010;57:163–168. doi: 10.1159/000321632. [DOI] [PubMed] [Google Scholar]
- de Luis DA, Aller R, Izaola O, Conde R. Metabolic syndrome and ALA54THR polymorphism of fatty acid-binding protein 2 in obese patients. Metabolism. 2010. in press . [DOI] [PubMed]
- Pihlajamaki J, Rissanen J, Heikkinen S, Karjalainen L. et al. Codon 54 polymorphism of the human intestinal fatty acid binding protein 2 gene is associated with dyslipidemias but not with insulin resistance in patients with familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol. 1997;17:1039–1044. doi: 10.1161/01.ATV.17.6.1039. [DOI] [PubMed] [Google Scholar]
- Qian Q, Kuo L, Yu YT, Rottman JN. A concise promoter region of the heart fatty acid-binding protein gene dictates tissue-appropriate expression. Circ Res. 1999;84:276–289. doi: 10.1161/01.RES.84.3.276. [DOI] [PubMed] [Google Scholar]
- Kawabe K, Saegusa H, Seto K, Urabe H. et al. Peroxisome proliferator-activated receptor alpha and its response element are required but not sufficient for transcriptional activation of the mouse heart-type fatty acid binding protein gene. Int J Biochem Cell Biol. 2005;37:1534–1546. doi: 10.1016/j.biocel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Gerbens F, Rettenberger G, Lenstra JA, Veerkamp JH. et al. Characterization, chromosomal localization, and genetic variation of the porcine heart fatty acid-binding protein gene. Mamm Genome. 1997;8:328–332. doi: 10.1007/s003359900433. [DOI] [PubMed] [Google Scholar]
- Offner GD, Troxler RF, Brecher P. Characterization of a fatty acid-binding protein from rat heart. J Biol Chem. 1986;261:5584–5589. [PubMed] [Google Scholar]
- Tanaka T, Hirota Y, Sohmiya K, Nishimura S. et al. Serum and urinary human heart fatty acid-binding protein in acute myocardial infarction. Clin Biochem. 1991;24:195–201. doi: 10.1016/0009-9120(91)90571-U. [DOI] [PubMed] [Google Scholar]
- Viswanathan K, Kilcullen N, Morrell C, Thistlethwaite SJ. et al. Heart-type fatty acid-binding protein predicts long-term mortality and re-infarction in consecutive patients with suspected acute coronary syndrome who are troponin-negative. J Am Coll Cardiol. 2010;55:2590–2598. doi: 10.1016/j.jacc.2009.12.062. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kim H, Kim SY, Lee HK. et al. Automated heart-type fatty acid-binding protein assay for the early diagnosis of acute myocardial infarction. Am J Clin Pathol. 2010;134:157–162. doi: 10.1309/AJCP0F6AXRCJMQQG. [DOI] [PubMed] [Google Scholar]
- Matsui Y, Satoh K, Mutsukura K, Watanabe T. et al. Development of an ultra-rapid diagnostic method based on heart-type fatty acid binding protein levels in the CSF of CJD patients. Cell Mol Neurobiol. 2010;30:991–999. doi: 10.1007/s10571-010-9529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerkamp JH, van Moerkerk HT, Prinsen CF, van Kuppevelt TH. Structural and functional studies on different human FABP types. Mol Cell Biochem. 1999;192:137–142. doi: 10.1023/A:1006866119264. [DOI] [PubMed] [Google Scholar]
- Zanotti G, Scapin G, Spadon P, Veerkamp JH. et al. Three-dimensional structure of recombinant human muscle fatty acid-binding protein. J Biol Chem. 1992;267:18541–18550. doi: 10.2210/pdb2hmb/pdb. [DOI] [PubMed] [Google Scholar]
- Young AC, Scapin G, Kromminga A, Patel SB. et al. Structural studies on human muscle fatty acid binding protein at 1. 4 A resolution: Binding interactions with three C18 fatty acids. Structure. 1994;2:523–534. doi: 10.1016/S0969-2126(00)00052-6. [DOI] [PubMed] [Google Scholar]
- Prinsen CF, Veerkamp JH. Fatty acid binding and conformational stability of mutants of human muscle fatty acid-binding protein. Biochem J. 1996;314:253–260. doi: 10.1042/bj3140253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HD, Kim LH, Park BL, Jung HS. et al. Polymorphisms in fatty acid-binding protein-3 (FABP3)--Putative association with type 2 diabetes mellitus. Hum Mutat. 2003;22:180. doi: 10.1002/humu.9168. [DOI] [PubMed] [Google Scholar]
- Ye MH, Chen JL, Zhao GP, Zheng MQ. et al. Associations of A-FABP and H-FABP markers with the content of intramuscular fat in Beijing-You chicken. Anim Biotechnol. 2010;21:14–24. doi: 10.1080/10495390903328116. [DOI] [PubMed] [Google Scholar]
- Gerbens F, van Erp AJ, Harders FL, Verburg FJ. et al. Effect of genetic variants of the heart fatty acid-binding protein gene on intramuscular fat and performance traits in pigs. J Anim Sci. 1999;77:846–852. doi: 10.2527/1999.774846x. [DOI] [PubMed] [Google Scholar]
- Switonski M, Stachowiak M, Cieslak J, Bartz M. et al. Genetics of fat tissue accumulation in pigs: A comparative approach. J Appl Genet. 2010;51:153–168. doi: 10.1007/BF03195724. [DOI] [PubMed] [Google Scholar]
- Lazary A, Kosa JP, Tobias B, Lazary J. et al. Single nucleotide polymorphisms in new candidate genes are associated with bone mineral density and fracture risk. Eur J Endocrinol. 2008;159:187–196. doi: 10.1530/EJE-08-0021. [DOI] [PubMed] [Google Scholar]
- Sul HS. Adipocyte differentiation and gene expression. Curr Opin Cell Biol. 1989;1:1116–1121. doi: 10.1016/S0955-0674(89)80059-6. [DOI] [PubMed] [Google Scholar]
- Vasseur-Cognet M, Lane MD. Trans-acting factors involved in adipogenic differentiation. Curr Opin Genet Dev. 1993;3:238–245. doi: 10.1016/0959-437X(93)90029-O. [DOI] [PubMed] [Google Scholar]
- Graves RA, Tontonoz P, Ross SR, Spiegelman BM. Identification of a potent adipocyte-specific enhancer: Involvement of an NF-1-like factor. Genes Dev. 1991;5:428–437. doi: 10.1101/gad.5.3.428. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Graves RA, Budavari AI. et al. mPPAR gamma 2: Tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- Krusinova E, Pelikanova T. Fatty acid binding proteins in adipose tissue: A promising link between metabolic syndrome and atherosclerosis? Diabetes Res Clin Pract. 2008;82(Suppl 2):S127–S134. doi: 10.1016/j.diabres.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Reese-Wagoner A, Thompson J, Banaszak L. Structural properties of the adipocyte lipid binding protein. Biochim Biophys Acta. 1999;1441:106–116. doi: 10.1016/S1388-1981(99)00154-7. [DOI] [PubMed] [Google Scholar]
- Makowski L, Brittingham KC, Reynolds JM, Suttles J. et al. The fatty acid-binding protein, aP2, coordinates macrophage cholesterol trafficking and inflammatory activity. Macrophage expression of aP2 impacts peroxisome proliferator-activated receptor gamma and I kappa B kinase activities. J Biol Chem. 2005;280:12888–12895. doi: 10.1074/jbc.M413788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui X, Li H, Zhou Z, Lam KS. et al. Adipocyte fatty acid-binding protein modulates inflammatory responses in macrophages through a positive feedback loop involving c-Jun NH2-terminal kinases and activator protein-1. J Biol Chem. 2010;285:10273–10280. doi: 10.1074/jbc.M109.097907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, Gustafsson T, Sundberg CJ, Norrbom J. et al. Fatty acid binding protein 4 in human skeletal muscle. Biochem Biophys Res Commun. 2006;346:125–130. doi: 10.1016/j.bbrc.2006.05.083. [DOI] [PubMed] [Google Scholar]
- Matarese V, Bernlohr DA. Purification of murine adipo-cyte lipid-binding protein. Characterization as a fatty acid- and retinoic acid-binding protein. J Biol Chem. 1988;263:14544–14551. [PubMed] [Google Scholar]
- Hellberg K, Grimsrud PA, Kruse AC, Banaszak LJ. et al. X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal. Protein Sci. 2010;19:1480–1489. doi: 10.1002/pro.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaLonde JM, Bernlohr DA, Banaszak LJ. X-ray crystallographic structures of adipocyte lipid-binding protein complexed with palmitate and hexadecane sulfonic acid. Properties of cavity binding sites. Biochemistry. 1994;33:4885–4895. doi: 10.1021/bi00182a017. [DOI] [PubMed] [Google Scholar]
- Xu Z, Bernlohr DA, Banaszak LJ. The adipocyte lipid-binding protein at 1.6-A resolution. Crystal structures of the apoprotein and with bound saturated and unsaturated fatty acids. J Biol Chem. 1993;268:7874–7884. [PubMed] [Google Scholar]
- Xu Z, Bernlohr DA, Banaszak LJ. Crystal structure of recombinant murine adipocyte lipid-binding protein. Biochemistry. 1992;31:3484–3492. doi: 10.1021/bi00128a024. [DOI] [PubMed] [Google Scholar]
- LaLonde JM, Bernlohr DA, Banaszak LJ. Adipocyte lipid-binding protein complexed with arachidonic acid. Titration calorimetry and X-ray crystallographic studies. J Biol Chem. 1994;269:25339–25347. doi: 10.2210/pdb1adl/pdb. [DOI] [PubMed] [Google Scholar]
- LiCata VJ, Bernlohr DA. Surface properties of adipocyte lipid-binding protein: Response to lipid binding, and comparison with homologous proteins. Proteins. 1998;33:577–589. doi: 10.1002/(SICI)1097-0134(19981201)33:4<577::AID-PROT10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Buelt MK, Shekels LL, Jarvis BW, Bernlohr DA. In vitro phosphorylation of the adipocyte lipid-binding protein (p15) by the insulin receptor. Effects of fatty acid on receptor kinase and substrate phosphorylation. J Biol Chem. 1991;266:12266–12271. [PubMed] [Google Scholar]
- Buelt MK, Xu Z, Banaszak LJ, Bernlohr DA. Structural and functional characterization of the phosphorylated adipocyte lipid-binding protein (pp15) Biochemistry. 1992;31:3493–3499. doi: 10.1021/bi00128a025. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS, Johnson RS, Distel RJ, Ellis R. et al. Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science. 1996;274:1377–1379. doi: 10.1126/science.274.5291.1377. [DOI] [PubMed] [Google Scholar]
- Lehmann F, Haile S, Axen E, Medina C. et al. Discovery of inhibitors of human adipocyte fatty acid-binding protein, a potential type 2 diabetes target. Bioorg Med Chem Lett. 2004;14:4445–4448. doi: 10.1016/j.bmcl.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Xu A, Wang Y, Xu JY, Stejskal D. et al. Adipocyte fatty acid-binding protein is a plasma biomarker closely associated with obesity and metabolic syndrome. Clin Chem. 2006;52:405–413. doi: 10.1373/clinchem.2005.062463. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu S, Niu T, Xu X. SNPHunter: A bioinformatic software for single nucleotide polymorphism data acquisition and management. BMC Bioinformatics. 2005;6:60. doi: 10.1186/1471-2105-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncman G, Erbay E, Hom X, De Vivo I. et al. A genetic variant at the fatty acid-binding protein aP2 locus reduces the risk for hypertriglyceridemia, type 2 diabetes, and cardiovascular disease. Proc Natl Acad Sci USA. 2006;103:6970–6975. doi: 10.1073/pnas.0602178103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damcott CM, Moffett SP, Feingold E, Barmada MM. et al. Genetic variation in fatty acid-binding protein-4 and peroxi-some proliferator-activated receptor gamma interactively influence insulin sensitivity and body composition in males. Metabolism. 2004;53:303–309. doi: 10.1016/j.metabol.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Khalyfa A, Bhushan B, Hegazi M, Kim J. et al. Fatty-acid binding protein 4 gene variants and childhood obesity: Potential implications for insulin sensitivity and CRP levels. Lipids Health Dis. 2010;9:18. doi: 10.1186/1476-511X-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tang J, Wang B, Song J. et al. FABP4: A novel candidate gene for polycystic ovary syndrome. Endocrine. 2009;36:392–396. doi: 10.1007/s12020-009-9228-5. [DOI] [PubMed] [Google Scholar]
- Bleck B, Hohoff C, Binas B, Rustow B. et al. Cloning and chromosomal localisation of the murine epidermal-type fatty acid binding protein gene (Fabpe) Gene. 1998;215:123–130. doi: 10.1016/S0378-1119(98)00262-5. [DOI] [PubMed] [Google Scholar]
- Madsen P, Rasmussen HH, Leffers H, Honore B. et al. Molecular cloning and expression of a novel keratinocyte protein (psoriasis-associated fatty acid-binding protein [PA-FABP]) that is highly up-regulated in psoriatic skin and that shares similarity to fatty acid-binding proteins. J Invest Dermatol. 1992;99:299–305. doi: 10.1111/1523-1747.ep12616641. [DOI] [PubMed] [Google Scholar]
- Siegenthaler G, Hotz R, Chatellard-Gruaz D, Jaconi S. et al. Characterization and expression of a novel human fatty acid-binding protein: The epidermal type (E-FABP) Biochem Biophys Res Commun. 1993;190:482–487. doi: 10.1006/bbrc.1993.1073. [DOI] [PubMed] [Google Scholar]
- Jenkins-Kruchten AE, Bennaars-Eiden A, Ross JR, Shen WJ. et al. Fatty acid-binding protein-hormone-sensitive lipase interaction. Fatty acid dependence on binding. J Biol Chem. 2003;278:47636–47643. doi: 10.1074/jbc.M307680200. [DOI] [PubMed] [Google Scholar]
- Hohoff C, Borchers T, Rustow B, Spener F. et al. Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein. Biochemistry. 1999;38:12229–12239. doi: 10.1021/bi990305u. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Gonzalez LH, Ludwig C, Hohoff C, Rademacher M. et al. Solution structure and backbone dynamics of human epidermal-type fatty acid-binding protein (E-FABP) Biochem J. 2002;364:725–737. doi: 10.1042/BJ20020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagens G, Masouye I, Augsburger E, Hotz R. et al. Calcium-binding protein S100A7 and epidermal-type fatty acid-binding protein are associated in the cytosol of human keratinocytes. Biochem J. 1999;339:419–427. doi: 10.1042/0264-6021:3390419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PH, Leygue ER, Murphy LC. Psoriasin (S100A7) Int J Biochem Cell Biol. 1998;30:567–571. doi: 10.1016/S1357-2725(97)00066-6. [DOI] [PubMed] [Google Scholar]
- Jing C, Beesley C, Foster CS, Chen H. et al. Human cutaneous fatty acid-binding protein induces metastasis by up-regulating the expression of vascular endothelial growth factor gene in rat Rama 37 model cells. Cancer Res. 2001;61:4357–4364. [PubMed] [Google Scholar]
- Maeda K, Uysal KT, Makowski L, Gorgun CZ. et al. Role of the fatty acid binding protein mal1 in obesity and insulin resistance. Diabetes. 2003;52:300–307. doi: 10.2337/diabetes.52.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owada Y, Suzuki I, Noda T, Kondo H. Analysis on the phenotype of E-FABP-gene knockout mice. Mol Cell Biochem. 2002;239:83–86. doi: 10.1023/A:1020524621933. [DOI] [PubMed] [Google Scholar]
- Li B, Reynolds JM, Stout RD, Bernlohr DA. et al. Regulation of Th17 differentiation by epidermal fatty acid-binding protein. J Immunol. 2009;182:7625–7633. doi: 10.4049/jimmunol.0804192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung DC, Wang Y, Xu A, Cheung SC. et al. Epidermal fatty-acid-binding protein: A new circulating biomarker associated with cardio-metabolic risk factors and carotid atherosclerosis. Eur Heart J. 2008;29:2156–2163. doi: 10.1093/eurheartj/ehn295. [DOI] [PubMed] [Google Scholar]
- Maekawa M, Iwayama Y, Arai R, Nakamura K. et al. Polymorphism screening of brain-expressed FABP7, 5 and 3 genes and association studies in autism and schizophrenia in Japanese subjects. J Hum Genet. 2010;55:127–130. doi: 10.1038/jhg.2009.133. [DOI] [PubMed] [Google Scholar]
- Estelle J, Perez-Enciso M, Mercade A, Varona L. et al. Characterization of the porcine FABP5 gene and its association with the FAT1 QTL in an Iberian by Landrace cross. Anim Genet. 2006;37:589–591. doi: 10.1111/j.1365-2052.2006.01535.x. [DOI] [PubMed] [Google Scholar]
- Birkenmeier EH, Rowe LB, Crossman MW, Gordon JI. et al. Ileal lipid-binding protein (Illbp) gene maps to mouse chromosome 11. Mamm Genome. 1994;5:805–806. doi: 10.1007/BF00292019. [DOI] [PubMed] [Google Scholar]
- Walz DA, Wider MD, Snow JW, Dass C. et al. The complete amino acid sequence of porcine gastrotropin, an ileal protein which stimulates gastric acid and pepsinogen secretion. J Biol Chem. 1988;263:14189–14195. [PubMed] [Google Scholar]
- Gantz I, Nothwehr SF, Lucey M, Sacchettini JC. et al. Gastrotropin: Not an enterooxyntin but a member of a family of cytoplasmic hydrophobic ligand binding proteins. J Biol Chem. 1989;264:20248–20254. [PubMed] [Google Scholar]
- Fujita M, Fujii H, Kanda T, Sato E. et al. Molecular cloning, expression, and characterization of a human intestinal 15-kDa protein. Eur J Biochem. 1995;233:406–413. doi: 10.1111/j.1432-1033.1995.406_2.x. [DOI] [PubMed] [Google Scholar]
- Grober J, Zaghini I, Fujii H, Jones SA. et al. Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene. Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J Biol Chem. 1999;274:29749–29754. doi: 10.1074/jbc.274.42.29749. [DOI] [PubMed] [Google Scholar]
- Barley NF, Taylor V, Shaw-Smith CJ, Chakravarty P. et al. Human ileal bile acid-binding protein promoter and the effects of CDX2. Biochim Biophys Acta. 2003;1630:138–143. doi: 10.1016/j.bbaexp.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Landrier JF, Thomas C, Grober J, Zaghini I. et al. The gene encoding the human ileal bile acid-binding protein (I-BABP) is regulated by peroxisome proliferator-activated receptors. Biochim Biophys Acta. 2005;1735:41–49. doi: 10.1016/j.bbalip.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Zaghini I, Landrier JF, Grober J, Krief S. et al. Sterol regulatory element-binding protein-1c is responsible for cholesterol regulation of ileal bile acid-binding protein gene in vivo. Possible involvement of liver-X-receptor. J Biol Chem. 2002;277:1324–1331. doi: 10.1074/jbc.M106375200. [DOI] [PubMed] [Google Scholar]
- Kramer W, Sauber K, Baringhaus KH, Kurz M. et al. Radiation-inactivation analysis of the Na+/bile acid co-transport system from rabbit ileum. Biochem J. 1995;306:241–246. doi: 10.1042/bj3060241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AW, van Moerkerk HT, Veerkamp JH. Ligand specificity and conformational stability of human fatty acid-binding proteins. Int J Biochem Cell Biol. 2001;33:865–876. doi: 10.1016/S1357-2725(01)00070-X. [DOI] [PubMed] [Google Scholar]
- Kurz M, Brachvogel V, Matter H, Stengelin S. et al. Insights into the bile acid transportation system: The human ileal lipid-binding protein-cholyltaurine complex and its comparison with homologous structures. Proteins. 2003;50:312–328. doi: 10.1002/prot.10289. [DOI] [PubMed] [Google Scholar]
- Kramer W, Sauber K, Baringhaus KH, Kurz M. et al. Identification of the bile acid-binding site of the ileal lipid-binding protein by photoaffinity labeling, matrix-assisted laser desorption ionization-mass spectrometry, and NMR structure. J Biol Chem. 2001;276:7291–7301. doi: 10.1074/jbc.M006877200. [DOI] [PubMed] [Google Scholar]
- Miller KR, Cistola DP. Titration calorimetry as a binding assay for lipid-binding proteins. Mol Cell Biochem. 1993;123:29–37. doi: 10.1007/BF01076472. [DOI] [PubMed] [Google Scholar]
- Capaldi S, Saccomani G, Fessas D, Signorelli M. et al. The X-ray structure of zebrafish (Danio rerio) ileal bile acid-binding protein reveals the presence of binding sites on the surface of the protein molecule. J Mol Biol. 2009;385:99–116. doi: 10.1016/j.jmb.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Tochtrop GP, DeKoster GT, Covey DF, Cistola DP. A single hydroxyl group governs ligand site selectivity in human ileal bile acid binding protein. J Am Chem Soc. 2004;126:11024–11029. doi: 10.1021/ja047589c. [DOI] [PubMed] [Google Scholar]
- Fisher E, Nitz I, Lindner I, Rubin D. et al. Candidate gene association study of type 2 diabetes in a nested case-control study of the EPIC-Potsdam cohort -- Role of fat assimilation. Mol Nutr Food Res. 2007;51:185–191. doi: 10.1002/mnfr.200600162. [DOI] [PubMed] [Google Scholar]
- Fisher E, Grallert H, Klapper M, Pfafflin A. et al. Evidence for the Thr79Met polymorphism of the ileal fatty acid binding protein (FABP6) to be associated with type 2 diabetes in obese individuals. Mol Genet Metab. 2009;98:400–405. doi: 10.1016/j.ymgme.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Shimizu F, Watanabe TK, Shinomiya H, Nakamura Y. et al. Isolation and expression of a cDNA for human brain fatty acid-binding protein (B-FABP) Biochim Biophys Acta. 1997;1354:24–28. doi: 10.1016/S0167-4781(97)00115-2. [DOI] [PubMed] [Google Scholar]
- Feng L, Heintz N. Differentiating neurons activate transcription of the brain lipid-binding protein gene in radial glia through a novel regulatory element. Development. 1995;121:1719–1730. doi: 10.1242/dev.121.6.1719. [DOI] [PubMed] [Google Scholar]
- Josephson R, Muller T, Pickel J, Okabe S. et al. POU transcription factors control expression of CNS stem cell-specific genes. Development. 1998;125:3087–3100. doi: 10.1242/dev.125.16.3087. [DOI] [PubMed] [Google Scholar]
- Bisgrove DA, Monckton EA, Packer M, Godbout R. Regulation of brain fatty acid-binding protein expression by differential phosphorylation of nuclear factor I in malignant glioma cell lines. J Biol Chem. 2000;275:30668–30676. doi: 10.1074/jbc.M003828200. [DOI] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): A novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Kuhar SG, Feng L, Vidan S, Ross ME. et al. Changing patterns of gene expression define four stages of cerebellar granule neuron differentiation. Development. 1993;117:97–104. doi: 10.1242/dev.117.1.97. [DOI] [PubMed] [Google Scholar]
- Xu LZ, Sanchez R, Sali A, Heintz N. Ligand specificity of brain lipid-binding protein. J Biol Chem. 1996;271:24711–24719. doi: 10.1074/jbc.271.40.24711. [DOI] [PubMed] [Google Scholar]
- Balendiran GK, Schnutgen F, Scapin G, Borchers T. et al. Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J Biol Chem. 2000;275:27045–27054. doi: 10.1074/jbc.M003001200. [DOI] [PubMed] [Google Scholar]
- Owada Y, Abdelwahab SA, Kitanaka N, Sakagami H. et al. Altered emotional behavioral responses in mice lacking brain-type fatty acid-binding protein gene. Eur J Neurosci. 2006;24:175–187. doi: 10.1111/j.1460-9568.2006.04855.x. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Toyota T, Owada Y, Hayashi T. et al. Fabp7 maps to a quantitative trait locus for a schizophrenia endophenotype. PLoS Biol. 2007;5:e297. doi: 10.1371/journal.pbio.0050297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treadwell JA. Integrative strategies to identify candidate genes in rodent models of human alcoholism. Genome. 2006;49:1–7. doi: 10.1139/g05-083. [DOI] [PubMed] [Google Scholar]
- Sanchez-Font MF, Bosch-Comas A, Gonzalez-Duarte R, Marfany G. Overexpression of FABP7 in Down syndrome fetal brains is associated with PKNOX1 gene-dosage imbalance. Nucleic Acids Res. 2003;31:2769–2777. doi: 10.1093/nar/gkg396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan V, Kaestner KH, Tennekoon GI. Structure of the mouse myelin P2 protein gene. J Neurochem. 1991;57:75–80. doi: 10.1111/j.1471-4159.1991.tb02101.x. [DOI] [PubMed] [Google Scholar]
- Majava V, Polverini E, Mazzini A, Nanekar R. et al. Structural and functional characterization of human peripheral nervous system myelin protein P2. PLoS One. 2010;5:e10300. doi: 10.1371/journal.pone.0010300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Bergfors T, Sedzik J, Unge T. et al. The three-dimensional structure of P2 myelin protein. EMBO J. 1988;7:1597–1604. doi: 10.1002/j.1460-2075.1988.tb02985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouresmaeili F, Khalili T, Jeddi Tehrani M, Bandehpour M. et al. A novel human lipid binding protein coding gene: PERF15, sequence and cloning. J Reprod Infertil. 2009;10:199–205. [PMC free article] [PubMed] [Google Scholar]
- Pouresmaeili F, Morales CR, Oko R. Molecular cloning and structural analysis of the gene encoding PERF 15 protein present in the perinuclear theca of the rat spermatozoa. Biol Reprod. 1997;57:655–659. doi: 10.1095/biolreprod57.3.655. [DOI] [PubMed] [Google Scholar]
- Oko R, Morales CR. A novel testicular protein, with sequence similarities to a family of lipid binding proteins, is a major component of the rat sperm perinuclear theca. Dev Biol. 1994;166:235–245. doi: 10.1006/dbio.1994.1310. [DOI] [PubMed] [Google Scholar]
- Kido T, Arata S, Suzuki R, Hosono T. et al. The testicular fatty acid binding protein PERF15 regulates the fate of germ cells in PERF15 transgenic mice. Dev Growth Differ. 2005;47:15–24. doi: 10.1111/j.1440-169x.2004.00775.x. [DOI] [PubMed] [Google Scholar]
- Selvaraj V, Asano A, Page JL, Nelson JL. et al. Mice lacking FABP9/PERF15 develop sperm head abnormalities but are fertile. Dev Biol. 2010;348:177–189. doi: 10.1016/j.ydbio.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R. et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M. et al. Jalview Version 2 -- A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]


