Abstract
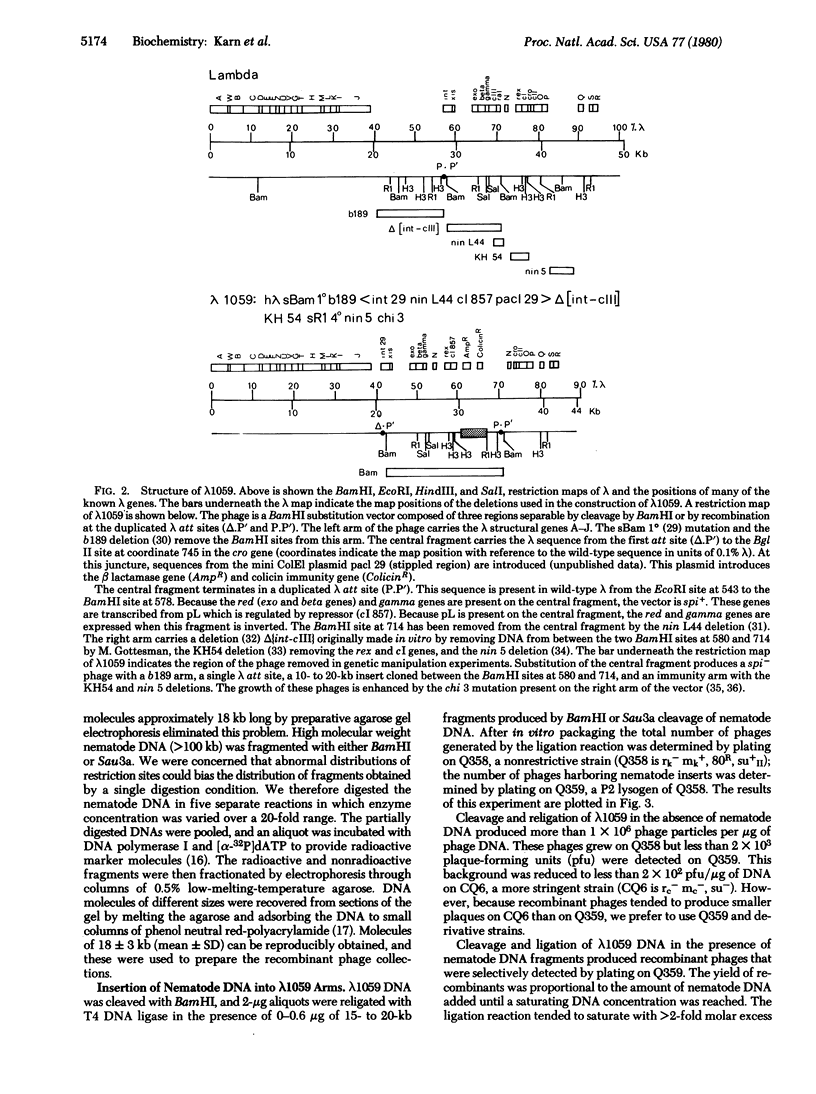
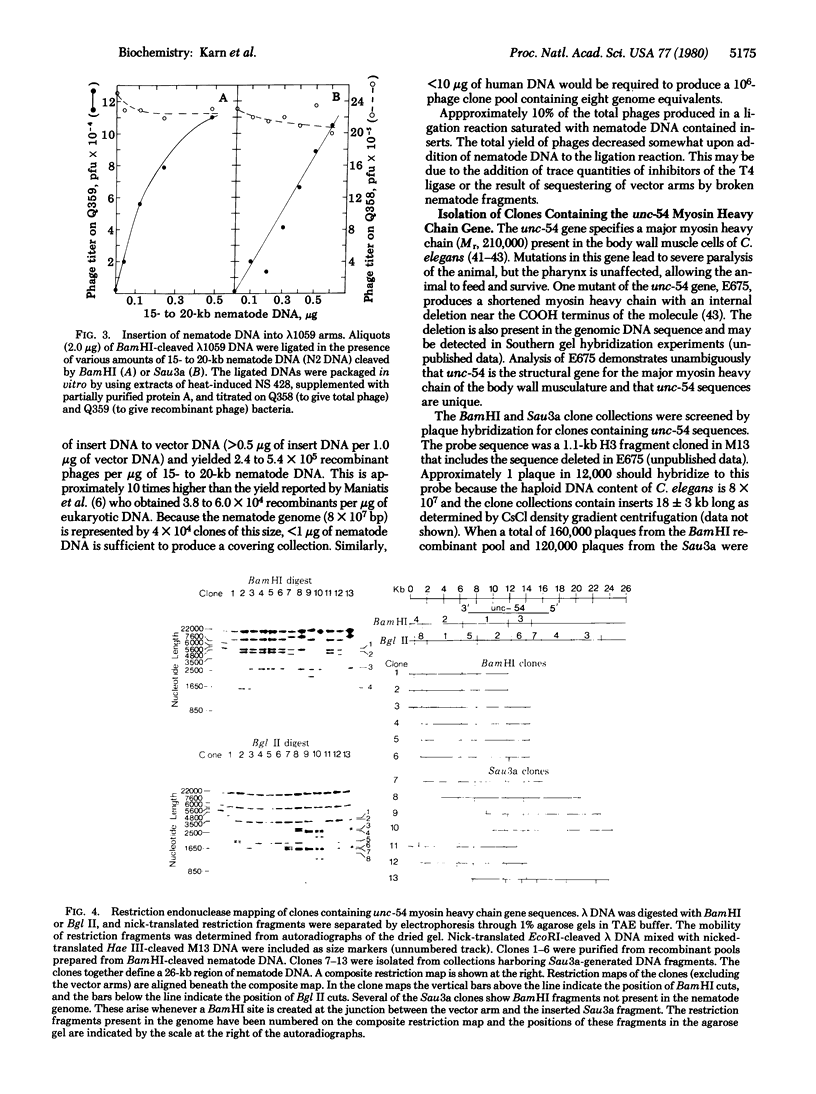
A simple method for generating phage collections representing eukaryotic genomes has been developed by using a novel bacteriophage lambda vector, lambda 1059. The phage is a BamHI substitution vector that accommodates DNA fragments 6-24 kilobases long. Production of recombinants in lambda 1059 requires deletion of the lambda red and gamma genes. The recombinants are therefore spi- and may be separated from the spi+ vector phages by plating on strains lysogenic for bacteriophage P2. Random fragments suitable for insertion into lambda 1059 are obtained by partial digestion of high molecular weight eukaryotic DNA with Sau3a. This restriction enzyme cleaves at the sequence G-A-T-C and leaves a 5'-tetranucleotide "sticky end." Because G-A-T-C extensions are also produced by BamHI cleavage, these fragments may be annealed directly to BamHI-cleaved lambda 1059. By using these methods, a set of clones covering the entire Caenorhabditis elegans genome was constructed. DNA segments which include the unc-54 myosin heavy chain gene have been isolated from this collection.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Blechl A. E., Denniston-Thompson K., Faber H. E., Richards J. E., Slightom J. L., Tucker P. W., Smithies O. Cloning human fetal gamma globin and mouse alpha-type globin DNA: preparation and screening of shotgun collections. Science. 1978 Dec 22;202(4374):1279–1284. doi: 10.1126/science.725603. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Fiandt M., Hass K. K., Twose P. A., Szybalski W. Deletions and insertions in the immunity region of coliphage lambda: revised measurement of the promoter-startpoint distance. Virology. 1974 Dec;62(2):458–471. doi: 10.1016/0042-6822(74)90407-3. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünemann H., Müller W. Base specific fractionation of double stranded DNA: affinity chromatography on a novel type of adsorbant. Nucleic Acids Res. 1978 Mar;5(3):1059–1074. doi: 10.1093/nar/5.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Court D., Sato K. Studies of novel transducing variants of lambda: dispensability of genes N and Q. Virology. 1969 Oct;39(2):348–352. doi: 10.1016/0042-6822(69)90060-9. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Parkinson J. S. Deletion mutants of bacteriophage lambda. 3. Physical structure of att-phi. J Mol Biol. 1971 Mar 14;56(2):403–423. doi: 10.1016/0022-2836(71)90473-6. [DOI] [PubMed] [Google Scholar]
- Duncan C. H., Wilson G. A., Young F. E. Biochemical and genetic properties of site-specific restriction endonucleases in Bacillus globigii. J Bacteriol. 1978 Apr;134(1):338–344. doi: 10.1128/jb.134.1.338-344.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist L. W., Weisberg R. A. A genetic analysis of the att-int-xis region of coliphage lambda. J Mol Biol. 1977 Apr;111(2):97–120. doi: 10.1016/s0022-2836(77)80117-4. [DOI] [PubMed] [Google Scholar]
- Epstein H. F., Waterston R. H., Brenner S. A mutant affecting the heavy chain of myosin in Caenorhabditis elegans. J Mol Biol. 1974 Dec 5;90(2):291–300. doi: 10.1016/0022-2836(74)90374-x. [DOI] [PubMed] [Google Scholar]
- Garapin A. C., Lepennec J. P., Roskam W., Perrin F., Cami B., Krust A., Breathnach R., Chambon P., Kourilsky P. Isolation by molecular cloning of a fragment in the split ovalbumin gene. Nature. 1978 Jun 1;273(5661):349–354. doi: 10.1038/273349a0. [DOI] [PubMed] [Google Scholar]
- Henderson D., Weil J. Recombination-deficient deletions in bacteriophage lambda and their interaction with chi mutations. Genetics. 1975 Feb;79(2):143–174. doi: 10.1093/genetics/79.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B., Murray K. Packaging recombinant DNA molecules into bacteriophage particles in vitro. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3259–3263. doi: 10.1073/pnas.74.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes L. H., Chang A. C., Houseman D., Cohen S. N. Isolation of histone genes from unfractionated sea urchin DNA by subculture cloning in E. coli. Nature. 1975 Jun 12;255(5509):533–538. doi: 10.1038/255533a0. [DOI] [PubMed] [Google Scholar]
- Kemp D. J., Cory S., Adams J. M. Cloned pairs of variable region genes for immunoglobulin heavy chains isolated from a clone library of the entire mouse genome. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4627–4631. doi: 10.1073/pnas.76.9.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein B., Murray K. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease I of Bacillus amyloliquefaciens H. J Mol Biol. 1979 Sep 15;133(2):289–294. doi: 10.1016/0022-2836(79)90537-0. [DOI] [PubMed] [Google Scholar]
- Lindahl G., Sironi G., Bialy H., Calendar R. Bacteriophage lambda; abortive infection of bacteria lysogenic for phage P2. Proc Natl Acad Sci U S A. 1970 Jul;66(3):587–594. doi: 10.1073/pnas.66.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A. R., Waterston R. H., Brenner S. An internal deletion mutant of a myosin heavy chain in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5336–5340. doi: 10.1073/pnas.74.12.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A. R., Waterston R. H., Fishpool R. M., Brenner S. Identification of the structural gene for a myosin heavy-chain in Caenorhabditis elegans. J Mol Biol. 1977 Jul;114(1):133–140. doi: 10.1016/0022-2836(77)90287-x. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Bruce S. A., Murray K. Molecular cloning of the DNA ligase gene from bacteriophage T4. II. Amplification and preparation of the gene product. J Mol Biol. 1979 Aug 15;132(3):493–505. doi: 10.1016/0022-2836(79)90271-7. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Two restriction endonucleases from Bacillus globiggi. Nucleic Acids Res. 1976 Jul;3(7):1747–1760. doi: 10.1093/nar/3.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1980 Jan 11;8(1):r63–r80. doi: 10.1093/nar/8.1.197-d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction endonucleases. CRC Crit Rev Biochem. 1976 Nov;4(2):123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Wilson G. A., Young F. E. Recognition sequence of specific endonuclease BamH.I from Bacillus amyloliquefaciens H. Nature. 1977 Jan 6;265(5589):82–84. doi: 10.1038/265082a0. [DOI] [PubMed] [Google Scholar]
- Salstrom J. S., Fiandt M., Szybalski W. N-independent leftward transcription in coliphage lambda: deletions, insertions and new promoters bypassing termination functions. Mol Gen Genet. 1979 Jan 10;168(2):211–230. doi: 10.1007/BF00431446. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Crasemann J. M., Stahl M. M. Rec-mediated recombinational hot spot activity in bacteriophage lambda. III. Chi mutations are site-mutations stimulating rec-mediated recombination. J Mol Biol. 1975 May 15;94(2):203–212. doi: 10.1016/0022-2836(75)90078-9. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Tiemeier D., Enquist L. In vitro packaging of a lambda Dam vector containing EcoRI DNA fragments of Escherichia coli and phage P1. Gene. 1977 May;1(3-4):255–280. doi: 10.1016/0378-1119(77)90049-x. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Weisberg R. Packaging of prophage and host DNA by coliphage lambda. Nature. 1975 Jul 10;256(5513):97–103. doi: 10.1038/256097a0. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Brenner S. The DNA of Caenorhabditis elegans. Genetics. 1974 May;77(1):95–104. doi: 10.1093/genetics/77.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussenbach J. S., Monfoort C. H., Schiphof R., Stobberingh E. E. A restriction endonuclease from Staphylococcus aureus. Nucleic Acids Res. 1976 Nov;3(11):3193–3202. doi: 10.1093/nar/3.11.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Cameron J. R., Davis R. W. Viable molecular hybrids of bacteriophage lambda and eukaryotic DNA. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4579–4583. doi: 10.1073/pnas.71.11.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman S. M., Tiemeier D. C., Seidman J. G., Peterlin B. M., Sullivan M., Maizel J. V., Leder P. Intervening sequence of DNA identified in the structural portion of a mouse beta-globin gene. Proc Natl Acad Sci U S A. 1978 Feb;75(2):725–729. doi: 10.1073/pnas.75.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil J., Cunningham R., Martin R., 3rd, Mitchell E., Bolling B. Characteristics of lambda p4, a lambda derivative containing 9 per cent excess DNA. Virology. 1972 Nov;50(2):373–380. doi: 10.1016/0042-6822(72)90388-1. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Wilson G. A., Young F. E. Isolation of a sequence-specific endonuclease (BamI) from Bacillus amyloliquefaciens H. J Mol Biol. 1975 Sep 5;97(1):123–125. doi: 10.1016/s0022-2836(75)80028-3. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]



