Abstract
In the dinoflagellate Amphidinium carterae, photoadaptation involves changes in the transcription of genes encoding both of the major classes of light-harvesting proteins, the peridinin chlorophyll a proteins (PCPs) and the major a/c-containing intrinsic light-harvesting proteins (LHCs). PCP and LHC transcript levels were increased up to 86- and 6-fold higher, respectively, under low-light conditions relative to cells grown at high illumination. These increases in transcript abundance were accompanied by decreases in the extent of methylation of CpG and CpNpG motifs within or near PCP- and LHC-coding regions. Cytosine methylation levels in A. carterae are therefore nonstatic and may vary with environmental conditions in a manner suggestive of involvement in the regulation of gene expression. However, chemically induced undermethylation was insufficient in activating transcription, because treatment with two methylation inhibitors had no effect on PCP mRNA or protein levels. Regulation of gene activity through changes in DNA methylation has traditionally been assumed to be restricted to higher eukaryotes (deuterostomes and green plants); however, the atypically large genomes of dinoflagellates may have generated the requirement for systems of this type in a relatively “primitive” organism. Dinoflagellates may therefore provide a unique perspective on the evolution of eukaryotic DNA-methylation systems.
In many respects dinoflagellates are an unusual group of chromophytic algae, their closest relatives being the apicomplexans (van der Peer et al., 1993). For example, uniquely among eukaryotes, their chromatin appears to be entirely devoid of histones (Rizzo, 1981, 1991) and is not organized in the normal nucleosomal structure (Herzog and Soyer, 1981). Light harvesting is mediated in dinoflagellates by two classes of proteins, the PCPs and the LHCs. PCP, a water-soluble protein containing the carotenoid peridinin, is unique to dinoflagellates and appears to be unrelated to any other light-harvesting protein (Norris and Miller, 1994), whereas the LHCs are related to the cab proteins of higher plants (Hiller et al., 1993) and their algal homologs (for review, see Grossman et al., 1990). Both PCPs and LHCs are present in multiple forms in dinoflagellates. The functional significance of this and the nature of the interaction between these two classes of light-harvesting proteins are completely unknown. Chromophyte chloroplasts appear to lack many of the mechanisms known to be central to photoadaptation in chlorophytes, such as lateral mobility of photosynthetic complexes and stacking/unstacking of thylakoids. One hypothesis is that the multiple forms of light-harvesting proteins fulfill related roles in dinoflagellates, i.e. that the synthesis of different PCP and LHC complements facilitates photoadaptation.
In Amphibinium carterae, both PCPs and LHCs are encoded by nuclear genes (Hiller et al., 1995; Sharples et al., 1996) and synthesized with N-terminal transit peptides directing chloroplast translocation (Norris and Miller, 1994; Hiller et al., 1995; Sharples et al., 1996). As in the case of Euglena gracilis, the A. carterae LHC mRNA encodes a polyprotein. The PCPs are encoded by discrete mRNAs, but at least some of the genes appear to be tandemly organized (Hiller et al., 1995).
At this time, almost nothing is known about the molecular basis of photoadaptation in dinoflagellates. PCP may account for up to 95% of the total cellular-soluble protein under low-light conditions (Prezelin, 1987), and there is some indirect evidence that PCP synthesis is light regulated at the level of transcription in Glenodinium sp. (Roman et al., 1988). As stated above, the LHCs are the dinoflagellate homologs of the cab proteins and, by analogy with higher plants (for review, see Anderson, 1986) and green algae (Kindle, 1986), light regulation is likely.
In green plants light-regulated transcription of the cab genes is mediated via phytochrome; response elements have been identified in the promoters of the cab genes (Terzaghi and Cashmore, 1995). However, phytochrome has been detected only in chlorophytes, implying that the regulation of synthesis of light-harvesting proteins may be achieved via alternative mechanisms in other algal groups. In the chrysophyte Giraudyopsis stellifer, synthesis of the chlorophyll a/c protein is light regulated at the level of transcription (Passaquet and Lichtl, 1995), whereas in E. gracilis, LHC synthesis is apparently regulated posttranscriptionally (for review, see Houlne and Schanz, 1993). How these effects are mediated in the apparent absence of the phytochrome system is unknown at this time.
Preliminary studies in our laboratory suggested the presence of significant levels of 5-MeC at CpG motifs in genomic DNA from the dinoflagellate A. carterae. Many studies suggest that a major function of this type of methylation in eukaryotes is in the regulation of gene expression (for review, see Lewis and Bird, 1991). Transcriptionally inactive regions of chromatin are generally highly meth-ylated at CpG motifs (Keshet et al., 1986; for review, see Lewis and Bird, 1991), whereas active chromatin is often undermethylated (Antequera et al., 1989; Kochanek et al., 1993). Moreover, for many genes CpG methylation status reflects transcriptional activity.
Although it is not clear whether methylation is a cause or a consequence of alterations in gene activity, a number of lines of evidence suggest that the effects of methylation may be mediated via modifications of chromatin structure. For example, introduced DNAs generally adopt chromatin structures specified by their methylation status (Keshet et al., 1986; Buschhausen et al., 1987). Methylation may affect chromatin structure by modifying nucleosome assembly through the action of methylcytosine-binding proteins (Nan et al., 1997) or by a combination of processes. Regions containing 5-MeC may preferentially bind histone H1 (Ball et al., 1983; Jost and Hofsteenge, 1992) and associate with histones H3 and H4 mainly in their nonacetylated forms (Tazi and Bird, 1990), presumably promoting more condensed forms of chromatin. Methylation-specific binding proteins (Meehan et al., 1989; Lewis et al., 1992) can also mediate changes in chromatin structure.
In summary, the evolution and roles of eukaryotic DNA-methylation systems and the interactions between three processes, methylation, alterations of chromatin structure, and changes in gene activity, are not well understood. In this respect, dinoflagellates may be highly informative because, although otherwise typical eukaryotes, they are unique in posessing permanently condensed chromosomes and in lacking histones (Rizzo, 1981, 1991) and nucleosomes (Herzog and Soyer, 1981).
To investigate the possible involvement of DNA methylation in the regulation of gene expression in dinoflagellates, we examined the methylation status of A. carterae PCP and LHC genes under different light conditions. Photoadaptation involved major changes in the levels of transcripts encoding both of these classes of light-harvesting proteins. DNA from A. carterae was normally highly methylated at CpG motifs and significantly methylated at CpNpG motifs. However, when grown under reduced light conditions, elevated mRNA levels were accompanied by decreases in the methylation of CpG and CpNpG motifs within or near PCP- and LHC-coding regions. Therefore, cytosine-methylation levels are nonstatic in A. carterae and may be influenced by environmental factors. However, chemically induced undermethylation was found to be insufficient for gene activation. These results provide the first evidence to our knowledge that the relationship between cytosine methylation and transcriptional activity, which has been observed for a large number of genes in a wide range of eukaryotes, does not require histones and nucleosomes.
MATERIALS AND METHODS
Source and Maintenance of Cultures
A unialgal culture of Amphidinium carterae (Cs-21) was obtained from the Microalgae Culture Collection (Commonwealth Scientific and Industrial Research Organization, Division of Fisheries, Hobart, Tasmania). Cells were grown axenically in (0.4 μm) filtered seawater (Millipore) supplemented with Provasoli's enrichment solution (Sigma). Generally, cultures (500 mL) were grown at 18°C in 1-L Erlenmeyer flasks without agitation until the density reached 0.5 to 3.0 × 106 cells mL−1. Cells were normally grown on a 14-h/10-h light-dark cycle at 80 to 100 μmol m−2 s−1 (two white, two warm, and two Grow-Lux tubes [Sylvania]), referred to here as high-light conditions. For low-light treatments, cells were grown under high-light conditions for 1 week before being transferred to either 18 to 20 μmol m−2 s−1 (moderate-light cultures; two Grow-Lux fluorescent tubes only) or < 2 μmol m−2 s−1 (low-light cultures) for 14 d.
DNA Extraction, Restriction, and Southern Analysis
To avoid possible interference by intrinsic circadian rhythms, cells were always harvested by centrifugation 6 h into the light phase of growth. Pellets were ground in liquid N2, and genomic DNA was isolated using the standard high-salt method (Dellaporta, 1983). Purified DNA was restricted and fractionated on 0.8 to 1.2% agarose gels. After Southern transfer to Hybond-N membranes (Amersham), blots were hybridized with 32P-labeled (Feinberg and Vogelstein, 1983) DNA probes. Bacteriophage λDNA was used as an internal control in restriction digests. Homologous probes for the LHC (pRGH201; Hiller et al., 1995) or the main form of the PCP genes (see figure 6 of Sharples et al., 1996) were kindly provided by R. Hiller (Macquarie University, Sydney, Australia). 16S-Like rDNA probes were generated from A. carterae DNA using universal PCR primers (Medlin et al., 1988). Heterologous hybridization probes (from the coral Acropora millepora) for the highly conserved ubiquitin CEP52 (ub52; Berghammer et al., 1996) and elongation factor 1α (Ef 1α) genes were provided by H. Berghammer (James Cook University, Townsville, Australia).
RNA Extraction, Transfer, and Hybridization
Poly(A+) RNA was extracted from approximately 107 cells with an mRNA purification kit (QuickPrep, Pharmacia), using the manufacturer's recommended protocol. Precipitated RNA was resuspended in Tris-EDTA buffer, quantified spectrophotometrically, and stored at −70°C. Aliquots of mRNA (0.25–1.0 μg) were applied to nylon membranes through a slotted template following the supplier's (Schleicher & Schuell) instructions. For the preparation of northern blots, 2.0-μg aliquots of mRNA were subjected to electrophoresis in 1% agarose/formaldehyde gels prior to transfer to nylon membranes in 20× SSC. Hybridizations (6× SSC, 0.1% SDS, and 2× Denhardt's solution at 60°C for 24 h) and three washes (0.5× SSC, and 0.1% SDS at 60°C for 20 min) were conducted as described by Sambrook et al. (1989).
RESULTS
PCP and LHC mRNA Levels Increase under Low-Light Conditions
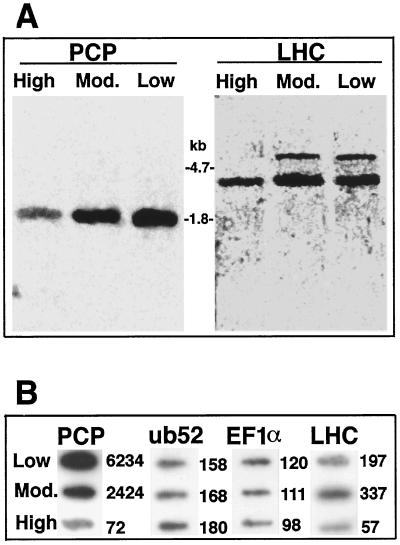
Northern analysis (Fig. 1A) and slot-blotting experiments (Fig. 1B) clearly demonstrated that the abundance of PCP and LHC mRNAs changed substantially in response to light conditions, whereas the level of the mRNAs corresponding to the housekeeping genes ub52 and EF1α did not vary significantly.
Figure 1.
PCP and LHC mRNA levels are increased under reduced light conditions. A, Northern analysis of mRNA (2 μg/lane) from A. carterae grown under various light conditions and probed with either the insert in pRGH201 (Hiller et al., 1995) for LHC or the part of the MFPCP gene encoding the mature protein (generated by PCR; see figure 6A of Sharples et al., 1996). B, Slot blots of mRNA from A. carterae grown under various light conditions (High, moderate [Mod.], or Low) probed with probes for PCP or LHC (as described in A) or for the housekeeping genes ub52 and EF1α. Numbers to the right of the blots are the corresponding standardized photon equivalents, determined by quantitative phosphor imaging.
Quantitative analyses indicated that the abundance of the PCP transcript was increased approximately 33- and 86-fold in cells grown in moderate- and low-light conditions, respectively, relative to cells grown under high-light conditions (Fig. 1B). Low-light conditions resulted in both quantitative and qualitative changes in the LHC mRNA population (Fig. 1A; right panel). Total LHC mRNA was approximately one order of magnitude more abundant in cells grown under lower illumination compared with cells grown under high-light conditions. Under lower illumination, two distinct LHC transcripts, approximately 3 and 6 kb in size, were detected, whereas the larger of these was not detected under high-light conditions.
In addition, significant differences in cell pigmentation were observed in response to light levels. A. carterae cells grown under full-light conditions were rather pale, whereas cultures grown under reduced illumination were intensely dark-brown pigmented. Four days after transfer from high-light conditions to either moderate- or low-light conditions, PCP protein levels increased from 2 to 50% of total soluble protein (data not shown).
PCP and LHC Loci Are Normally Hypermethylated at CpG Motifs and Their Methylation Status Is Modified by Light Conditions
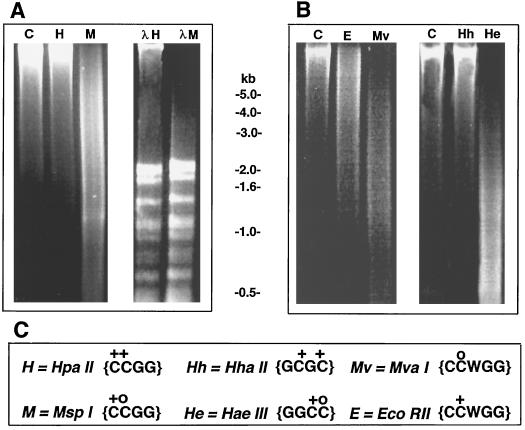
Preliminary experiments indicated a substantial level of cytosine methylation in the genome of A. carterae. DNA prepared by Dellaporta's (1983) method was digestible by a range of methylation-insensitive restriction endonucleases, including MvaI and HaeIII. However, the same DNA preparations were highly resistant to a range of CpG-methylation-sensitive restriction endonucleases, including HpaII and HhaI (Fig. 2). The presence of diffusible inhibitors of digestion in the DNA preparations was excluded by the use of bacteriophage λDNA as an internal control (see Fig. 2A, right panel). These results therefore imply that A. carterae DNA is hypermethylated at CpG motifs, as has been reported for a few higher plants (Gruenbaum et al., 1981) and algae (Jarvis et al. 1992).
Figure 2.
DNA from A. carterae is highly resistant to digestion by restriction enzymes that are sensitive to CpG methylation. Aliquots (10 μg) of genomic DNA prepared from cells grown under high-light conditions were digested with a 6-fold excess of the restriction enzymes indicated and subjected to electrophoresis on 1% (w/v) agarose, and the gels were stained with ethidium bromide. Control lanes (C) were treated exactly as digests, except that restriction enzymes were not added. Lanes λH and λM correspond to restriction digests of A. carterae DNA to which 3 μg of bacteriophage λDNA was added. C, Methylation sensitivities and recognition specificities of the endonucleases used. “+” indicates that digestion with the restriction endonuclease is inhibited by a 5-MeC at that position. “o” indicates that digestion with the restriction endonuclease was not inhibited by a 5-MeC at that position.
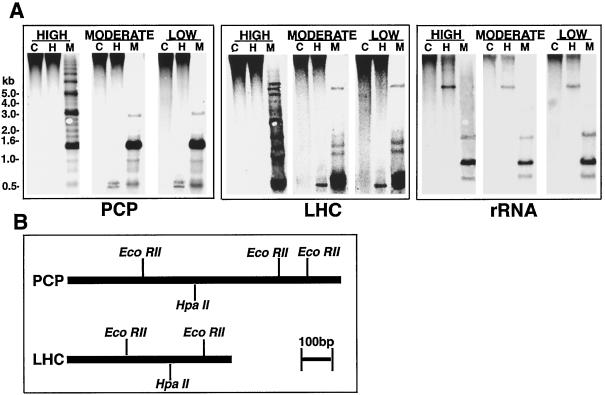
To establish the extent of cytosine methylation at PCP and LHC loci, and to determine whether methylation levels varied under different light regimes, we hybridized Southern blots of restricted DNA from cells grown under various levels of illumination using homologous probes. High-light-grown A. carterae cells exhibited high levels of methylation at CpG motifs within HpaII restriction fragments homologous to either PCP, LHC, or rDNA probes (Fig. 3). When grown under different light intensities, the methylation status of both PCP and LHC loci was variable at some CpG motifs (Fig. 3A, left and center panels). By contrast, we found no evidence for changes in the methylation status of the 16S-like rDNA in response to light levels (Fig. 3A, right panel).
Figure 3.
A. carterae PCP and LHC loci are normally hypermethylated at CpG motifs and their methylation status is modified by light conditions. A, Southern analysis of PCP, LHC, and rDNA loci in cells grown under different light conditions using the restriction enzymes HpaII (H) and MspI (M); C, controls. The sizes indicated to the left of the figure correspond to positions of DNA size standards. Note that the apparently repeating pattern seen for the MspI digest of high-light-grown cells (A, left panel) is consistent with the hypothesis that several of the PCP loci are tandemly organized (Sharples et al., 1996). B, Restriction maps of single A. carterae PCP (based on figure 6A of Sharples et al., 1996; bp 35–1110)- and LHC (based on figure 1B of Hiller et al., 1995, bp 10–600)-coding sequences that were used as probes.
Under high-light conditions, all PCP and LHC loci appear to be fully methylated at HpaII sites (CCGG); the single high-Mr hybridization signal indicates that 5-MeC methylation occurs at every HpaII site, in all homologous loci, and in every cell present in the culture. Under lower light conditions, the presence of low-Mr (< 1000 bp) hybridizing fragments implied significant undermethylation at these same sites in both PCP and LHC loci, but not in rDNA loci. These patterns suggest a homogeneous and complete demethylation event, which is likely to involve restriction sites near or within the corresponding coding regions. However, the remaining strong hybridization signals in the high-Mr (> 10.0 kb) range suggest that either only a few members of the total cell population or a limited subset of PCP and LHC genes undergo light-related demethylation at the CpG motifs analyzed.
Heterogenous DNA-Methylation Changes at CpNpG Motifs
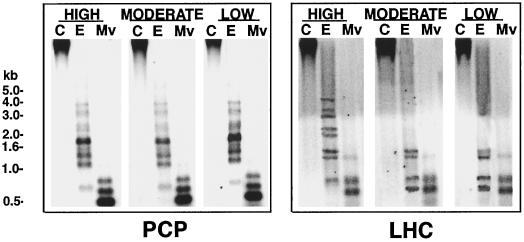
The complex and variable band patterns given by MspI digests (see Fig. 3, left and center panel) implied significant methylation at CpNpG as well as at CpG motifs in A. carterae. Although MspI is insensitive to methylation at the second C in its recognition sequence (CCGG), it is sensitive to methylation at the first C. Whereas CpG methylation at PCP and LHC loci appears to be relatively homogenous (see above), the wide range of restriction fragment sizes (10.0–0.3 kb) detected in high-light-grown A. carterae implied that CpNpG methylation was considerably more heterogeneous, either among loci or among cells. Light-related changes in CpNpG methylation in MspI sites at PCP and LHC loci were pronounced and appeared to affect the majority of cells and homologous loci.
To further investigate CpNpG methylation levels, we probed Southern blots generated using a second pair of restriction enzymes, EcoRII and MvaI, which have the same recognition sequence (CCWGG), but differ in that the former is sensitive to CpNpG methylation, whereas the latter is not. In this case, changes in EcoRII-specific CpNpG-methylation were observed for LHC loci only (Fig. 4).
Figure 4.
CpNpG methylation at LHC loci is variable in A. carterae. The figure shows Southern analysis of DNA from cells grown under various light conditions digested with either EcoRII (E; sensitive to CpNpG methylation) or MvaI (Mv; an EcoRII isoschizomer insensitive to CpNpG methylation) hybridized with the corresponding probe. Lanes C, Controls.
Chemically Induced Undermethylation Does Not Alter PCP Transcript Levels
Since demethylation events near or within the PCP- and LHC-coding regions correlated with increases in the abundance of the corresponding transcripts, it was of immediate interest to determine if transcription patterns could be altered by artificial manipulation of 5-MeC levels. 5-MeC undermethylation was induced by treating cells with 5-azacytidine or ethionine. 5-Azacytidine is a base analog that is both an inhibitor of many methylases and, once incorporated, cannot be methylated at the 5-position. Ethionine is a Met analog that inhibits Met-adenosyl-transferase. Culturing A. carterae under high-light conditions in the presence of either of these inhibitors considerably increased the susceptibility of DNA to cleavage by HpaII (Fig. 5A), confirming that cytosine methylation was responsible for the observed inhibition of restriction digestion.
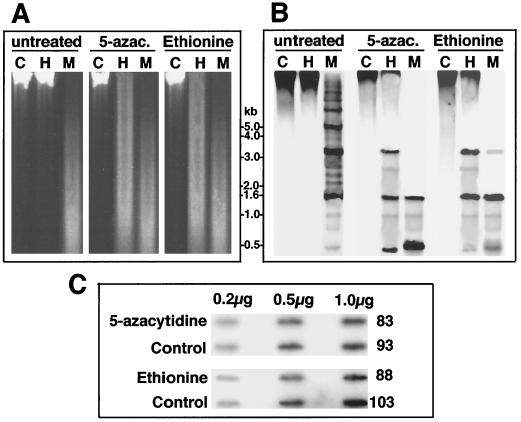
Figure 5.
Chemically induced undermethylation is not sufficient to activate transcription of PCP genes in A. carterae. A. carterae cultures in the exponential phase of growth were transferred to medium supplemented with either 5-azacytidine (200 μm) or ethionine (300 μm), and grown under high-light conditions for 6 weeks, after which DNA was extracted and digested with HpaII (H) or MspI (M) prior to electrophoresis (A), and blotting and hybridization with the PCP probe (B). Lanes C, Controls. C, Slot blots of mRNA prepared from cells treated in the same manner and hybridized with the PCP probe. Numbers to the right of the blots are the corresponding standardized photon equivalents determined by quantitative phosphor imaging. Note that exposure of A. carterae to these compounds resulted in reductions in growth rates of around 35 to 50%, but appeared to otherwise have no adverse effects on morphology or development of the organisms (data not shown). Transfer of treated cells back into normal medium resulted in an immediate return to normal growth rates.
After growth in the presence of 5-azacytidine, significant reductions in CpG and CpNpG methylation were observed (data for PCP loci are shown in Fig. 5B). However, levels of the corresponding mRNAs did not differ significantly between controls and inhibitor treatments (Fig. 5C).
The use of azacytidine and ethionine treatment in methylation studies has sometimes been criticized on the grounds that these agents can cause pathological (i.e. unrepresentative) changes in some organisms (e.g. Juetterman et al., 1994). However, although exposure to either inhibitor reduced the growth rate of A. carterae by around 35 to 50% (data not shown), there appeared to be no adverse effects on morphology or development. Motility appeared to be unaltered in the presence of inhibitors, and effects on growth rates were fully reversible; transfer of azacytidine-treated A. carterae back into normal medium resulted in an immediate return to normal growth rates. Treated and untreated cells were indistinguishable in terms of pigmentation, and levels of both total mRNA and soluble proteins (data not shown) were shown not to differ significantly between treatments. In summary, although we cannot completely exclude chemically induced artifacts, our results strongly suggest that simply reducing the extent of 5-MeC within or near PCP genes does not appear to affect transcription.
DISCUSSION
Light-Regulated Transcription of PCP and LHC Genes
Transcription of LHC and PCP genes is light regulated in A. carterae, and it appears likely that this is the major means by which levels of the corresponding proteins are controlled. This is consistent with indirect evidence from in vitro translation studies, which implied that changes in PCP protein levels correlated with mRNA abundance in Glenodinium sp. (Roman et al., 1988).
The size of the PCP transcript was consistent with a previous estimate (Hiller et al., 1995). The larger (6 kb) form of the A. carterae LHC mRNA detected here corresponds to the previously estimated size (Hiller et al., 1995). Although lower-Mr forms of the LHC mRNA have previously been observed (Hiller et al., 1995), these appeared to be very minor components. The A. carterae LHC-coding sequences are relatively heterogeneous (Hiller et al., 1993, 1995), and the use of different LHC probes in the previous and present studies may account for this difference; the probes have approximately 70% identity, which, under the conditions employed, may have had a significant effect on the results of northern analysis. The functional significance of the qualitative shift in the LHC mRNA population with light intensity is presently unclear. The LHC mRNA encodes a polyprotein, with the 6.1-kb form encoding possibly up to 10 LHC protein units (Hiller et al., 1995). If there is only one LHC locus, we might predict that the proteins encoded by the 3′ end of the mRNA might be required only under reduced illumination. Alternatively, multiple loci under independent regulation may encode the 3- and 6-kb forms of the LHC mRNA. Analyses of the genomic loci will be required to resolve these alternatives.
CpG and CpNpG Methylation Patterns in A. carterae May Be Influenced by Environmental Factors
Although no significant differences were observed between cells grown under moderate- and low-light conditions, when A. carterae cells grown under high-light conditions were shifted to lower light intensities, demethylation occurred at CpG and CpNpG motifs within or proximal to the LHC- and PCP-coding regions. CpG demethylation appears to be partial, affecting a limited number of meth-ylatable sites or loci, a minor proportion of cells, or a combination of both. By contrast, light-induced CpNpG demethylation events appear to be more pronounced and uniform. Consideration of the available sequence data for A. carterae PCP (Sharples et al., 1996) and LHC (Hiller et al., 1995) implies that whereas changes in the methylation status of CpG motifs may be localized at or near the coding sequence, CpNpG methylation changes are likely to occur over larger regions, affecting sequences distant to those transcribed or occurring in tandemly arranged multiple copy loci. The existence of tandemly organized PCP loci has been recently documented for Gonyaulax polyedra (Le et al., 1997) and has been previously proposed for A. carterae (Sharples et al., 1996). Our apparently repeating pattern seen for the MspI digest of high-light-grown cells (Fig. 3A, left panel) is consistent with this proposal.
There are many precedents for environmentally influenced changes in methylation patterns for endogenous genes (e.g. Burn et al., 1993; Galaud, 1993) or transgenes (e.g. Meyer et al., 1992); for example, light-induced de-methylation of distant regulatory sites correlated with increased levels of PEPCase mRNA levels in Zea mays (Langdale et al., 1991). However, we were unable to find precedents for methylation changes affecting endogenous genes involved in light harvesting.
Decreased Methylation Is Not Sufficient to Activate Transcription of PCP and LHC Genes
The question of whether DNA methylation states are required for gene activity or represent consequences of genomic changes involved in the activation of chromatin is controversial. Although treatment with 5-azacytidine or ethionine reduced the extent of methylation of PCP genes, these changes were not accompanied by increased transcript levels, nor did the corresponding protein levels increase (data not shown). This indicates that simply reducing overall levels of methylation is not sufficient to activate PCP transcription in A. carterae. As in our study, 5-azacytidine treatment did not reactivate methylated regions in Z. mays (Brown, 1989); however, there are numerous reports of the opposite effect, 5-azacytidine-treatment-mediated reactivation of T-DNA in plants (Klaas et al., 1989; Kilby et al., 1992) and of a hepatitis-C cDNA transgene in mice (Kato et al., 1996) and chemically mediated demethylation initiating flowering in Arabidopsis thaliana and Thlaspi arvense (Burn et al., 1993).
There are a number of possible explanations for the failure of chemically induced undermethylation to activate A. carterae PCP genes. Transcription may require demethylation of specific CpG motifs that are not sufficiently affected by azacytidine or ethionine treatment. Alternatively, demethylation may be necessary but not sufficient for transcription activation, or demethylation may be a consequence rather than a cause of gene activity (Selker, 1990).
The Evolution and Possible Roles of Dinoflagellate DNA-Methylation Systems
Our studies establish the presence of significant and variable levels of CpG and CpNpG methylation in dinoflagellates, suggesting the presence of a higher-plant-like methyltransferase(s) (Theiss et al., 1987). Our data are consistent with the hypothesis that methylation systems may have arisen as a means of noise reduction in organisms with large genomes (Bestor, 1990; Bird, 1995; Jablonka and Regev, 1995). Dinoflagellates have atypically large genomes, ranging from 2 to 3 pg in A. carterae to > 200 pg in G. polyedra (Holm-Hansen, 1969; Rizzo, 1987), compared with other algae at the same apparent level of complexity (e.g. < 0.2 pg for the green algae Chlamydomonas and Chlorella; Cattolico and Gibbs, 1975; Holm-Hansen, 1969). This suggests that major amounts of heterochromatin may be present in dinoflagellates, perhaps generating a requirement for methylation systems to maintain this in a transcriptionally inactive state.
Although, in principle, dinoflagellates are attractive model organisms to study the relationship between cytosine methylation and gene activity, there are a number of major hurdles that must be overcome before their full potential can be realized. All dinoflagellate genes characterized to date are members of complex families. In addition to the multiplicity of PCPs and LHCs that have been documented for a number of dinoflagellates, there are thought to be at least 1000 copies of the gene encoding the luciferin-binding protein in G. polyedra (Machabee et al., 1994), and certainly tens and probably hundreds of copies of the rbcL gene in Symbiodinium microadriaticum (R. Rowan, personal communication). None of these complex multigene families has been fully characterized, and in every case probes for single genes are likely to cross-hybridize. Thus, it is not possible to relate demethylation events to active copies of genes.
Changes in the DNA-methylation status of 5′-regulatory sequences are also likely to be involved in regulation (e.g. Langdale et al., 1991); however, with the exception of one copy (of the approximately 1000 copies) of a G. polyedra LBP gene (Lee et al., 1993) and one copy (of the suggested 5000 copies) of a G. polyedra PCP gene (Le et al., 1997), sequence data are not available for any dinoflagellate promoters. Further progress in understanding the significance of DNA methylation in dinoflagellate gene regulation requires the characterization of genes or gene families and the corresponding promoters to a level that will permit the development of highly specific probes for single loci.
ACKNOWLEDGMENT
We thank Dr. R. Hiller for providing cloned A. carterae PCP and LHC cDNAs.
Abbreviations:
- 5-MeC
5-methylcytosine
- LHC(s)
chlorophyll a/c-containing intrinsic light-harvesting protein(s)
- PCP(s)
peridinin chlorophyll a protein(s)
Footnotes
This research was supported by grants from the Australian Research Council and a James Cook University merit research grant. M.R.t.L. acknowledges receipt of an Australian Research Council postdoctoral fellowship.
LITERATURE CITED
- Anderson JM. Photoregulation of the composition, function, and structure of thylakoid membranes. Annu Rev Plant Physiol. 1986;37:93–136. [Google Scholar]
- Antequera F, McLeod D, Bird AP. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989;58:509–517. doi: 10.1016/0092-8674(89)90431-5. [DOI] [PubMed] [Google Scholar]
- Ball DJ, Gross DS, Garrard WT. 5-Methylcytosine is localized in nucleosomes that contain histone H1. Proc Natl Acad Sci USA. 1983;80:5490–5494. doi: 10.1073/pnas.80.18.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghammer H, Hayward D, Harrison P, Miller DJ. Nucleotide sequence of ub52 from the cnidarian Acropora millepora reveals high evolutionary conservation. Gene. 1996;178:195–197. doi: 10.1016/0378-1119(96)00353-8. [DOI] [PubMed] [Google Scholar]
- Bester TH. DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Phil Trans R Soc Lond. 1990;326:179–187. doi: 10.1098/rstb.1990.0002. [DOI] [PubMed] [Google Scholar]
- Bird A. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- Brown PTH. DNA methylation in plants and its role in tissue culture. Genome. 1989;31:717–729. [Google Scholar]
- Burn JE, Bagnall DJ, Metzger JD, Dennis ES, Peacock WJ. DNA methylation, vernalization and the initiation of flowering. Proc Natl Acad Sci USA. 1993;90:287–291. doi: 10.1073/pnas.90.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhausen G, Wittog B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattolico RA, Gibbs SP. Rapid filter method for the microfluorometric analysis of DNA. Anal Biochem. 1975;69:572–582. doi: 10.1016/0003-2697(75)90162-1. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Galaud JP. Isolation, sequencing and analysis of the expression of Bryonia calmodulin after mechanical perturbation. Plant Mol Biol. 1993;23:839–846. doi: 10.1007/BF00021538. [DOI] [PubMed] [Google Scholar]
- Grossmann A, Mandori A, Snyder D. Light harvesting proteins of diatoms: their relationship to the chlorophyll a/b proteins of higher plants and their mode of transport into the plastids. Mol Gen Genet. 1990;224:91–100. doi: 10.1007/BF00259455. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Cedar H, Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981;292:860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Herzog M, Soyer MO. Distinctive features of dinoflagellate chromatin: absence of nucleosomes in a primitive species Prorocentrum micans E. Eur J Cell Biol. 1981;23:295–302. [PubMed] [Google Scholar]
- Hiller RG, Wrench PM, Gooley AP, Shoebridge G, Breton J. The major intrinsic light-harvesting protein of Amphidinium carterae: characterisation and relation to other light harvesting proteins. Photochem Photobiol. 1993;57:125–131. doi: 10.1111/j.1751-1097.1993.tb02267.x. [DOI] [PubMed] [Google Scholar]
- Hiller RG, Wrench PM, Sharples FP. The light harvesting chlorophyll a-c-binding protein of dinoflagellates: a putative polyprotein. FEBS Lett. 1995;363:175–178. doi: 10.1016/0014-5793(95)00297-m. [DOI] [PubMed] [Google Scholar]
- Holm-Hansen O. Algae: amounts of DNA and organic carbon in single cells. Science. 1969;163:87–88. doi: 10.1126/science.163.3862.87. [DOI] [PubMed] [Google Scholar]
- Houlne G, Schanz R. Expression of polyproteins in Euglena. Crit Rev Plant Sci. 1993;12:1–17. [Google Scholar]
- Jablonka E, Regev A. Gene number, methylation and biological complexity. Trends Genet. 1995;11:383–382. doi: 10.1016/s0168-9525(00)89117-9. [DOI] [PubMed] [Google Scholar]
- Jarvis EE, Dunahay TG, Brown LM. DNA nucleotide composition and methylation in several species of microalgae. J Pycol. 1992;28:356–362. [Google Scholar]
- Jost J-P, Hofsteenge J. The repressor MDBP-2 is a member of the histone H1 family that binds preferentially in vitro and in vivo to methylated nonspecific DNA sequences. Proc Natl Acad Sci USA. 1992;89:9499–9503. doi: 10.1073/pnas.89.20.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juetterman E, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA methylation. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Ahmed M, Yamamoto T, Takahashi H, Oohara M, Ikeda T, Aida Y, Katsuki M, Arakawa Y, Shikata T and others. Inactivation of hepatitis C virus cDNA transgene by hyper-methylation in transgenic mice. Arch Virol. 1996;141:951–958. doi: 10.1007/BF01718169. [DOI] [PubMed] [Google Scholar]
- Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Kilby NJ, Ottoline Leyser HM, Furner IJ. Promoter methylation and progressive transgene inactivation in Arabidopsis. Plant Mol Biol. 1992;20:103–112. doi: 10.1007/BF00029153. [DOI] [PubMed] [Google Scholar]
- Kindle KL. Expression of a gene for light-harvesting chlorophyll a/b-binding protein in Chlamydamonas reinhardtii: effects of light and acetate. Plant Mol Biol. 1986;9:547–563. doi: 10.1007/BF00020532. [DOI] [PubMed] [Google Scholar]
- Klaas M, John MC, Crowell DN, Amasino RM. Rapid induction of genomic methylation and T-DNA gene expression in plant cells by 5-azacytosine derivatives. Plant Mol Biol. 1989;12:413–423. doi: 10.1007/BF00017581. [DOI] [PubMed] [Google Scholar]
- Kochanek S, Renz D, Doerfler W. Differences in the accessibility of methylated and unmethylated DNA to DNase I. Nucleic Acids Res. 1993;21:5843–5845. doi: 10.1093/nar/21.25.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA, Taylor WC, Nelson T. Cell specific accumulation of maize phophoenolpyruvate carboxylase is correlated with demethylation at a specific site > 3 kb upstream of the gene. Mol Gen Genet. 1991;225:49–55. doi: 10.1007/BF00282641. [DOI] [PubMed] [Google Scholar]
- Le QH, Markovic P, Hastings JW, Jovine RV, Morse D. Structure and organization of the peridinin-chlorophyll a-binding protein gene in Gonyaulax polyedra. Mol Gen Genet. 1997;255:595–604. doi: 10.1007/s004380050533. [DOI] [PubMed] [Google Scholar]
- Lee DH, Mittag M, Aczekan S, Morse D, Hasting JW. Molecular cloning and genomic organisation of a gene for luciferin-binding protein from the dinoflagellate Gonyaulax polyedra. J Biol Chem. 1993;12:8842–8850. [PubMed] [Google Scholar]
- Lewis J, Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991;285:155–159. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localisation of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Machabee S, Wall IL, Morse D. Expression and organisation of a dinoflagellate gene family. Plant Mol Biol. 1994;25:23–31. doi: 10.1007/BF00024195. [DOI] [PubMed] [Google Scholar]
- Medlin L, Elwood JH, Stickel S, Sogin ML. The characterisation of enzymatically amplified 16s-like ribosomal RNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner E, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Meyer P, Linn F, Heidmann, Meyer H, Niedenhoff I, Saedler H. Endogenous and environmental factors influence 35S promoter methylation of a maize A1 gene construct in transgenic petunia and its colour phenotype. Mol Gen Genet. 1992;231:345–352. doi: 10.1007/BF00292701. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Norris BJ, Miller DJ. Nucleotide sequence of a cDNA clone encoding the precursor of the peridinin-chlorophyll a-binding protein from the dinoflagellate Symbiodinium sp. Plant Mol Biol. 1994;24:673–677. doi: 10.1007/BF00023563. [DOI] [PubMed] [Google Scholar]
- Passaquet C, Lichtl C. Molecular study of a light-harvesting apoprotein of Giraudyopsis stellifer (Chrysophyceae) Plant Mol Biol. 1995;29:135–148. doi: 10.1007/BF00019125. [DOI] [PubMed] [Google Scholar]
- Prezelin BB. Photosynthetic physiology of dinoflagellates. In: Taylor FJR, editor. The Biology of Dinoflagellates: Botanical Monographs, Vol 20. Oxford, UK: Blackwell Scientific Publications; 1987. pp. 174–223. [Google Scholar]
- Rizzo PJ. Comparative aspects of basic chromatin proteins in dinoflagellates. BioSystems. 1981;14:433–443. doi: 10.1016/0303-2647(81)90048-4. [DOI] [PubMed] [Google Scholar]
- Rizzo PJ. Biochemistry of the dinoflagellate nucleus. In: Taylor FJR, editor. The Biology of Dinoflagellates: Botanical Monographs, Vol 20. Oxford, UK: Blackwell Scientific Publications; 1987. pp. 143–173. [Google Scholar]
- Rizzo PJ. The enigma of the dinoflagellate chromosome. J Protozool. 1991;38:246–252. [Google Scholar]
- Roman JS, Govind NS, Triplett EL, Prezelin BB. Light regulation of peridinin-chlorophyll a-protein (PCP) complexes in the dinoflagellate, Glenodinium sp. Plant Physiol. 1988;88:594–599. doi: 10.1104/pp.88.3.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Selker EU. DNA methylation and chromatin structure: a view from below. TIBS. 1990;15:103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]
- Sharples FP, Wrench PM, Hiller RG. Two distinct forms of the peridinin-chlorophyll a-protein from Amphidinium carterae. Biochim Biophys Acta. 1996;1276:117–123. doi: 10.1016/0005-2728(96)00066-7. [DOI] [PubMed] [Google Scholar]
- Tazi J, Bird A. DNA methylation and chromatin structure. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Theiss G, Schleicher R, Schimpf-Weiland G, Follmann H. DNA methylation in wheat: purification and properties of DNA methyltransferase. Eur J Biochem. 1987;167:89–96. doi: 10.1111/j.1432-1033.1987.tb13307.x. [DOI] [PubMed] [Google Scholar]
- van de Peer Y, Neefs J-M, de Rijk P, de Wachter R. Evolution of eukaryotes deduced from small ribosomal subunit RNA sequences. Biochem System Ecol. 1993;21:43–55. [Google Scholar]