Abstract
Alternative pre-mRNA splicing is a common way of gene expression regulation in metazoans. The selective use of specific exons can be modulated in response to various manipulations that alter Ca++ signals, particularly in neurons. A number of splicing factors have also been found to be controlled by Ca++ signals. Moreover, pre-mRNA elements have been identified that are essential and sufficient to mediate Ca++-regulated splicing, providing model systems for dissecting the involved molecular components. In neurons, this regulation likely contributes to the fine-tuning of neuronal properties.
Keywords: Alternative splicing, Ca++ signal, Pre-mRNA element, Splicing factor, Gene expression, Neuron, Brain, Ion channel, Neurodegenerative disease, Protein diversity, Fine-tuning
1. Introduction
Ca++ signalling is important for a variety of cellular processes such as muscle contraction, hormone secretion and gene transcription. In neurons, Ca++-regulation of gene expression is known to be critical for the long-lasting changes associated with learning and memory [1]. Accumulating evidence supports that Ca++ signalling also controls the alternative splicing of precursor messenger RNA (pre-mRNA) transcripts (Fig. 1). Examples of this regulation have been identified in myotube cultures, endocrine cells, and mostly, in neuronal cells (Table 1).
Fig. 1.
Diagram of Ca++ signal-regulation of alternative splicing. Shown in the middle is a cassette exon (e2) with two flanking introns (a and b) and constitutive exons (e1 and e3), with exonic and intronic cis-acting pre-mRNA elements as splicing enhancers (green, +) or silencers (red, −) and trans-acting splicing factors (ovals) indicated. Without signal stimulation (above), the pre-mRNA is spliced in two pathways (I and II) with a given ratio leading to the inclusion (I) or exclusion (II) of the alternative exon e2 and the respective lariats. With signal induction, like Ca++ (below), one pathway (here for example, II, heavier arrows) over the other pathway (I) is favored in a particular cell to promote the production of one variant mRNA. Through this regulation, Ca++ signals will change the relative ratio of the variant mRNA or protein isoforms of different properties. (?: mostly unknown intermediate steps).
Table 1.
List of alternative exons regulated by Ca++ signals
The list is according to gene name/description in alphabetical order, followed by other properties including exon, stimuli, inhibitor, protein function, reference, etc. PPase: protein phosphatase. NA: not available. For species, c: chick, h: human, r: rat, m: mouse. For more exons, see Xie et al., RNA’05 [107], Lee et al., Plos Biol.’07 [105] and An et al., Plos Biol.’07 [84].
The complex structure and function of the brain underlie the requirement for a high level of proteomic complexity. How can the human genome, with only the approximately 25,000 protein-coding genes [2], encode enough diverse proteins to achieve the high level of complexity to ensure, for example, that individual neurons are precisely “wired” with many others and that their functions are finely tuned? It is now clear that protein diversity can be greatly increased through alternative pre-mRNA splicing [3–5]. This is a particularly common way for the regulation of gene expression among neuronal genes and likely contributes to the fine-tuning of neuronal functions [6–8].
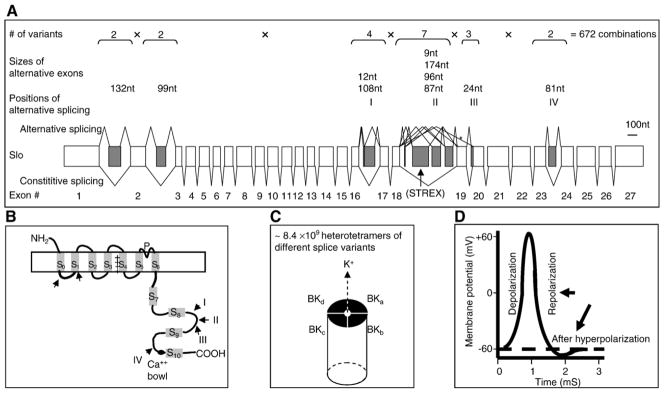
The pre-mRNA of most eukaryotic genes contains both expressed regions (exons) that will be included in the mature mRNA and regions that will be excluded (called intragenic regions, i.e. introns) [9]. Removal of introns and joining of exons happen in the cell nucleus through pre-mRNA splicing [10–12]. After introns were discovered, Walter Gilbert predicted in his short essay “Why genes in pieces” in Nature: “…the splicing need not be a hundred percent efficient; changes in sequence can alter the process so that base pairing and splicing occurs only some of the time” [9]. Soon afterward, alternative transcripts from a common precursor RNA were discovered [13]. Since then, more and more eukaryotic genes have been found to undergo alternative splicing [6,14,15]. Currently, the estimated percentage of human genes with alternative splicing is more than 50% [16,17]. In some cases, a single pre-mRNA transcript has the potential to generate a large number of protein isoforms, for example, transcripts of the Dscam, Neurexin and Slo genes [4,18–38] (Fig. 2).
Fig. 2.
An example of the generation of diverse protein functions through alternative splicing of genes important in neuronal functions: complex alternative splicing patterns of the middle exons of the mammalian BK potassium channel gene Slo (GenBank accession #: U23767). In A, the exon/intron organization of the Slo gene and alternative splicing positions, exon (boxes) sizes and possible number of variant transcripts produced from each position are indicated above the gene, with the exon numbers below it. The total number of channel subunits produced from all possible combinations of alternative splicing is 2×2×4×7×3×2=672, as indicated. Position of the STREX exon is indicated with an arrow. The length (nt) bar is for exon sizes. In B, positions where the alternative exons are inserted relative to the protein domains are indicated with arrows. S: transmembrane (or putative) domains. P: channel pore. Black oval: Ca++ bowl. The S4 voltage-sensitive domain is labelled with “+”s. (C) A channel heterotetramer of four different variant subunits a, b, c and d, with the total number of such heterotetramers indicated above the channel (see text for calculations). (D) Diagram of an action potential with depolarization, repolarization and afterhyperpolarization phases indicated. BK channels participate in the last two phases (arrows).
The consequences of alternative splicing include altered mRNA stability or subcellular localization and the addition or deletion of specific protein sequences. Functional differences among protein isoforms range from subtle modulations to on/off switches or antagonistic effects [39–41]. Defects in alternative splicing factors or aberrant inclusion of alternative exons can result in genetic diseases [42–47]. During the Cambrian “explosion” of metazoans, alternative splicing likely contributed to the expansion of proteomes [48]. Therefore, this is an important control in the transmission of genetic information from DNA to proteins. Currently with the huge amount of genomic information available from various genome sequencing projects, it is becoming imperative to understand how alternative splicing is controlled, particularly by cell signals, and to elucidate the role of this form of regulation in normal cell function and human genetic diseases.
Splicing happens in two transesterification steps within a large spliceosome complex containing more than 100 proteins assembled stepwise onto pre-mRNA introns [12,49]. During alternative splicing, the spliceosome assembly is altered so that a splice site (s) is optionally used depending on the cell type, developmental stage or sex, resulting in the inclusion or exclusion of alternative exon sequences in the mature mRNA (Fig. 1). Studies with tissue-or sex-specific exons identified both cis-acting pre-mRNA elements and trans-acting factors that control alternative splicing [15]. Depending on their location and effects on splicing, the RNA elements are called intronic or exonic splicing enhancers (ISE or ESE) or silencers (ISS or ESS). Many of these elements bind specific trans-acting factors (mostly proteins, but in some cases RNA) that enhance (enhancer) or inhibit (repressor) the assembly of the constitutive splicing factors. Well characterized examples of enhancer factors include most members of the arginine/serine-rich SR family that bind purine-rich ESE elements [50], and repressors include the heterogeneous ribonucleoprotein particle proteins hnRNP A1 and hnRNP I that bind purine-rich elements containing UAGG motifs and pyrimidine-rich elements containing UUCU, UCUU or CUCU motifs, respectively [51,52]. There are also elements/factors that function in a location-dependent way as either an enhancer or repressor, for example, the CA-repeat binding protein hnRNP L [53], the UGCAUG-binding proteins FOX-1 and -2 [54–61], and the YCAY-binding protein Nova-1 [62–66]. Binding of the trans-acting factors to the elements is thought to control mostly the early stages of spliceosome assembly before the first transesterification step [15]. For example, PTB prevents the binding of U2AF65, a factor of the E (early) complex, through competition or looping out the target exons [67–69]. In mammalian systems, splicing regulation is generally dependent on the combinatorial effects of multiple pre-mRNA elements and trans-acting factors [70–72] (Fig. 1). The relative levels of positive and negative regulatory factors determine the generation of variant mRNA isoforms in specific cell types [73–75]. Brain-enriched splicing factors include Nova-1 and -2 [62–66], FOX-1 and -2 [54–61], PTBP2 (nPTB) and neuronal Hu proteins [76–83].
In addition to cell type, developmental stage and sex-specific regulation, alternative splicing can also be dynamically regulated in response to extracellular stimuli such as cytokines, hormones or neurotransmitters [24,84–88], adding a further dimension to the control of the flow of genetic information [86]. Examining the inducible alternative splicing events will help understand their roles in cell functions as well as provide systems where the dynamic changes of spliceosomal components can be tracked down to facilitate the mechanistic studies of splicing. Moreover, understanding the molecular basis of this process will help develop ways to reverse the aberrant splicing events in genetic diseases using extracellular factors. However, it is not clear in most cases how alternative splicing is controlled by external stimuli and intracellular signalling pathways. Ca++-regulation of alternative splicing is among the regulatory events being intensively studied.
The following sections on the evidence for Ca++-regulation of alternative splicing will be focused on the regulated alternative exons, the splicing factors and the pre-mRNA elements. It should be noted that both constitutive and alternative splicing are coupled with transcription and this topic has been covered in recent reviews [89–92]. Promoter-dependent regulation of alternative splicing by Ca++ stimuli has also been observed [84]. Readers are referred to these reviews and original papers for this aspect [84,89–92].
2. Alternative exons regulated by Ca++ signals
Table 1 lists some of the alternative exons whose inclusion in the mature mRNA can be changed upon stimulation by extra-cellular factors that activate Ca++ signalling pathways.
These factors include the ionophore ionomycin [93], the neurotransmitter glutamate or its analogue NMDA (N-methyl-D-aspartate) [94–98], the muscarinic acetylcholine receptor agonist pilocarpine (inducing seizure) [99], the ionotropic glutamate receptor agonist kainate, the inhibitor of the sarco/endoplasmic reticulum Ca2+ ATPase thapsigargin [100–102], and depolarizing concentrations of KCl [84,98,101,103–106] (for a more complete list of the pharmacological agents and their descriptions, see Table 2). These stimuli can either promote or repress the inclusion of an exon (Table 1). Importantly, their effects on splicing can be inhibited by the L-type Ca++ channel blockers verapamil, nifedipine or nimodipine [95,96,102,103,107], by the NMDA receptor antagonist AP5 or MK-801 [84,95,96,103], by the Ca++/calmodulin-dependent protein kinase inhibitors KN93 or KN62 [84,95,96,104,105], or by the cell permeable Ca++ chelator BAPTA-AM [101,108]. Together, these observations strongly support the regulation of alternative pre-mRNA splicing by variations in intracellular Ca++ levels.
Table 2.
An alphabetical list of pharmacological agents/treatments referred to in this paper
| Chemical/treatment | Effect on Ca++ signalling |
|---|---|
| Acetylcholine | Agonist of acetylcholine receptor, increase Ca++ influx |
| AP5 | Antagonist of NMDA receptor, reduce Ca++ influx |
| BAPTA-AM | A cell permeable Ca++ chelator, reduce cytosolic Ca++ |
| CdCl2 | A voltage-sensitive Ca++ channel blocker |
| FK506 | A calcineurin inhibitor |
| Glutamate | Agonist of glutamate receptor, increase Ca++ influx through ionotropic glutamate receptors |
| Ionomycin | Ionophore, increase intracellular Ca++ |
| Ischemia | increase intracellular Ca++ |
| Kainate | Ionotropic glutamate receptor agonist, increase Ca++ influx |
| KCl (25–50 mM) | Induce Ca++ influx through Ca++ channels |
| KN62 | CaM kinase inhibitor |
| KN93 | CaM kinase inhibitor |
| MK-801 | Antagonist of NMDA receptor, reduce Ca++ influx |
| Nifedipine | L-type calcium channel blocker |
| Nimodipine | L-type calcium channel blocker |
| NMDA | Agonist of NMDA receptor, increase Ca++ influx |
| Pilocarpine | Agonist of muscarinic acetylcholine receptor, induce seizure, increase Ca++ |
| Thapsigargin | Inhibitor of the sarco/endoplasmic reticulum Ca++ ATPase, elevate cytosolic Ca++ |
| Verapamil | L-type calcium channel blocker |
Most of these regulated exons are in genes known to be important for neuronal functions. Examples include the neural cell adhesion molecule (NCAM) in axon guidance [102,109–113], the synapse-associated protein of 25 kDa (SNAP25), NMDA receptor type I (NMDAR1), BK and Ca++ channels and neurexin in synaptic function [8,96,108,114–123], the intracellular plasma-membrane calcium-pump isoforms 2 and 4 (PMCA2 and 4) and inositol triphosphate receptor type 1 (IP3R1) in Ca++ homeostasis [124–132], the caspase ICH-1 in neuronal survival/death [98,101,133], and the splicing factors Tra2 beta1 and Ania-6 in pre-mRNA splicing [99,106,134–136] (Table 1).
Inclusion of these alternative exons regulates the subcellular localization of proteins or their functions [25,26,119,120, 127–129,123,125,134,137–141], ranging from subtle changes such as current kinetics of ion channels to on or off switches in the sensitivity to protein kinases or hormones [26,114,139, 142–144] (Table 1). These observations suggest that the regulation of alternative splicing by Ca++ signals has effects on the expression of genes involved in a variety of cellular functions, from cell adhesion molecules at the synapse, ion channels in the cellular endoplasmic membrane, caspase in the cytosol as well as splicing factors inside the nucleus.
Of the various Ca++ signals that regulate splicing, those through the CaMK IV pathway have been the most intensively studied [104,105,107]. A prime example of the effect on splicing by this pathway is the BK channel STREX exon, one of the more than 10 alternative exons of the vertebrate Slo gene [19,21–35] (Fig. 2A). The BK channels contain both Ca++ and voltage-sensitive domains and are important in coupling Ca++ transients and electrical properties of excitable cells by participating in the repolarization and after hyperpolarization of action potentials [122] (Fig. 2B–D). These channels are important in neurotransmitter release [145], motor control [146], alcohol sensitivity [147–149], blood pressure and the fine-tuning of hearing frequencies [150,151]. The STREX-encoded peptide lies in the Ca++-sensitivity domain at the COOH-terminal [30,152]. Inclusion of this exon confers higher voltage and Ca++ sensitivity on the channel [24,29–31,153], inhibition by PKA and oxidation [142,154], and sensitivity to glucocorticoids [143]. The exon is included in endocrine cells and neurons and enriched in the high frequency region of cochleae [22–24,84,107], contributing to the fine-tuning of hearing frequencies in birds and turtles [23,31]. Its inclusion is also regulated by steroid hormones in animals and by membrane depolarization in cultured cells [24,104,107,155–157]. Thus, the STREX exon inclusion is inducible by external stimuli and the regulation may modulate multiple properties of the BK channels.
CaMK IV regulation of the STREX exon inclusion has been examined in both cultured cells and in mice. In rat pituitary GH3 cells and cerebellar neurons, addition of depolarizing concentrations of KCl leads to a decrease in transcripts containing the STREX exon [104,107] (Fig. 3). This effect can be specifically blocked by nifedipine [107], suggesting that L-type Ca++ channels mediate the depolarization-induced alternative splicing (Fig. 3). The splicing change is also completely blocked by KN93 [104], a CaM kinase inhibitor. Moreover, the repression of STREX exon inclusion is only seen with overexpressed constitutively active CaMK IV but not CaMK I or II [104]. Conversely, increased inclusion of the STREX exon was observed in the cerebella of CaMK IV+/− heterozygote mice compared to that of the wild type [107]. These findings support the proposal that membrane depolarization activates the L-type Ca++ channels and CaMK IV to repress STREX exon inclusion.
Fig. 3.
Coupling of Ca++ signals with pre-mRNA elements that are sufficient to mediate Ca++-regulated alternative splicing. Shown is a diagram of the pathway from membrane depolarization to the CaRRE and the UAGG motifs. In several neuronal or endocrine cells as indicated, membrane depolarization induces Ca++ signals through L-type Ca++ channels, likely activating CaMK IV, which targets pre-mRNA elements CaRRE1 and 2, probably also the UAGG motif, to control the inclusion of the exons, for example, STREX exon of Slo or exons 5 and 21 of NMDAR1. The changed exon compositions will later translate into proteins with different electrical properties (STREX, exon 5) or subcellular localizations (exon 21) to incorporate into the ion channels in cell membranes, to impact cellular electrical properties. In the pre-mRNA, the branch point (A), polypyrimidine tract (Yn) and 3′ AG (ag), and the exons (boxes) and introns (lines) are indicated. The CaRREs or the UAGG motif is shown as red bars. CaRRE1 was found in the upstream intron of the STREX exon and the NMDAR1 exon 5, and CaRRE1, 2 and UAGG motifs in the NMDAR1 exon 21. hnRNPA1 and the unknown factors are as red ovals. The “?” next to the hnRNP A1 pathway is to reflect the fact that this regulation requires CaM kinases but an effect by CaMK IV is not demonstrated yet.
Besides the Slo gene, similar regulation of the alternative splicing of NMDAR1 and other genes by membrane depolarization/CaMK IV has also been observed [84,104,105,107].
Another Ca++ signalling component calcineurin also appears to have a role in the control of alternative splicing. The inclusion of the 175 nt CII exon of the PMCA4 gene is reduced by KCl and NMDA in rat granule neurons [97]. This reduction can be prevented by FK506, a calcineurin inhibitor [97]. These data suggest that Ca++-dependent protein phosphatases and dephosphorylation events are required for depolarization-induced splicing in these cells.
For a single exon, its regulation by Ca++ signals can be different, depending on cell types or Ca++ signal stimuli. For example, inclusion of the STREX exon is repressed by depolarization in GH3 cells and cerebellar neurons but enhanced in cortical neurons [84,104,107]. In the Ania-6 gene, the retention of intron 6′ is promoted by glutamate but repressed by KCl depolarization [94], although both glutamate and KCl induce Ca++ signals and activate CaM kinases [158]. Thus, the control of alternative splicing by Ca++ signals is cell- and Ca++ signal-inducer dependent. Since the pre-mRNA elements are the same in a cell or among different cell types, except when RNA editing occurs [159], it is possible that diverse factors upstream of the RNA elements are involved [73–75], as is the case in the control of gene transcription [158].
These examples of Ca++ signal-regulated alternative splicing are found in several species including chick, rat, mouse and human, suggesting that this regulation is evolutionarily conserved.
Taken together, these observations from many independent studies strongly support that Ca++ signals control alternative splicing. This regulation controls the expression of a wide range of genes likely involving diverse factors, and is evolutionarily conserved.
However, most of these observations did not provide direct evidence for the involvement of the splicing regulatory components including the essential splicing factors and cis-acting pre-mRNA elements. To address this, in the following two sections, I will review literature on the splicing factors and pre-mRNA elements that are targeted by Ca++ signals.
3. Splicing factors regulated by Ca++ signals
Of the cis-acting elements in alternative splicing, most that have been studied to date are bound by protein factors, although, as in constitutive splicing, RNA secondary structures and transacting small RNAs have also been described [12,160–162]. Protein splicing factors reportedly regulated by Ca++ signals at various levels are listed in Table 3.
Table 3.
List of RNA binding proteins/splicing factors regulated by Ca++ signals
| Splicing factor | Ca++ signal-inducer/inhibitor/partner | Splicing factor change/interaction | Earliest time splicing change detected | Cell/extract | Reference |
|---|---|---|---|---|---|
| CPSF30 | CaM | Inhibited RNA binding activity | NA | A. thaliana | Delaney et al. [192] |
| hnRNP A1 | Depolarization | Increased nuclear protein level | 12 h | Cortical culture | An and Grabowski [84] |
| hnRNP A2 | CaM | Binds CaM column, inhibited phosphorylation by Casein kinase 2, colocalization with CaM in nucleus | NA | Rat liver | Bosser et al. [190] |
| hnRNP C | CaM | Binds CaM column, inhibited phosphorylation by Casein kinase 2, colocalization with CaM in nucleus | NA | Rat liver | Bosser et al. [190] |
| hnRNP H3 | Depolarization | Repressed exon inclusion | 24 h | Cortical culture | An and Grabowski [84] |
| PP2A | CaMK IV | Dephosphorylated Thr196 of CaMK IV and inhibited gene transcription | NA | In vitro | Tokumitsu et al. [185], Anderson et al. [187] |
| RNPS1 | Depolarization | Repressed exon inclusion | 6 h | Cortical culture | An and Grabowski [84] |
| SAP145 | CaM | Binds CaM column | NA | NA | Agell et al. [191] |
| Tra2 beta1 | Thapsigargin | Cytoplasmic accumulation | 1 h | Cortical culture | Daoud et al. [106] |
The list is according to the protein name/description in alphabetical order. NA: not available.
The protein level of splicing factors can be controlled by Ca++ signals. In some cases, it takes several hours for the splice variant mRNA to change significantly, and it has been shown that de novo protein synthesis is required [97,163,164]. Interestingly, the nuclear level of the hnRNP A1 protein is increased in depolarized neurons in regulating the exon 21 (or called C1 cassette exon) of NMDAR1 [72,84]. The increase is expected to contribute to the exclusion of exon 21 upon depolarization. Therefore, control of the protein level of splicing factors by membrane depolarization and the downstream Ca++ signalling is one way for Ca++ signals to regulate alternative splicing.
The alternative splicing of splicing factors themselves can also be regulated by Ca++ signals. For example, the inclusion of the alternative exons of hnRNP H3 and RNPS1 is specifically repressed by depolarization in cortical neurons [84], in contrast to the inclusion of the hnRNP A1 alternative exon 8, which is barely changed. The Ania-6 protein (also called cyclin L1) contains arginine/serine-rich (RS) domains that are characteristic of SR protein splicing factors and regulates alternative splicing [136,165]. The retention of the intron 6′ of Ania-6 is controlled by glutamate and depolarization in striatal neurons [94,95]. In another example, the exon III of the Tra2 beta1 is repressed in the brain during pilocarpine-induced seizure [99]. Since the alternative splicing of both Ania-6 and Tra2 beta1 is likely to alter their splicing activity [94,134,136,166], their own splicing regulation by Ca++ stimuli is expected to alter their target gene splicing as well.
Relocalization of splicing factors in response to Ca++ signals has also been observed, as in the cases of other cell signals [167–170]. In cortical neurons, thapsigargin or ischemia treament induces Tra2 beta1 relocalization from the nucleus to the cytoplasm [99]. This translocation is accompanied by an increased inclusion of a 61 nt cassette exon of the ICH-1 gene.
Phosphorylation of splicing factors has been described in many other cases [85,87,171,172]. In Ca++-regulated splicing, the CaM kinase inhibitor KN93 inhibits the splicing changes of the Slo gene [84,104]. Whether this is due to inhibition of the phosphorylation of a splicing factor remains unclear.
Phosphatases, including PP1 [173–180], PP2C-gamma [181,182], PP2A [178,183], and the pol II CTD phosphatase FCP1 [184], also interact with splicing factors or are known to control splicing. Of these, PP2A binds CaMK IV in a signalling complex [185,186]. Moreover, PP2A is also shown to inactivate CaMK IV by directly dephosphorylating the Thr196 of CaMK IV [187]. Since the CaMK IV activity is essential for its role in the control of splicing [104,105,107], it is possible that the PP2A dephosphorylation of CaMK IV at Thr196 has an effect on CaMK IV-regulated alternative splicing.
Protein methylation has also been shown to control splicing factors or splicing [188,189]. Currently there is no evidence available to support a role for methylation in Ca++-regulated alternative splicing.
Can Ca++ or calmodulin (CaM) directly interact with the splicing machinery? In an experiment using calmodulin-affinity chromatography, several RNA binding proteins including hnRNP A2 and C were identified [190]. The binding of CaM to these splicing factors inhibits their phosphorylation by Casein kinase II [190]. SAP145 [191], a member of the spliceosome-associated proteins and a subunit of SF3 (b) [49], was also identified as binding to a CaM affinity column. In another report, polyadenylation specificity factor 30 (CPSF30) of Arabidopsis thaliana was found to bind calmodulin [192], which inhibits the RNA binding activity of CPSF30. In HeLa nuclear extracts, CPSF couples transcription with splicing [193]. Therefore, it will be interesting to know whether varying Ca++ concentrations and CaM can control any specific alternative splicing event through direct interaction with the factors that regulate splicing.
Taken together, the current evidence indicates that Ca++ signalling can control or interact with components of the splicing machinery. The regulation of hnRNP A1 by membrane depolarization to control the inclusion of exon 21 of the NMDAR1 gene through the UAGG motifs is particularly well characterized. In most other cases, however, a direct molecular link between the regulation of the splicing factors by Ca++ signalling and a change in RNA splicing remains to be demonstrated.
4. Pre-mRNA elements responsive to Ca++ signals in the control of alternative splicing
Since most mammalian exons are controlled in a combinatorial way by multiple positive and negative regulatory elements (Fig. 1), the role of a single RNA element in splicing regulation is usually isolated from its endogeneous gene context and tested in a heterologous mini-gene, as has been done in studying the control of gene transcription [194,195]. This approach has also proved useful in studying the regulation of splicing by the CaM kinase pathway.
Two CaMK IV-responsive RNA elements (CaRREs) have been isolated [104,105,107] (Fig. 3), which, when transferred to a heterologous gene, confer CaMK IV regulation on an otherwise constitutive exon. Similarly, a UAGG motif is also shown to mediate depolarization-regulated alternative splicing [84], which is inhibited by the CaM kinase inhibitor KN93. These observations allow the coupling of depolarization/CaMK IV with short RNA sequence elements, providing regulatory systems much simpler than the endogeneous genes for characterizing the components in-between the stimuli and RNA elements.
From the example of the regulation of NMDAR1 exon 21, it appears that multiple elements, either the same (UAGG) or different (both CaRRE1 and 2), can function within the same exon to respond to Ca++ signals to control alternative splicing [84,104,105]. However, it is unclear whether this is a feature common to the regulation of other exons.
Actually, search in alternative splicing databases or genomes for these depolarization/CaMK IV-responsive RNA elements identified different groups of exons and target genes [84,105,107]. This raises questions whether the target genes of a particular element are involved in specific cellular functions and whether each element has its preferred Ca++ stimulus, since CaMK IV can be activated by different stimuli [196–199].
As mentioned earlier, a trans-acting factor has been identified in the case of the UAGG motifs of the NMDAR1 exon 21. This motif binds hnRNPA1 to repress the 5′ splice site and exon inclusion [72,84]. Interestingly, the nuclear level of the hnRNP A1 protein is increased in depolarized neurons [84]. Further evidence with loss-of-function of hnRNP A1 will help verify its role in the inducible regulation of alternative splicing of endogenous genes in neurons.
Taken together, these studies couple Ca++ signals with specific pre-mRNA elements. This coupling isolates single RNA elements from the complex influences of their surrounding elements in the endogenous gene transcripts thus providing simple model systems for studying Ca++-regulation of alternative splicing.
5. The impact of Ca++-regulated alternative splicing on neuronal functions and diseases
As shown in Table 1 and references therein, alternative exons controlled by Ca++ signalling can affect a broad range of neuronal functions. Based on data from these Ca++ signal-regulated splicing events and work from others [7,8], I will speculate on the effect of Ca++-regulated alternative splicing on neuronal functions and diseases.
5.1. Fine-tuning of neuronal functions
One fascinating property of neurons, for which the underlying molecular basis is not fully understood, is the fine-tuning of their functions. Interestingly, the functions of many neuronal genes are diversified and optimized through alternative splicing [6,8,18,200]. The large number of splice variants contributes greatly to the diversity of the protein functions, especially when functional proteins such as ion channels can be composed of heteromultimers of different splice variants. Take for example the production of 672 splice variants in all possible combinations of the mammalian BK channel subunits (Fig. 2). If any four variants are chosen to create a tetramer channel without considering their relative positions inside the channel, the total number of all possible combinations of every four variants can be calculated according to the algebra formula for “Combination” . Thus, theoretically about eight billion heterotetramers of BK channels can be formed in total, all composed of four different variant subunits. Note that this calculation is still an underestimate since it has not considered the relative subunit positions of a channel, homotetramers and combinations of homo- and hetero-subunits. With this large number of ion channels, a high resolution spectrum of BK channel properties emerges (Fig. 4A). For example, for a Ca++ sensitivity range of 0–100 μM, the spectrum will be a much smoother gradient of Ca++ sensitivities formed by different channels when there are eight billion than when only one or ten channel variants are available. The fine-tuning of electrical properties is exemplified in turtle and bird cochlear hair cells where the BK channel kinetics is “tuned” along the tonotopic map [151]. Differential alternative splicing of the Slo gene along the map is also observed and proposed to contribute to the fine-tuning of hearing frequencies [151]. By analogy, the electrical and other properties of neurons could also be finely tuned this way.
Fig. 4.
Speculations on the impact of Ca++-regulated splicing on protein and neuronal functions. (A) On the fine-tuning of the Ca++ sensitivity of ion channels (for example, the BK channels). With Ca++ sensitivities between 0 to 100 μM (top), and increasing number of different ion channels (right) available through alternative splicing, the spectrum of the Ca++ sensitivity is becoming increasingly refined with narrower intervals (suppose the Ca++ concentration intervals are even). For simplicity, only the effect of up to 1000 variant channels is shown, with more indicated by “… …”. (B) Modulation of splice variant ratios beyond cell-specific expression of a limited number of variants by cell signals such as Ca++. Shown are two splice variants expressed in a cell with n% for variant 1 and (100-n)% for variant 2 when there is no signal. This relative percentage can be shifted smoothly if varying strengths of Ca++ stimuli are applied to modulate the splicing resulting in a spectrum of variant percentages ranging from 0% to 100% for each variant. (C) Diagram of the possible contribution of Ca++/CaMK IV-regulated alternative splicing to changes in neuronal electrical properties during electrophysiological memory. Changed electrical firing during the 1st and 2nd round stimulations is indicated with a different amplitude and frequency. This change requires gene expression through CaMK IV. The control of alternative splicing of ion channels by CaMK IV to either promote or repress the inclusion of certain exons is expected to contribute to the changes in ion channel subunit compositions and therefore the firing properties.
Greater diversity of protein properties could be further obtained by varying the strength of the signals controlling alternative splicing, particularly for individual cells where only a limited set of splice variants is often observed. For example, when only two variants are produced in a cell and their relative levels are dependent on the strength of a Ca++ stimulus (Fig. 4B), varying the stimulus strength will induce a gradient of the relative levels of the two variant proteins leading to the formation of heterotetrameric channels with varying ratios of variant subunits and the generation of electrical currents of higher diversity than with only two homotetramers. In fact, splicing changes can be KCl concentration-dependent [84,105], and different ratios of two BK channel subunits generate channel current kinetics in-between the two homotetramer channels [19,201]. This way, alternative splicing regulation by Ca++ signals will greatly contribute to the fine-tuning of electrical properties of individual neurons.
5.2. Autoregulation and adaptive changes
As shown in Table 1, the alternative splicing of several Ca++ channel genes is regulated by Ca++ signals and some exons of ion channel genes are regulated by their own channel ligands or by membrane depolarization. For example, the inclusion of the exons 5 and 21 of NMDAR1 is controlled by NMDA or glutamate, and the STREX exon of the BK channel by membrane depolarization [103,104]. Inclusion of these exons has known effects on the functions or localization of the specific channels [24,29,30,39,140,141,202]. Therefore, a given first round stimulus (for example, depolarization during electrical stimulation) could cause a change in the ion channel subunits via alternative splicing (Fig. 4C). This change will provide neurons with a different set of variant ion channels to respond to a second-round identical stimulus. The second-round stimulation will now have different effects on the neuronal electrical properties because of the different channels formed from the first round stimulation-induced alternative splicing. Such self-regulatory and adaptive changes are interesting when considering the most important functions of neurons and the brain: learning and memory, particularly electrophysiological memory that requires CaMK IV [203].
5.3. Neuronal death and neurodegenerative diseases
Aberrant alternative splicing of several genes or deficiencies in alternative splicing factors have been implicated in neuronal diseases [43]. Ca++ is also involved in neuronal diseases [204–206]. In normal neurons, Ca++ levels are under homeostatic control. However, when the balance is tilted, deficient neuronal functions or diseases develop [204]. For example, when excess Ca++ ions are present, neuronal toxicity occurs leading to neuron death or contributing to neurodegenerative diseases [204,206]. Inhibition of CaMK IV also leads to neuron death in cell cultures [207], and CaMK IV deficiency in mice results in loss of learning or memory or in less drug tolerance [208–211]. The ion channels involved in Ca++ toxicity including the AMPA and NMDA receptors undergo alternative splicing [202,204], and inclusion of the NMDAR1 exons 5 and 21 is controlled by membrane depolarization and CaMK IV [84,104,105,107,212]. In addition, the ICH-1S variant induced by thapsigargin or ischemia prevents cell death [213,214]. Therefore, it will be interesting to determine whether the regulation by Ca++ signals of these and other ion channels and apoptosis-related genes is important for Ca++ homeostasis in normal cells or imbalance during neuronal death and in the development of neurodegenerative diseases.
6. Perspectives
Even with the progress described above, much remains to be learned about Ca++ control of alternative splicing and its role in cellular, particularly neuronal, functions and diseases. For example, what is the nature of the molecular link between CaMK IV and the trans-acting splicing factors that function through the CaRRE elements? What is the physiological impact of a specific stimulus through the regulation of a group of alternative exons? How important is this inducible alternative splicing in neuronal and whole brain functions such as learning and memory? Understanding the molecular basis of this regulation will help us to address these questions.
In addition to the traditional molecular, cellular, biochemical and genetic approaches for the examination of individual genes or proteins, several recent developments could be helpful in answering these questions. Firstly, large-scale proteomic approaches may help identify splicing factors and their posttranslational modifications induced by Ca++ signals. Secondly, whole genome RNA interference libraries have been developed recently and applied to whole genome loss-of-function screening of genes [215–222]. Screening for alternative splicing factors by RNAi has already been carried out in fly cells [215,216]. Hopefully, the now available mammalian RNAi libraries could also be applied for functional screening of genes essential in inducible alternative splicing. Thirdly, splicing-sensitive microarrays and whole genome exon arrays [64,78,82,83,223–225], combined with loss-of-functions of splicing factors (such as knockout or RNAi), will allow the identification of a group of exons/genes controlled by a particular factor during Ca++-regulation of splicing. Lastly, bioinformatics approaches with highly predictive RNA elements could also help identify a group of exons regulated by Ca++ signals, as has been demonstrated for the alternative splicing factor Nova-1 [62]. These recent developments will facilitate the study of Ca++-regulated splicing and will allow a more accurate assessment of the physiological impact of a particular splicing factor/RNA element through regulating the alternative splicing of a specific functional group of genes.
In summary, Ca++ signal-regulated alternative splicing modulates the function of a variety of genes that have important roles in cellular, particularly neuronal, functions and diseases. Recent studies have identified downstream splicing factors and coupled Ca++ signals with pre-mRNA elements. Further detailed analyses of individual elements/exons using model systems shall help to bring the molecular basis of this regulation into focus. Future studies with genomewide coverage of alternative exons and splicing factors shall help in our understanding of the impact of splicing regulation on cellular functions and its role in the development of diseases.
Acknowledgments
I thank Mary Lynn Duckworth and Jean Paterson for editing the manuscript. Work in my lab is supported by an operating grant (MOP#68919) and a New Investigator Salary Award from the Canadian Institutes of Health Research (CIHR), by a research grant (#016355) from the National Cancer Institute of Canada (NCIC), and by Manitoba Health Research Council and Canada Foundation for Innovation (CFI) funds.
References
- 1.West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci U S A. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.I.H.G.S. Consortium, Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 3.Roberts GC, Smith CW. Alternative splicing: combinatorial output from the genome. Curr Opin Chem Biol. 2002;6:375–383. doi: 10.1016/s1367-5931(02)00320-4. [DOI] [PubMed] [Google Scholar]
- 4.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 5.Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 6.Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 7.Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Lipscombe D. Neuronal proteins custom designed by alternative splicing. Curr Opin Neurobiol. 2005;15:358–363. doi: 10.1016/j.conb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 10.Berget SM, Moore C, Sharp PA. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 12.Burge CB, Tuschl T, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland R, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor Laboratory Press; New York: 1999. pp. 525–560. [Google Scholar]
- 13.Berk AJ, Sharp PA. Structure of the adenovirus 2 early mRNAs. Cell. 1978;14:695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- 14.Nadal-Ginard B, Smith CW, Patton JG, Breitbart RE. Alternative splicing is an efficient mechanism for the generation of protein diversity: contractile protein genes as a model system. Adv Enzyme Regul. 1991;31:261–286. doi: 10.1016/0065-2571(91)90017-g. [DOI] [PubMed] [Google Scholar]
- 15.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;27:27. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 16.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 18.Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 19.Lagrutta A, Shen KZ, North RA, Adelman JP. Functional differences among alternatively spliced variants of slowpoke, a Drosophila calcium-activated potassium channel. J Biol Chem. 1994;269:20347–20351. [PubMed] [Google Scholar]
- 20.Shen KZ, Lagrutta A, Davies NW, Standen NB, Adelman JP, North RA. Tetraethylammonium block of Slowpoke calcium-activated potassium channels expressed in Xenopus oocytes: evidence for tetrameric channel formation, Pfluegers. Arch Eur J Physiol. 1994;426:440–445. doi: 10.1007/BF00388308. [DOI] [PubMed] [Google Scholar]
- 21.Tseng-Crank J, Foster CD, Krause JD, Mertz R, Godinot N, DiChiara TJ, Reinhart PH. Cloning, expression, and distribution of functionally distinct Ca(2+)-activated K+ channel isoforms from human brain. Neuron. 1994;13:1315–1330. doi: 10.1016/0896-6273(94)90418-9. [DOI] [PubMed] [Google Scholar]
- 22.Ferrer J, Wasson J, Salkoff L, Permutt MA. Cloning of human pancreatic islet large conductance Ca(2+)-activated K+ channel (hSlo) cDNAs: evidence for high levels of expression in pancreatic islets and identification of a flanking genetic marker. Diabetologia. 1996;39:891–898. doi: 10.1007/BF00403907. [DOI] [PubMed] [Google Scholar]
- 23.Jones EMC, Laus C, Fettiplace R. Identification of Ca2+-activated K+ channel splice variants and their distribution in the turtle cochlea. Proc R Soc Lond, B Biol Sci. 1998;265:685–692. doi: 10.1098/rspb.1998.0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie J, McCobb DP. Control of alternative splicing of potassium channels by stress hormones. Science (Washington D C) 1998;280:443–446. doi: 10.1126/science.280.5362.443. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Tian L, MacDonald SH, McClafferty H, Hammond MS, Huibant JM, Ruth P, Knaus HG, Shipston MJ. Functionally diverse complement of large conductance calcium- and voltage-activated potassium channel (BK) alpha-subunits generated from a single site of splicing. J Biol Chem. 2005;280:33599–33609. doi: 10.1074/jbc.M505383200. [DOI] [PubMed] [Google Scholar]
- 26.Zarei MM, Zhu N, Alioua A, Eghbali M, Stefani E, Toro L. A novel MaxiK splice variant exhibits dominant-negative properties for surface expression. J Biol Chem. 2001;276:16232–16239. doi: 10.1074/jbc.M008852200. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Chang Y, Reinhart PH, Sontheimer H. Cloning and characterization of glioma BK, a novel BK channel isoform highly expressed in human glioma cells. J Neurosci. 2002;22:1840–1849. doi: 10.1523/JNEUROSCI.22-05-01840.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korovkina VP, Fergus DJ, Holdiman AJ, England SK. Characterization of a novel 132-bp exon of the human maxi-K channel. Am J Physiol, Cell Physiol. 2001;281:C361–C367. doi: 10.1152/ajpcell.2001.281.1.C361. [DOI] [PubMed] [Google Scholar]
- 29.Shipston MJ, Duncan RR, Clark AG, Antoni FA, Tian L. Molecular components of large conductance calcium-activated potassium (BK) channels in mouse pituitary corticotropes. Mol Endocrinol. 1999;13:1728–1737. doi: 10.1210/mend.13.10.0355. [DOI] [PubMed] [Google Scholar]
- 30.Saito M, Nelson C, Salkoff L, Lingle CJ. A cysteine-rich domain defined by a novel exon in a Slo variant in rat adrenal chromaffin cells and PC12 cells. J Biol Chem. 1997;272:11710–11717. doi: 10.1074/jbc.272.18.11710. [DOI] [PubMed] [Google Scholar]
- 31.Ramanathan K, Michael TH, Jiang GJ, Hiel K, Fuchs PA. A molecular mechanism for electrical tuning of cochlear hair cells. Science (Washington D C) 1999;283:215–217. doi: 10.1126/science.283.5399.215. [DOI] [PubMed] [Google Scholar]
- 32.Rosenblatt KP, Sun ZP, Heller S, Hudspeth AJ. Distribution of Ca2+-activated K+ channel isoforms along the tonotopic gradient of the chicken’s cochlea. Neuron. 1997;19:1061–1075. doi: 10.1016/s0896-6273(00)80397-9. [DOI] [PubMed] [Google Scholar]
- 33.Navaratnam DS, Bell TJ, Tu TD, Cohen EL, Oberholtzer JC. Differential distribution of Ca2+-activated K+ channel splice variants among hair cells along the tonotopic axis of the chick cochlea. Neuron. 1997;19:1077–1085. doi: 10.1016/s0896-6273(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 34.Butler A, Tsunoda S, McCobb DP, Wei A, Salkoff L. MSlo, a complex mouse gene encoding “maxi” calcium-activated potassium channels. Science (Washington D C) 1993;261:221–224. doi: 10.1126/science.7687074. [DOI] [PubMed] [Google Scholar]
- 35.McCobb DP, Fowler NL, Featherstone T, Lingle CJ, Salkoff LB. The human “BK” calcium-activated K channel gene and cDNA’s from arterial smooth muscle. J Gen Physiol. 1994;104:11A–12A. doi: 10.1152/ajpheart.1995.269.3.H767. [DOI] [PubMed] [Google Scholar]
- 36.Brandle U, Frohnmayer S, Krieger T, Zenner HP, Ruppersberg JP, Maassen MM. Expression of Ca(2+)-activated K(+) channel subunits and splice variants in the rat cochlea. Hear Res. 2001;161:23–28. doi: 10.1016/s0378-5955(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 37.Kukuljan M, Taylor A, Chouinard H, Olguin P, Rojas CV, Ribera AB. Selective regulation of xSlo splice variants during Xenopus embryogenesis. J Neurophysiol. 2003;90:3352–3360. doi: 10.1152/jn.00398.2003. [DOI] [PubMed] [Google Scholar]
- 38.Quirk JC, Reinhart PH. Identification of a novel tetramerization domain in large conductance K(ca) channels. Neuron. 2001;32:13–23. doi: 10.1016/s0896-6273(01)00444-5. [DOI] [PubMed] [Google Scholar]
- 39.Shipston MJ. Alternative splicing of potassium channels: a dynamic switch of cellular excitability. Trends Cell Biol. 2001;11:353–358. doi: 10.1016/s0962-8924(01)02068-2. [DOI] [PubMed] [Google Scholar]
- 40.Wu JY, Tang H, Havlioglu N. Alternative pre-mRNA splicing and regulation of programmed cell death. Prog Mol Subcell Biol. 2003;31:153–185. doi: 10.1007/978-3-662-09728-1_6. [DOI] [PubMed] [Google Scholar]
- 41.Revil T, Shkreta L, Chabot B. Pre-mRNA alternative splicing in cancer: functional impact, molecular mechanisms and therapeutic perspectives. Bull Cancer. 2006;93:909–919. [PubMed] [Google Scholar]
- 42.Hammes A, Guo JK, Lutsch G, Leheste JR, Landrock D, Ziegler U, Gubler MC, Schedl A. Two splice variants of the Wilms’ tumor 1 gene have distinct functions during sex determination and nephron formation. Cell. 2001;106:319–329. doi: 10.1016/s0092-8674(01)00453-6. [DOI] [PubMed] [Google Scholar]
- 43.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 44.Novoyatleva T, Tang Y, Rafalska I, Stamm S. Pre-mRNA missplicing as a cause of human disease. Prog Mol Subcell Biol. 2006;44:27–46. doi: 10.1007/978-3-540-34449-0_2. [DOI] [PubMed] [Google Scholar]
- 45.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 46.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 47.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 48.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 50.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chabot B, LeBel C, Hutchison S, Nasim FH, Simard MJ. Heterogeneous nuclear ribonucleoprotein particle A/B proteins and the control of alternative splicing of the mammalian heterogeneous nuclear ribonucleoprotein particle A1 pre-mRNA. Prog Mol Subcell Biol. 2003;31:59–88. doi: 10.1007/978-3-662-09728-1_3. [DOI] [PubMed] [Google Scholar]
- 52.Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M, Smith CW. Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- 53.Hui J, Hung LH, Heiner M, Schreiner S, Neumuller N, Reither G, Haas SA, Bindereif A. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fukumura K, Kato A, Jin Y, Ideue T, Hirose T, Kataoka N, Fujiwara T, Sakamoto H, Inoue K. Tissue-specific splicing regulator Fox-1 induces exon skipping by interfering E complex formation on the downstream intron of human F1{gamma} gene. Nucleic Acids Res. 2007;35:5303–5311. doi: 10.1093/nar/gkm569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol Cell Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, Hou VC, Lo AJ, Short SA, Chasis JA, Winkelmann JC, Conboy JG. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- 59.Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33:714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 63.Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc Natl Acad Sci U S A. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, Zeeberg BR, Kane D, Weinstein JN, Blume J, Darnell RB. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 65.Buckanovich RJ, Posner JB, Darnell RB. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- 66.Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 67.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 70.Modafferi EF, Black DL. Combinatorial control of a neuron-specific exon. RNA (New York) 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 72.Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 74.Hanamura A, Caceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 76.Chan RC, Black DL. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ashiya M, Grabowski PJ. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA (New York) 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 78.Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polydorides AD, Okano HJ, Yang YYL, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci U S A. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu H, Hasman RA, Barron VA, Luo G, Lou H. A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators. Mol Biol Cell. 2006;17:5105–5114. doi: 10.1091/mbc.E06-02-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.An P, Grabowski PJ. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol. 2007;5:e36. doi: 10.1371/journal.pbio.0050036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 86.Stamm S. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum Mol Genet. 2002;11:2409–2416. doi: 10.1093/hmg/11.20.2409. [DOI] [PubMed] [Google Scholar]
- 87.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev, Mol Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 88.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev, Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 89.Cramer P, Srebrow A, Kadener S, Werbajh S, de la Mata M, Melen G, Nogues G, Kornblihtt AR. Coordination between transcription and pre-mRNA processing. FEBS Lett. 2001;498:179–182. doi: 10.1016/s0014-5793(01)02485-1. [DOI] [PubMed] [Google Scholar]
- 90.Reed R. Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 91.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 92.Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 93.Ogimoto M, Katagiri T, Hasegawa K, Mizuno K, Yakura H. Induction of CD45 isoform switch in murine B cells by antigen receptor stimulation and by phorbol myristate acetate and ionomycin. Cell Immunol. 1993;151:97–109. doi: 10.1006/cimm.1993.1224. [DOI] [PubMed] [Google Scholar]
- 94.Berke JD, Sgambato V, Zhu PP, Lavoie B, Vincent M, Krause M, Hyman SE. Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron. 2001;32:277–287. doi: 10.1016/s0896-6273(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 95.Sgambato V, Minassian R, Nairn AC, Hyman SE. Regulation of ania-6 splice variants by distinct signaling pathways in striatal neurons. J Neurochem. 2003;86:153–164. doi: 10.1046/j.1471-4159.2003.01816.x. [DOI] [PubMed] [Google Scholar]
- 96.Choi JY, Beaman-Hall CM, Vallano ML. Granule neurons in cerebellum express distinct splice variants of the inositol trisphosphate receptor that are modulated by calcium. Am J Physiol, Cell Physiol. 2004;287:C971–C980. doi: 10.1152/ajpcell.00571.2003. [DOI] [PubMed] [Google Scholar]
- 97.Guerini D, Wang X, Li L, Genazzani A, Carafoli E. Calcineurin controls the expression of isoform 4CII of the plasma membrane Ca(2+) pump in neurons. J Biol Chem. 2000;275:3706–3712. doi: 10.1074/jbc.275.5.3706. [DOI] [PubMed] [Google Scholar]
- 98.Guerini D, Garcia-Martin E, Gerber A, Volbracht C, Leist M, Merino CG, Carafoli E. The expression of plasma membrane Ca2+ pump isoforms in cerebellar granule neurons is modulated by Ca2+ J Biol Chem. 1999;274:1667–1676. doi: 10.1074/jbc.274.3.1667. [DOI] [PubMed] [Google Scholar]
- 99.Daoud R, Da Penha Berzaghi M, Siedler F, Hubener M, Stamm S. Activity-dependent regulation of alternative splicing patterns in the rat brain. Eur J Neurosci. 1999;11:788–802. doi: 10.1046/j.1460-9568.1999.00486.x. [DOI] [PubMed] [Google Scholar]
- 100.Vigues S, Gastaldi M, Chabret C, Massacrier A, Cau P, Valmier J. Regulation of calcium channel alpha(1A) subunit splice variant mRNAs in kainate-induced temporal lobe epilepsy. Neurobiol Dis. 1999;6:288–301. doi: 10.1006/nbdi.1999.0248. [DOI] [PubMed] [Google Scholar]
- 101.Zacharias DA, Strehler EE. Change in plasma membrane Ca2(+)-ATPase splice-variant expression in response to a rise in intracellular Ca2+ Curr Biol. 1996;6:1642–1652. doi: 10.1016/s0960-9822(02)70788-4. [DOI] [PubMed] [Google Scholar]
- 102.Rafuse VF, Landmesser L. Contractile activity regulates isoform expression and polysialylation of NCAM in cultured myotubes: involvement of Ca2+ and protein kinase C. J Cell Biol. 1996;132:969–983. doi: 10.1083/jcb.132.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vallano ML, Beaman-Hall CM, Benmansour S. Ca2+ and pH modulate alternative splicing of exon 5 in NMDA receptor subunit 1. Neuroreport. 1999;10:3659–3664. doi: 10.1097/00001756-199911260-00036. [DOI] [PubMed] [Google Scholar]
- 104.Xie J, Black DL. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 2001;410:936–939. doi: 10.1038/35073593. [DOI] [PubMed] [Google Scholar]
- 105.Lee JA, Xing Y, Nguyen D, Xie J, Lee CJ, Black DL. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 2007;5:e40. doi: 10.1371/journal.pbio.0050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Daoud R, Mies G, Smialowska A, Olah L, Hossmann KA, Stamm S. Ischemia induces a translocation of the splicing factor tra2-beta 1 and changes alternative splicing patterns in the brain. J Neurosci. 2002;22:5889–5899. doi: 10.1523/JNEUROSCI.22-14-05889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie J, Jan C, Stoilov P, Park J, Black DL. A consensus CaMK IV-responsive RNA sequence mediates regulation of alternative exons in neurons. RNA. 2005;11:1825–1834. doi: 10.1261/rna.2171205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rozic-Kotliroff G, Zisapel N. Ca2+-dependent splicing of neurexin IIalpha. Biochem Biophys Res Commun. 2007;352:226–230. doi: 10.1016/j.bbrc.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 109.Hanson MG, Landmesser LT. Normal patterns of spontaneous activity are required for correct motor axon guidance and the expression of specific guidance molecules. Neuron. 2004;43:687–701. doi: 10.1016/j.neuron.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 110.Walsh FS, Doherty P. Neural cell adhesion molecules of the immunoglobulin superfamily: role in axon growth and guidance. Annu Rev Cell Dev Biol. 1997;13:425–456. doi: 10.1146/annurev.cellbio.13.1.425. [DOI] [PubMed] [Google Scholar]
- 111.Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10:19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- 112.Owens GC, Edelman GM, Cunningham BA. Organization of the neural cell adhesion molecule (N-CAM) gene: alternative exon usage as the basis for different membrane-associated domains. Proc Natl Acad Sci U S A. 1987;84:294–298. doi: 10.1073/pnas.84.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Polo-Parada L, Bose CM, Plattner F, Landmesser LT. Distinct roles of different neural cell adhesion molecule (NCAM) isoforms in synaptic maturation revealed by analysis of NCAM 180 kDa isoform-deficient mice. J Neurosci. 2004;24:1852–1864. doi: 10.1523/JNEUROSCI.4406-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chaudhuri D, Chang SY, DeMaria CD, Alvania RS, Soong TW, Yue DT. Alternative splicing as a molecular switch for Ca2+/calmodulin-dependent facilitation of P/Q-type Ca2+ channels. J Neurosci. 2004;24:6334–6342. doi: 10.1523/JNEUROSCI.1712-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hepp R, Dupont JL, Aunis D, Langley K, Grant NJ. NGF enhances depolarization effects on SNAP-25 expression: induction of SNAP-25b isoform. Neuroreport. 2001;12:673–677. doi: 10.1097/00001756-200103260-00011. [DOI] [PubMed] [Google Scholar]
- 116.Craig AM, Kang Y. Neurexin–neuroligin signaling in synapse development. Curr Opin Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lise MF, El-Husseini A. The neuroligin and neurexin families: from structure to function at the synapse. Cell Mol Life Sci. 2006;63:1833–1849. doi: 10.1007/s00018-006-6061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dean C, Dresbach T. Neuroligins and neurexins: linking cell adhesion, synapse formation and cognitive function. Trends Neurosci. 2006;29:21–29. doi: 10.1016/j.tins.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 119.Sorensen JB, Nagy G, Varoqueaux F, Nehring RB, Brose N, Wilson MC, Neher E. Differential control of the releasable vesicle pools by SNAP-25 splice variants and SNAP-23. Cell. 2003;114:75–86. doi: 10.1016/s0092-8674(03)00477-x. [DOI] [PubMed] [Google Scholar]
- 120.Bark C, Bellinger FP, Kaushal A, Mathews JR, Partridge LD, Wilson MC. Developmentally regulated switch in alternatively spliced SNAP-25 isoforms alters facilitation of synaptic transmission. J Neurosci. 2004;24:8796–8805. doi: 10.1523/JNEUROSCI.1940-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nakanishi S, Nakajima Y, Masu M, Ueda Y, Nakahara K, Watanabe D, Yamaguchi S, Kawabata S, Okada M. Glutamate receptors: brain function and signal transduction. Brain Res Brain Res Rev. 1998;26:230–235. doi: 10.1016/s0165-0173(97)00033-7. [DOI] [PubMed] [Google Scholar]
- 122.Salkoff L, Butler A, Ferreira G, Santi C, Wei A. High-conductance potassium channels of the SLO family. Nat Rev Neurosci. 2006;7:921–931. doi: 10.1038/nrn1992. [DOI] [PubMed] [Google Scholar]
- 123.Jurkat-Rott K, Lehmann-Horn F. The impact of splice isoforms on voltage-gated calcium channel alpha1 subunits. J Physiol. 2004;554:609–619. doi: 10.1113/jphysiol.2003.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Strehler EE, Treiman M. Calcium pumps of plasma membrane and cell interior. Curr Mol Med. 2004;4:323–335. doi: 10.2174/1566524043360735. [DOI] [PubMed] [Google Scholar]
- 125.Strehler EE, Zacharias DA. Role of alternative splicing in generating isoform diversity among plasma membrane calcium pumps. Physiol Rev. 2001;81:21–50. doi: 10.1152/physrev.2001.81.1.21. [DOI] [PubMed] [Google Scholar]
- 126.Garcia ML, Strehler EE. Plasma membrane calcium ATPases as critical regulators of calcium homeostasis during neuronal cell function. Front Biosci. 1999;4:D869–D882. doi: 10.2741/garcia. [DOI] [PubMed] [Google Scholar]
- 127.Chicka MC, Strehler EE. Alternative splicing of the first intracellular loop of plasma membrane Ca2+-ATPase isoform 2 alters its membrane targeting. J Biol Chem. 2003;278:18464–18470. doi: 10.1074/jbc.M301482200. [DOI] [PubMed] [Google Scholar]
- 128.Hill JK, Williams DE, LeMasurier M, Dumont RA, Strehler EE, Gillespie PG. Splice-site A choice targets plasma-membrane Ca2+-ATPase isoform 2 to hair bundles. J Neurosci. 2006;26:6172–6180. doi: 10.1523/JNEUROSCI.0447-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Keeton TP, Burk SE, Shull GE. Alternative splicing of exons encoding the calmodulin-binding domains and C termini of plasma membrane Ca(2+)-ATPase isoforms 1, 2, 3, and 4. J Biol Chem. 1993;268:2740–2748. [PubMed] [Google Scholar]
- 130.Keeton TP, Shull GE. Primary structure of rat plasma membrane Ca(2+)-ATPase isoform 4 and analysis of alternative splicing patterns at splice site A. Biochem J. 1995;306(Pt 3):779–785. doi: 10.1042/bj3060779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nucifora FC, Jr, Li SH, Danoff S, Ullrich A, Ross CA. Molecular cloning of a cDNA for the human inositol 1,4,5-trisphosphate receptor type 1, and the identification of a third alternatively spliced variant. Brain Res Mol Brain Res. 1995;32:291–296. doi: 10.1016/0169-328x(95)00089-b. [DOI] [PubMed] [Google Scholar]
- 132.Nakagawa T, Okano H, Furuichi T, Aruga J, Mikoshiba K. The subtypes of the mouse inositol 1,4,5-trisphosphate receptor are expressed in a tissue-specific and developmentally specific manner. Proc Natl Acad Sci U S A. 1991;88:6244–6248. doi: 10.1073/pnas.88.14.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Deshmukh M, Vasilakos J, Deckwerth TL, Lampe PA, Shivers BD, Johnson EM., Jr Genetic and metabolic status of NGF-deprived sympathetic neurons saved by an inhibitor of ICE family proteases. J Cell Biol. 1996;135:1341–1354. doi: 10.1083/jcb.135.5.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stoilov P, Daoud R, Nayler O, Stamm S. Human tra2-beta1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum Mol Genet. 2004;13:509–524. doi: 10.1093/hmg/ddh051. [DOI] [PubMed] [Google Scholar]
- 135.Jiang Z, Tang H, Havlioglu N, Zhang X, Stamm S, Yan R, Wu JY. Mutations in tau gene exon 10 associated with FTDP-17 alter the activity of an exonic splicing enhancer to interact with Tra2 beta. J Biol Chem. 2003;278:18997–19007. doi: 10.1074/jbc.M301800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dickinson LA, Edgar AJ, Ehley J, Gottesfeld JM. Cyclin L is an RS domain protein involved in pre-mRNA splicing. J Biol Chem. 2002;277:25465–25473. doi: 10.1074/jbc.M202266200. [DOI] [PubMed] [Google Scholar]
- 137.Goellner GM, DeMarco SJ, Strehler EE. Characterization of PISP, a novel single-PDZ protein that binds to all plasma membrane Ca2+-ATPase b-splice variants. Ann N Y Acad Sci. 2003;986:461–471. doi: 10.1111/j.1749-6632.2003.tb07230.x. [DOI] [PubMed] [Google Scholar]
- 138.Jiang ZH, Wu JY. Alternative splicing and programmed cell death. Proc Soc Exp Biol Med. 1999;220:64–72. doi: 10.1046/j.1525-1373.1999.d01-11.x. [DOI] [PubMed] [Google Scholar]
- 139.Soong TW, DeMaria CD, Alvania RS, Zweifel LS, Liang MC, Mittman S, Agnew WS, Yue DT. Systematic identification of splice variants in human P/Q-type channel alpha1(2.1) subunits: implications for current density and Ca2+-dependent inactivation. J Neurosci. 2002;22:10142–10152. doi: 10.1523/JNEUROSCI.22-23-10142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ehlers MD, Tingley WG, Huganir RL. Regulated subcellular distribution of the NR1 subunit of the NMDA receptor. Science. 1995;269:1734–1737. doi: 10.1126/science.7569904. [DOI] [PubMed] [Google Scholar]
- 141.Okabe S, Miwa A, Okado H. Alternative splicing of the C-terminal domain regulates cell surface expression of the NMDA receptor NR1 subunit. J Neurosci. 1999;19:7781–7792. doi: 10.1523/JNEUROSCI.19-18-07781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tian L, Duncan RR, Hammond MS, Coghill LS, Wen H, Rusinova R, Clark AG, Levitan IB, Shipston MJ. Alternative splicing switches potassium channel sensitivity to protein phosphorylation. J Biol Chem. 2001;276:7717–7720. doi: 10.1074/jbc.C000741200. [DOI] [PubMed] [Google Scholar]
- 143.Tian L, Hammond MS, Florance H, Antoni FA, Shipston MJ. Alternative splicing determines sensitivity of murine calcium-activated potassium channels to glucocorticoids. J Physiol. 2001;537:57–68. doi: 10.1111/j.1469-7793.2001.0057k.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wagner LE, II, Betzenhauser MJ, Yule DI. ATP binding to a unique site in the type-1 S2-inositol 1,4,5-trisphosphate receptor defines susceptibility to phosphorylation by protein kinase A. J Biol Chem. 2006;281:17410–17419. doi: 10.1074/jbc.M601340200. [DOI] [PubMed] [Google Scholar]
- 145.Wang ZW, Saifee O, Nonet ML, Salkoff L. SLO-1 potassium channels control quantal content of neurotransmitter release at the C. elegans neuromuscular junction. Neuron. 2001;32:867–881. doi: 10.1016/s0896-6273(01)00522-0. [DOI] [PubMed] [Google Scholar]
- 146.Sausbier M, Arntz C, Bucurenciu I, Zhao H, Zhou XB, Sausbier U, Feil S, Kamm S, Essin K, Sailer CA, Abdullah U, Krippeit-Drews P, Feil R, Hofmann F, Knaus HG, Kenyon C, Shipston MJ, Storm JF, Neuhuber W, Korth M, Schubert R, Gollasch M, Ruth P. Elevated blood pressure linked to primary hyperaldosteronism and impaired vasodilation in BK channel-deficient mice. Circulation. 2005;112:60–68. doi: 10.1161/01.CIR.0000156448.74296.FE. [DOI] [PubMed] [Google Scholar]
- 147.Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, McIntire SL. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- 148.Liu J, Asuncion-Chin M, Liu P, Dopico AM. CaM kinase II phosphorylation of slo Thr107 regulates activity and ethanol responses of BK channels. Nat Neurosci. 2006;9:41–49. doi: 10.1038/nn1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Crowder CM. Ethanol targets: a BK channel cocktail in C. elegans. Trends Neurosci. 2004;27:579–582. doi: 10.1016/j.tins.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 150.Brenner R, Perez GJ, Bonev AD, Eckman DM, Kosek JC, Wiler SW, Patterson AJ, Nelson MT, Aldrich RW. Vasoregulation by the beta1 subunit of the calcium-activated potassium channel (Comment In: Nature. 2000 Oct 19;407(6806):845, 847–8 UI: 20509330) Nature. 2000;407:870–876. doi: 10.1038/35038011. [DOI] [PubMed] [Google Scholar]
- 151.Fettiplace R, Fuchs PA. Mechanisms of hair cell tuning. Annu Rev Physiol. 1999;61:809–834. doi: 10.1146/annurev.physiol.61.1.809. [DOI] [PubMed] [Google Scholar]
- 152.Wei AD, Solaro C, Lingle C, Salkoff L. Calcium sensitivity of BK-type KCa channels determined by a separable domain. Neuron. 1994;13:671–681. doi: 10.1016/0896-6273(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 153.Jones EMC, Gray-Keller M, Fettiplace R. The role of Ca2+-activated K+ channel spliced variants in the tonotopic organization of the turtle cochlea. J Physiol (Cambridge) 1999;518:653–665. doi: 10.1111/j.1469-7793.1999.0653p.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Erxleben C, Everhart AL, Romeo C, Florance H, Bauer MB, Alcorta DA, Rossie S, Shipston MJ, Armstrong DL. Interacting effects of N-terminal variation and strex exon splicing on slo potassium channel regulation by calcium, phosphorylation, and oxidation. J Biol Chem. 2002;277:27045–27052. doi: 10.1074/jbc.M203087200. [DOI] [PubMed] [Google Scholar]
- 155.Lai G-J, McCobb DP. Opposing actions of adrenal androgens and glucocorticoids on alternative splicing of Slo potassium channels in bovine chromaffin cells. Proc Natl Acad Sci U S A. 2002;99:7722–7727. doi: 10.1073/pnas.112619799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McCobb DP, Hara Y, Lai GJ, Mahmoud SF, Flugge G. Subordination stress alters alternative splicing of the Slo gene in tree shrew adrenals. Horm Behav. 2003;43:180–186. doi: 10.1016/s0018-506x(02)00010-7. [DOI] [PubMed] [Google Scholar]
- 157.Zhu N, Eghbali M, Helguera G, Song M, Stefani E, Toro L. Alternative splicing of Slo channel gene programmed by estrogen, progesterone and pregnancy. FEBS Lett. 2005;579:4856–4860. doi: 10.1016/j.febslet.2005.07.069. [DOI] [PubMed] [Google Scholar]
- 158.Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science (Washington D C) 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- 159.Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 160.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 161.Kishore S, Stamm S. Regulation of alternative splicing by snoRNAs. Cold Spring Harbor Symp Quant Biol. 2006;71:329–334. doi: 10.1101/sqb.2006.71.024. [DOI] [PubMed] [Google Scholar]
- 162.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Metheny LJ, Skuse GR. NF1 mRNA isoform expression in PC12 cells: modulation by extrinsic factors. Exp Cell Res. 1996;228:44–49. doi: 10.1006/excr.1996.0297. [DOI] [PubMed] [Google Scholar]
- 164.Lynch KW, Weiss A. A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Mol Cell Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen HH, Wang YC, Fann MJ. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol Cell Biol. 2006;26:2736–2745. doi: 10.1128/MCB.26.7.2736-2745.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Beil B, Screaton G, Stamm S. Molecular cloning of htra2-beta-1 and htra2-beta-2, two human homologs of tra-2 generated by alternative splicing. DNA Cell Biol. 1997;16:679–690. doi: 10.1089/dna.1997.16.679. [DOI] [PubMed] [Google Scholar]
- 167.van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Caceres JF. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Allemand E, Guil S, Myers M, Moscat J, Caceres JF, Krainer AR. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc Natl Acad Sci U S A. 2005;102:3605–3610. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc Natl Acad Sci U S A. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ma S, Liu G, Sun Y, Xie J. Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochim Biophys Acta-Mol Cell Res. 2007;1773:912–923. doi: 10.1016/j.bbamcr.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 171.Misteli T. RNA splicing: what has phosphorylation got to do with it? Curr Biol. 1999;9:R198–R200. doi: 10.1016/s0960-9822(99)80128-6. [DOI] [PubMed] [Google Scholar]
- 172.Stojdl DF, Bell JC. SR protein kinases: the splice of life. Biochem Cell Biol. 1999;77:293–298. [PubMed] [Google Scholar]
- 173.Chalfant CE, Rathman K, Pinkerman RL, Wood RE, Obeid LM, Ogretmen B, Hannun YA. De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. J Biol Chem. 2002;277:12587–12595. doi: 10.1074/jbc.M112010200. [DOI] [PubMed] [Google Scholar]
- 174.Misteli T, Spector DL. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol Biol Cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hirano K, Erdodi F, Patton JG, Hartshorne DJ. Interaction of protein phosphatase type 1 with a splicing factor. FEBS Lett. 1996;389:191–194. doi: 10.1016/0014-5793(96)00577-7. [DOI] [PubMed] [Google Scholar]
- 176.Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, Burghes AH, Bollen M, Stamm S. Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing. Hum Mol Genet. 2007;17:52–70. doi: 10.1093/hmg/ddm284. [DOI] [PubMed] [Google Scholar]
- 177.Shi Y, Manley JL. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol Cell. 2007;28:79–90. doi: 10.1016/j.molcel.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 178.Shi Y, Reddy B, Manley JL. PP1/PP2A phosphatases are required for the second step of pre-mRNA splicing and target specific snRNP proteins. Mol Cell. 2006;23:819–829. doi: 10.1016/j.molcel.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 179.Boudrez A, Beullens M, Groenen P, Van Eynde A, Vulsteke V, Jagiello I, Murray M, Krainer AR, Stalmans W, Bollen M. NIPP1-mediated interaction of protein phosphatase-1 with CDC5L, a regulator of pre-mRNA splicing and mitotic entry. J Biol Chem. 2000;275:25411–25417. doi: 10.1074/jbc.M001676200. [DOI] [PubMed] [Google Scholar]
- 180.Guil S, Caceres JF. Stressful splicing. Mol Cell. 2007;28:180–181. doi: 10.1016/j.molcel.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 181.Murray MV, Kobayashi R, Krainer AR. The type 2C Ser/Thr phosphatase PP2Cgamma is a pre-mRNA splicing factor. Genes Dev. 1999;13:87–97. doi: 10.1101/gad.13.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Allemand E, Hastings ML, Murray MV, Myers MP, Krainer AR. Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat Struct Mol Biol. 2007;14:630–638. doi: 10.1038/nsmb1257. [DOI] [PubMed] [Google Scholar]
- 183.Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Licciardo P, Amente S, Ruggiero L, Monti M, Pucci P, Lania L, Majello B. The FCP1 phosphatase interacts with RNA polymerase II and with MEP50 a component of the methylosome complex involved in the assembly of snRNP. Nucleic Acids Res. 2003;31:999–1005. doi: 10.1093/nar/gkg197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tokumitsu H, Brickey DA, Glod J, Hidaka H, Sikela J, Soderling TR. Activation mechanisms for Ca-2+/calmodulin-dependent protein kinase IV: identification of brain CaM-kinase IV kinase. J Biol Chem. 1994;269:28640–28647. [PubMed] [Google Scholar]
- 186.Westphal RS, Anderson KA, Means AR, Wadzinski BE. A signaling complex of Ca2+-calmodulin-dependent protein kinase IV and protein phosphatase 2A. Science (Washington D C) 1998;280:1258–1261. doi: 10.1126/science.280.5367.1258. [DOI] [PubMed] [Google Scholar]
- 187.Anderson KA, Noeldner PK, Reece K, Wadzinski BE, Means AR. Regulation and function of the calcium/calmodulin-dependent protein kinase IV/protein serine/threonine phosphatase 2A signaling complex. J Biol Chem. 2004;279:31708–31716. doi: 10.1074/jbc.M404523200. [DOI] [PubMed] [Google Scholar]
- 188.Yun CY, Fu XD. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J Cell Biol. 2000;150:707–718. doi: 10.1083/jcb.150.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xu YX, Hirose Y, Zhou XZ, Lu KP, Manley JL. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003;17:2765–2776. doi: 10.1101/gad.1135503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bosser R, Faura M, Serratosa J, Renau-Piqueras J, Pruschy M, Bachs O. Phosphorylation of rat liver heterogeneous nuclear ribonucleo-proteins A2 and C can be modulated by calmodulin. Mol Cell Biol. 1995;15:661–670. doi: 10.1128/mcb.15.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Agell N, Aligue R, Alemany V, Castro A, Jaime M, Pujol MJ, Rius E, Serratosa J, Taules M, Bachs O. New nuclear functions for calmodulin. Cell Calcium. 1998;23:115–121. doi: 10.1016/s0143-4160(98)90109-9. [DOI] [PubMed] [Google Scholar]
- 192.Delaney KJ, Xu R, Zhang J, Li QQ, Yun KY, Falcone DL, Hunt AG. Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 2006;140:1507–1521. doi: 10.1104/pp.105.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 194.Chandler VL, Maler BA, Yamamoto KR. DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell. 1983;33:489–499. doi: 10.1016/0092-8674(83)90430-0. [DOI] [PubMed] [Google Scholar]
- 195.Karin M, Haslinger A, Holtgreve H, Richards RI, Krauter P, Westphal HM, Beato M. Characterization of DNA sequences through which cadmium and glucocorticoid hormones induce human metallothionein-IIA gene. Nature. 1984;308:513–519. doi: 10.1038/308513a0. [DOI] [PubMed] [Google Scholar]
- 196.Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: A Ca-2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- 197.Marshall J, Dolan BM, Garcia EP, Sathe S, Tang X, Mao Z, Blair LA. Calcium channel and NMDA receptor activities differentially regulate nuclear C/EBPbeta levels to control neuronal survival. Neuron. 2003;39:625–639. doi: 10.1016/s0896-6273(03)00496-3. [DOI] [PubMed] [Google Scholar]
- 198.Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, Wayman GA, Storm DR, Soderling TR, Goodman RH. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 199.Deisseroth K, Heist EK, Tsien RW. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature (London) 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- 200.Gray AC, Raingo J, Lipscombe D. Neuronal calcium channels: splicing for optimal performance. Cell Calcium. 2007;42:409–417. doi: 10.1016/j.ceca.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tian L, Coghill LS, McClafferty H, MacDonald SH, Antoni FA, Ruth P, Knaus HG, Shipston MJ. Distinct stoichiometry of BKCa channel tetramer phosphorylation specifies channel activation and inhibition by cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 2004;101:11897–11902. doi: 10.1073/pnas.0402590101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 203.Ahn S, Ginty DD, Linden DJ. A late phase of cerebellar long-term depression requires activation of CaMKIV and CREB. Neuron. 1999;23:559–568. doi: 10.1016/s0896-6273(00)80808-9. [DOI] [PubMed] [Google Scholar]
- 204.Tanaka H, Grooms SY, Bennett MV, Zukin RS. The AMPAR subunit GluR2: still front and center-stage. Brain Res. 2000;886:190–207. doi: 10.1016/s0006-8993(00)02951-6. [DOI] [PubMed] [Google Scholar]
- 205.Ghosh A, Greenberg ME. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 206.Kwak S, Weiss JH. Calcium-permeable AMPA channels in neurodegenerative disease and ischemia. Curr Opin Neurobiol. 2006;16:281–287. doi: 10.1016/j.conb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 207.See V, Boutillier A-L, Bito H, Loeffler J-P. Calcium/calmodulin-dependent protein kinase type IV (CaMKIV) inhibits apoptosis induced by potassium deprivation in cerebellar granule neurons. FASEB J. 2001;15:134–144. doi: 10.1096/fj.00-0106com. [DOI] [PubMed] [Google Scholar]
- 208.Wei F, Qiu CS, Liauw J, Robinson DA, Ho N, Chatila T, Zhuo M. Calcium calmodulin-dependent protein kinase IV is required for fear memory. Nat Neurosci. 2002;5:573–579. doi: 10.1038/nn0602-855. [DOI] [PubMed] [Google Scholar]
- 209.Kang H, Sun LD, Atkins CM, Soderling TR, Wilson MA, Tonegawa S. An important role of neural activity-dependent CaMKIV signaling in the consolidation of long-term memory. Cell. 2001;106:771–783. doi: 10.1016/s0092-8674(01)00497-4. [DOI] [PubMed] [Google Scholar]
- 210.Blaeser F, Sanders MJ, Truong N, Ko S, Wu LJ, Wozniak DF, Fanselow MS, Zhuo M, Chatila TA. Long-term memory deficits in Pavlovian fear conditioning in Ca2+/calmodulin kinase kinase alpha-deficient mice. Mol Cell Biol. 2006;26:9105–9115. doi: 10.1128/MCB.01452-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ko SW, Jia Y, Xu H, Yim SJ, Jang DH, Lee YS, Zhao MG, Toyoda H, Wu LJ, Chatila T, Kaang BK, Zhuo M. Evidence for a role of CaMKIV in the development of opioid analgesic tolerance. Eur J Neurosci. 2006;23:2158–2168. doi: 10.1111/j.1460-9568.2006.04748.x. [DOI] [PubMed] [Google Scholar]
- 212.Mu Y, Otsuka T, Horton AC, Scott DB, Ehlers MD. Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron. 2003;40:581–594. doi: 10.1016/s0896-6273(03)00676-7. [DOI] [PubMed] [Google Scholar]
- 213.Jiang ZH, Zhang WJ, Rao Y, Wu JY. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc Natl Acad Sci U S A. 1998;95:9155–9160. doi: 10.1073/pnas.95.16.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 215.Park JW, Graveley BR. Use of RNA interference to dissect the roles of trans-acting factors in alternative pre-mRNA splicing. Methods. 2005;37:341–344. doi: 10.1016/j.ymeth.2005.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci U S A. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Paddison PJ, Caudy AA, Sachidanandam R, Hannon GJ. Short hairpin activated gene silencing in mammalian cells. Methods Mol Biol. 2004;265:85–100. doi: 10.1385/1-59259-775-0:085. [DOI] [PubMed] [Google Scholar]
- 218.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 219.Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O’Shaughnessy A, Gnoj L, Scobie K, Chang K, Westbrook T, Cleary M, Sachidanandam R, McCombie WR, Elledge SJ, Hannon GJ. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 220.Ito M, Kawano K, Miyagishi M, Taira K. Genome-wide application of RNAi to the discovery of potential drug targets. FEBS Lett. 2005;579:5988–5995. doi: 10.1016/j.febslet.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 221.Boiko AD, Porteous S, Razorenova OV, Krivokrysenko VI, Williams BR, Gudkov AV. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 2006;20:236–252. doi: 10.1101/gad.1372606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 223.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 224.Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, Davies C, Williams A, Turpaz Y. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]