Abstract
Resistance to platinum-based therapies arises by multiple mechanisms, including by alterations to cell-cycle kinases that mediate G2/M phase arrest. In this study, we conducted parallel high-throughput screens for microRNAs (miRNAs) that could restore sensitivity to cisplatin-resistant cells, and we screened for kinases targeted by miRNAs that mediated cisplatin resistance. Overexpression of the cell cycle kinases WEE1 and CHK1 occurred commonly in cisplatin-resistant cells. miRNAs in the miR-15/16/195/424/497 family were found to sensitize cisplatin-resistant cells to apoptosis by targeting WEE1 and CHK1. Loss-of-function and gain-of function studies demonstrated that miR-15 family members controlled the expression of WEE1 and CHK1. Supporting these results, we found that in the presence of cisplatin altering expression of miR-16 or related genes altered cell cycle distribution. Our findings reveal critical regulation of miRNAs and their cell cycle-associated kinase targets in mediating resistance to cisplatin.
Introduction
Cisplatin [cis-diamminedichloroplatinum(II)] and its analogs carboplatin and oxaliplatin are used to treat a range of malignancies including testicular, ovarian, head and neck, bladder, esophageal, and small cell lung cancer (1–4). Due to acquired or intrinsic resistance, tumors are not eliminated by treatment with the notable exception of testicular cancer (3–5). Cisplatin resistance studied in cultured cancer cells often results from a cellular defense mechanism that results in a highly complex pleiotropic phenotype that confers resistance by reducing apoptosis, up-regulating DNA damage repair mechanisms, altering cell cycle checkpoints, and disrupting assembly of the cytoskeleton (4). These alterations to the cytoskeleton disrupt cellular protein trafficking, and redirect transporters away from the cell surface. This results in cells that are not only permanently resistant to cisplatin but also show cross-resistance to other compounds that usually enter into cells via uptake transporters (such as methotrexate and toxic metals). The pleiotropic mechanisms underlying cisplatin resistance are well described, but poorly understood in their entirety and in terms of clinical significance (4, 6).
In cisplatin resistant (CP-r) cancer cells, cell cycle mechanisms allow the cell to compensate for drug toxicity (3, 6–8). When cancer cells are treated with a Pt-based chemotherapeutic, the drug forms Pt-DNA adducts that are generally believed to be responsible for cell death (4, 9). The lesion formed is recognized, activating DNA repair or apoptotic pathways (2, 7, 9). The classical cellular response to cisplatin is G2 arrest, allowing the cell to repair critical DNA damage before moving forward with mitosis. Cells that attempt to progress through G2 without sufficient repair undergo apoptosis (2, 9, 10). The length of time spent in G2 is therefore dependent on the extent of DNA platination (11). Entrance into G2 and the G2/M checkpoint is mediated by kinase-controlled signaling pathways (12). Multiple kinases have been shown to have an effect on sensitivity to chemotherapy, in part by regulating cell cycle checkpoints (13–18).
MicroRNAs (miRNAs) are small non-coding regulatory factors 18 to 25 nucleotides in length. miRNAs affect gene expression by complementary pairing with the 3′-untranslated region of mRNAs leading to translational repression, by either degradation or sequestration of mRNA. miRNAs have crucial roles in diverse biological processes such as cell differentiation and phenotypic stabilization, and have also been shown to be involved in tumor growth and response to chemotherapy (19–21). The extent of miRNA involvement in cisplatin resistance is not understood, but studies have begun to identify miRNA-mRNA targets involved in sensitizing or causing resistance, providing potential new targets and mechanisms as treatment options (21–26).
We hypothesized that miRNAs play a role in mediating cisplatin resistance through multiple pathways, including regulation of kinases critical to cell cycle control. In order to address this hypothesis, we designed a high-throughput screen with two purposes: 1) to identify miRNAs that confer sensitivity to cisplatin in resistant cells by mimicking miRNA, and 2) to identify kinases that increase the sensitivity of cisplatin-resistant cells when silenced. Kinase hits were validated by siRNA experiments, examined for expression in cell lines, and tested for involvement in CP-r with small molecule kinase inhibitors. miRNA hits from the screen that were predicted to target kinases identified by the screen were validated using loss-of-function/gain-of-function cell survival curves and Western blots to demonstrate a connection between miRNA activity and kinase regulation. We also tested our screen by selecting a miRNA that was not identified in our screen but was predicted to target the top kinase hit.
Materials and Methods
Cell lines and cell culture
The parental human epidermoid carcinoma cell line KB-3-1, and two cisplatin-resistant sub-lines, KB-CP.5 and KB-CP20 were employed for this study. KB-CP.5 cells were originally selected in a single step in 0.5 μg cisplatin/mL (1.6 μM) in our laboratory, as described previously (27, 28). KB-CP20 cells were selected by stepwise increases to 20 μg of cisplatin/mL of medium (66.7 μM), as described previously (29, 30). The CP-r cells were maintained in the presence of cisplatin, but it was removed from growth medium three days prior to all experiments. All cell lines were grown as monolayer cultures at 37°C in 5% CO2, using Dulbecco’s modified Eagle medium (DMEM) with 4.5 g/L glucose (Invitrogen, Carlsbad, CA), supplemented with L-glutamine, penicillin, streptomycin and 10% fetal bovine serum (BioWhittaker, Walkersville, MD). Cisplatin was purchased from Sigma (St. Louis, MO). Cross-resistance of cisplatin-resistant cells to cisplatin is confirmed on a regular (at least monthly) basis, using cell viability assays as described herein.
High-throughput screen
KB-CP.5 (resistant) cells were screened with both human kinome siRNA (Ambion SilencerSelect) and human miRNA mimic (Qiagen) libraries. The kinome library targets 704 genes with 3 siRNAs per gene arrayed in individual wells of 384 well plates (see Supplemental Table 2 for a full list of genes targeted). The miRNA mimic library is based on Sanger miRBase 13.0 and contains 875 mimics (Supplemental Table 3). For transfection, siRNA or miRNA mimic (0.8 pmol) was spotted to 384 well plates (Corning 3570) and 0.1 μL of Lipofectamine RNAiMax (Invitrogen) was then added in 20 μL of serum free media. This mixture was incubated at ambient temperature for 30 minutes prior to adding cells (1,200) in 20 μL of DMEM containing 20% FBS, yielding final transfection mixtures with 20 nM siRNA or miRNA mimic. Transfected cells were incubated for 24 h at 37 °C prior to addition of cisplatin or vehicle only (PBS). Cells were incubated with compound for 72 h prior to assaying cell growth (CellTiter Glo, Promega).
All screening plates had a full column (16 wells) of both negative (Ambion SilencerSelect Negative Control #2) and positive control (PLK1 siRNA, Ambion, s448, target sequence AACCAUUAACGAGCUGCUUAA) siRNAs. Positive control served to assess transfection efficiency, whereas the median value of each plate’s negative control column was used to normalize corresponding sample wells. For vehicle only plates, negative control wells received PBS alone. For compound treated plates, negative control wells received cisplatin (EC5 or EC30) in PBS. Candidate miRNAs were primarily selected by examining the ratio between the negative control normalized viability of each mimic in the presence and absence of cisplatin. Mimics ranking highest in terms of these ratios at both doses of cisplatin (>2.5 standard deviations from the median ratio value, ≈ among the top 3%) were prioritized for follow-up (12 mimics). A similar approach was used to select the top kinases, focusing on those with multiple siRNAs among the top actives at both doses (>2.5 standard deviations from the median, ≈ among the top 3%). Genes selected at both doses included ATR, CHEK1, and WEE1. Six additional siRNA sequences (Qiagen) were tested for each of the top genes in the context of a cisplatin dose response (1000 μM to 0 μM) (details of siRNAs in Supplemental Materials and Methods). Knock-down by additional siRNAs was confirmed by RT-PCR.
Small molecule kinase inhibitors
Cells were treated with the following kinase inhibitors: SB 218070 against Chk1, MK 1775 against Wee1, and PD 407824 against Wee1 and Chk1 (Tocris Bioscience, Minneapolis, MN).
MTT assay
Cell survival was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells were seeded at a density of 5,000 cells per well in 96-well plates and incubated at 37°C in humidified 5% CO2 for 24 h. Serially diluted cisplatin was added to give the intended final concentrations. Cells were then incubated an additional 72 h, and the MTT assay was performed according to the manufacturer’s instructions (Molecular Probes, Eugene, OR). Absorbance values were determined at 570 nm on a Spectra Max 250 spectrophotometer (Molecular Devices, Sunnyvale, CA). All MTT assays were performed three times in triplicate. The 50% inhibitory concentration (IC50) values were defined as the drug concentrations required to reduce cellular proliferation to 50% of the untreated control well and are reported here as such.
Flow cytometry for apoptosis and cell cycle analysis
KB-3-1 and KB-CP.5 cells were transfected with siNegative control, miR-16 mimic, or inhibitor. After 24 h, KB-3-1 cells were treated with 5 μM cisplatin and KB-CP.5 cells with 25 μM cisplatin. Cells were harvested in PBS 72 h post cisplatin treatment. For evaluation of apoptosis vs. viable populations, cells were washed with PBS and processed with Alexa Flour 488 annexin V/Dead Cell Apoptosis Kit with Alexa Flour 488 annexin V and PI for flow cytometry, as described by the manufacturer (Invitrogen, Eugene, OR). Flow cytometry analysis was performed with FlowJo v.7.6.4. For cell cycle analysis, cells were washed with PBS and fixed overnight in ice-cold 70% ethanol and stored at 4°C. After 24 h, cells were centrifuged and re-suspended in 100 U RNAse (Sigma-Aldrich, St. Louis, MO), and incubated at 37°C for 20 min. Propidium iodide (Invitrogen) solution (500 μL, 50 ug/mL in DPBS) was added to each tube and incubated in the dark at 4°C overnight. Flow cytometry was performed and analysis was completed with ModFit LT. All data is in triplicate and presented as a percent mean +/− SD.
RNA extraction, cDNA preparation, and quantitative real-time (RT)-PCR
Cisplatin was removed 72 h prior to experiments. RNA was harvested from KB-3-1 and KB-CP.5 cells using the mirVana miRNA Isolation Kit (Ambion, Carlsbad, CA). RNA integrity and concentration were measured using an NP-1000 Spectrometer to measure absorbance (Thermo Scientific, Wilmington, DE). cDNA was prepared from 20ng total RNA following the manufacturer’s instructions. RT-PCR was run using Exiqon probes on a 7500 fast real-time PCR system (Applied Biosystems) (details of probes in Supplemental Materials and Methods). The miRNA data was normalized to U6 RNA. Expression level was calculated by the ΔΔCt method.
Target prediction
miRNAs predicted to target kinases identified by the high throughput screen were identified by using miRanda (http://www.microrna.org/microrna/home.do) and TargetScanHuman (http://www.targetscan.org/) algorithms.
Transfection of miRNA mimics and inhibitors
Transfection was performed with RNAiMax (Invitrogen, Carlsbad, CA) (2.5 μL/mL). The pre-designed miRIDIAN miRNA mimics and inhibitors (details of inhibitors in Supplemental Experimental Procedures), and AllStars Negative Control (mock) (Qiagen) were plated with cells on day 1 at 10 nM. The transfection was performed as described by the manufacturer.
Preparation of cell lysates, quantification of protein, and Western blot analysis
Cell samples were obtained by trypsin digestion followed by two washes in PBS. Protein was extracted using Cell Extraction Buffer (Invitrogen) with 50 μL/mL Protease Inhibitor Cocktail (Sigma). Protein concentrations were determined using Bio-Rad Protein Assay based on the Bradford method as per the manufacturer’s instructions. Fifty-μg protein samples were loaded onto a NuPage 3–8% TA gel (Invitrogen) and run at 125 V. Transfer to nitrocellulose membranes was performed using an iBlot Gel Transfer Device (Invitrogen) at 20 V for 7 min. Membranes were blocked in 5% non-fat milk for 1 h, exposed to primary antibody overnight at 4 °C, and HRP-tagged secondary antibody for 1 h at room temperature (antibodies and conditions listed in Supplemental Experimental Conditions). Blots were incubated with Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA) and developed on Amersham Hyperfilm ECL (GE Healthcare, Buckinghamshire, UK).
Statistical Analysis
We used GraphPad Prism 5 software for graphs and statistics. All data are expressed as mean ± SD. Comparisons among groups were analyzed using Analysis of Variance (ANOVA). Comparisons between different groups were analyzed using Two-way ANOVA. P values <0.05 were considered significant differences.
Results
A high-throughput screen identifies miRNA and kinases that can sensitize cell lines to cisplatin
We developed a high-throughput screen (HTS) to identify miRNAs that when mimicked (increasing abundance) could sensitize CP-r KB-CP.5 cells to cisplatin (Supplementary Fig. S1). KB-CP.5 cells were screened with miRNA mimics (Sanger miRBase 13.0), and then challenged with cisplatin at IC5 (5 μM) and IC30 (25 μM) doses. After 72 h, cell viability was determined using a Promega CellTiter-Glo luminescent assay that reports on cell survival based on cellular ATP levels in each well. Conditions that sensitized the resistant cells (indicated by low cell number compared with cells transfected with negative control siRNA) were examined (Supplementary Table S1, available online). Twelve miRNA mimics were shared among the top hits for both the EC5 and EC30 doses, inducing > 2-fold change at the EC30 dose (Table 1, Supplementary Table S3). Five of these mimics belong to the miR-15/16/195/424/497 family. This family shares an identical seed sequence (a seven nucleotide sequence that complements and selects target mRNA for sequestering or degradation). miR-16-1* and miR-424* are part of the family but are matured from the 3′ strand of the hairpin loop of the pre-miRNA and therefore are a perfect complement to the family seed sequence. miR-16 transfection conferred a 5-fold increase in cisplatin sensitivity, and its family member miR-15a had a 3.4-fold increase providing two of the top three hits in the primary screen.
Table 1.
miRNA mimics found to sensitize cisplatin-resistant CP.5 cells to cisplatin in the high-throughput screen
| % Proliferation, miRNA mimic | % Proliferation, mimic + IC30 cisplatin | Fold change in % cell proliferation | |
|---|---|---|---|
| miR-16 | 71.3 | 14 | 5.1 |
| miR-503 | 82.9 | 23.7 | 3.5 |
| miR-15a | 69.5 | 20.3 | 3.4 |
| miR-29b-2* | 80.3 | 24.8 | 3.2 |
| miR-634 | 100 | 31.2 | 3.2 |
| miR-17 | 98.5 | 31.6 | 3.1 |
| miR-424* | 79.5 | 25.6 | 3.1 |
| miR-1262 | 67.1 | 24.5 | 2.7 |
| miR-16-1* | 104.1 | 43.8 | 2.4 |
| miR-15b | 101.7 | 43.3 | 2.4 |
| miR-29b | 112.3 | 51.4 | 2.2 |
| miR-101 | 102.6 | 48.7 | 2.1 |
miRNAs were considered to be hits if they were active at both concentrations of cisplatin (IC30 and IC5 doses). Active miRNA mimics identified are ranked in order of the fold-change sensitization. miR-16, miR-15a, miR-424*, miR-16-1*, and miR-15b all belong to the same family.
As part of the HTS, in parallel we evaluated a kinase library in order to determine which kinases had a significant effect on cisplatin resistance in our cell lines. Here, we screened for kinases that, when silenced, conferred sensitivity to CP-r cells. To accomplish this, the kinome siRNA (Ambion SilencerSelect, 704 genes, 3 siRNAs per gene) library was run under the same conditions as those used for the miRNA mimics (CP.5 cells with two low toxicity cisplatin doses, IC5 and IC30), with three siRNAs being tested for each kinase. Selected candidates had at least 2 of 3 siRNAs scoring at the top in terms of sensitizing CP-r cells to cisplatin at both doses (Table 2, Supplementary Table S4). The three strongest candidates were WEE1, ATR and CHEK1.
Table 2.
Kinase siRNAs found to sensitize cisplatin-resistant CP.5 cells to cisplatin
| siRNA target | % Proliferation, siRNA | % Proliferation, siRNA + IC30 cisplatin | Fold change in % survival | Predicted targets of miRNA from screen |
|---|---|---|---|---|
| WEE1 | 69.4 | 13.7 | 5.1 | miR-15a/15b/16/16-1*/424*, miR-17, miR-503 |
| ATR | 109.6 | 28.4 | 3.9 | None |
| CHEK1 | 75.6 | 19.9 | 3.8 | miR-15a/15b/16/16-1*/424*, miR-503 |
Silenced genes were considered significant if more than 1 siRNA against a given gene significantly sensitized CP.5 cells to cisplatin. Cell growth (proliferation) was determined as the percentage of siRNA-treated surviving cells compared with siNegative cells. The listed values correspond to the highest fold change observed for a given gene. miRNA hits predicted to target kinases using the TargetScanHuman and miRanda programs are listed.
ATR, WEE1, and CHEK1 expression and function confer resistance to cisplatin while interference by RNAi or small molecule inhibitors increases sensitivity
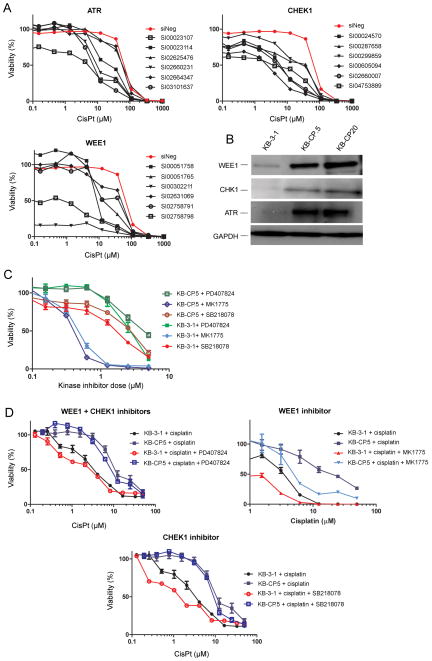
In order to confirm that the top three kinase hits from the screen affect cisplatin resistance, an additional 6 siRNAs for each gene were used in a follow-up dose-response validation (Fig. 1). Four siRNAs against ATR accorded a 3 to 10-fold increase in sensitivity to cisplatin. Six siRNAs against CHEK1 and six against WEE1 increased sensitivity 2 to 14-fold, indicating that these kinases play an important role in inhibiting cell killing by cisplatin (Fig. 1A). Knock-down of additional siRNAs was confirmed by RT-PCR (Supplementary Figure S4).
Figure 1. Validation of high-throughput kinase hits.
A, Follow-up dose response analyses using additional siRNAs confirms sensitization mediated by the knockdown of ATR, CHK1, and WEE1 in KB-CP.5 cells compared to siNegative treated cells. B, Western blots showing normal expression of kinases. ATR, WEE1, and CHK1 have increased expression in resistant lines (KB-CP.5 and KB-CP20) compared to sensitive (KB-3-1). The increase in expression is dependent on the level of resistance. C, Small molecule kinase inhibitors PD 407824 (CHK1 and WEE1), MK 1775 (WEE1), and SB 218078 (CHK1) have concentration-dependent toxic effect on sensitive and resistant cell lines. D, Cells were treated with sub-toxic doses of kinase inhibitors (10nM) and challenged with cisplatin (50 μL to 0 μL). KB-3-1 sensitive cells had a 1.2 – 3.9 fold decrease in survival in cisplatin when the kinase function was inhibited with small molecule inhibitors. KB-CP.5 resistant cells showed less of an effect at 1.2 to 1.9 fold increase in sensitivity to cisplatin. CisPt = cisplatin.
To confirm that ATR, WEE1, and CHK1 are involved in cisplatin resistance, we examined the expression of ATR, WEE1, and CHK1 protein in KB-3-1 (sensitive), KB-CP.5 (resistant), and KB-CP20 (highly resistant) cells (Fig. 1B). All three kinases showed an increase in expression in the CP-r lines compared to parental KB-3-1 cells. In addition, KB-CP20 cells showed an increase in expression compared to KB-CP.5, indicating that the expression of ATR, WEE1, and CHK1 correlate with the level of cisplatin resistance in each cell line, though whether the increased protein levels result in greater activity has not been determined. As ATR was not predicted as a target for miRNA identified in our screen, it was not studied further.
We sought to determine whether inhibiting WEE1 and CHK1 kinase activity could sensitize resistant lines and modulate resistance. To this end, we evaluated the response of the KB-3-1 and KB-CP.5 lines to the following small molecule kinase inhibitors: MK 1775 (WEE1), PD 407824 (WEE1/CHK1), and SB 218078 (CHK1) (Fig. 1C). All inhibitors demonstrated cytotoxicity towards both the KB-3-1 and KB-CP.5 lines. MK 1775 (WEE1) provided the strongest effect, killing 50% of the cell population (IC50) at 0.4 ± 0.1 μM irrespective of cell line (Table 3). KB-CP.5 cells showed cross-resistance to SB 218078 (CHK1, KB-3-1 IC50 = 1.7 ± 0.2 μM, KB-CP.5 IC50 = 2.8 ± 0.2 μM) and PD 407824 (CHK1/WEE1, KB-3-1 IC50 = 2.7 ± 0.5 μM, KB-CP.5 IC50 = 4.4 ± 0.6 μM).
Table 3.
Cytotoxicity (IC50) values for small molecule kinase inhibitors (KIs) against KB-3-1 and KB-CP.5 cells, with and without cisplatin
| Cell type | Kinase inhibitor (KI) | KI IC50 (μM) | Subtoxic dose (nM) | Target | Cisplatin IC50(μM) | Cisplatin IC50(μM) + KI | Sensitization |
|---|---|---|---|---|---|---|---|
| KB-3-1 | PD 407824 | 2.7 ± 0.5 | 10 | Wee 1, Chek 1 | 5.1 ± 0.9 | 4.3 ± 0.6 | 1.2 |
| KB-CP.5 | PD 407824 | 4.4 ± 0.6 | 10 | Wee 1, Chek 1 | 12.6 ± 3.6 | 9.1 ± 0.1 | 1.4 |
| KB-3-1 | MK 1775 | 0.4 ± 0.1 | 100 | Wee 1 | 5.1 ± 0.9 | 0.8 ± 0.3 | 6.3 |
| KB-CP.5 | MK 1775 | 0.5 ± 0.1 | 100 | Wee 1 | 12.6 ± 3.6 | 5.1 ± 0.4 | 2.5 |
| KB-3-1 | SB 218078 | 1.7 ± 0.2 | 10 | Chek 1 | 5.1 ± 0.9 | 1.3 ± 0.01 | 3.9 |
| KB-CP.5 | SB 218078 | 2.8 ± 0.2 | 10 | Chek 1 | 12.6 ± 3.6 | 10.2 ± 0.5 | 1.2 |
Cells were treated with KIs alone in order to determine a dose shown to kill 50% of the cell population (IC50), and to identify sub-toxic doses. Cell lines were then treated with cisplatin alone or cisplatin with a subtoxic dose of KIs. Sensitization is calculated as the fold-difference between the cisplatin IC50 divided by KI treated cells. A value > 1 signifies an increase in sensitivity.
Next, KB-3-1 and resistant KB-CP.5 cells were examined to determine whether inhibiting WEE1 or CHK1 sensitized them to cisplatin treatment. This was achieved by treating cells with a sub-toxic dose of each kinase inhibitor; MK 1775 (Wee1, 100 nM), PD 407824 (WEE1/CHK1, 10 nM), and SB 218078 (CHK1, 10 nM). Kinase inhibitors were used at a subtoxic dose to ensure the assay identified cell death dependent on cisplatin and not the inhibitors themselves. Inhibiting the function of kinases produced an increased sensitivity to cisplatin, albeit to a lesser degree than occurred with the siRNA (Fig. 1D), with MK 1775 (WEE1) being the most effective. It may be that the kinase inhibitors did not fully inhibit kinases at sub-toxic doses, leading to the lesser effect compared with siRNA. Sensitive KB-3-1 cells showed a greater degree of sensitization than resistant KB-CP.5 cells (Table 3). This may be due to the lower expression of WEE1 and CHK1 in KB-3-1 cells, as there is a lower concentration of kinase to inhibit with an identical dose. We cannot rule out, however, that the CP-r phenotype restricts cell entry of the kinase inhibitors. These data indicate that resistance to cisplatin includes increased expression of kinases responsible for G2/M checkpoint regulation, and inhibiting these same kinases sensitizes CP-r cells. The CP-r phenotype confers cross-resistance to kinase inhibitors against WEE1 and CHK1.
CHK1 and WEE1 are predicted to be targets of miR-15a/15b/16/16-1*/424*
Using TargetScanHuman and miRanda, online algorithm programs for the prediction of miRNA targets, we found that the miR-15/16/195/424/497 family identified in the miRNA mimic screen is predicted to target WEE1 and CHEK1, two of the hits provided in the kinome RNAi screen (Table 2). This is consistent with our hypothesis that a miRNA will down-regulate expression of genes associated with cisplatin resistance, leading to loss of phenotype and increased sensitivity to cisplatin (Figure 1). We therefore hypothesize that miR-15a/15b/16/16-1*/424* regulation of CHEK1 and WEE1 is causative for cisplatin resistance. miR-155 has been shown previously to target WEE1 kinase (31), but it was not identified by our screen. In evaluating the connection between the screen hits, miR-155 was incorporated to give us insight into the selectivity of the miRNA screen.
miR-15a/15b/16/16-1*/424*and miR-155 is decreased in cisplatin-resistant cells
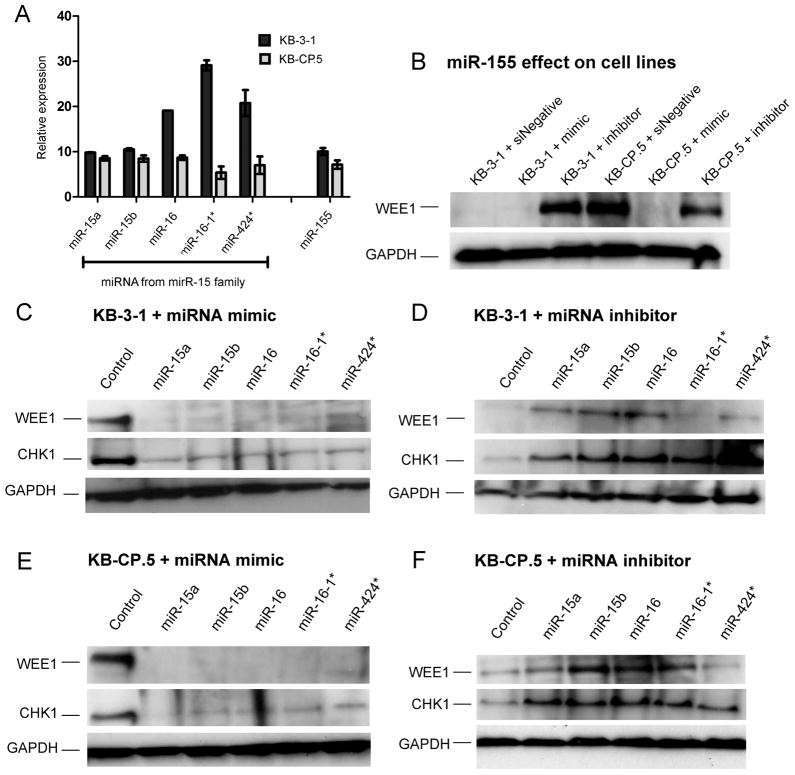
RT-PCR was used to determine the endogenous expression of miRNAs in KB-3-1 and KB-CP.5 cell lines (Figure 3A). ΔCT values were obtained using U6 snRNA as an internal control (32). miR-16-1* was the most abundant family member in KB-3-1 cells followed by miR-16 and miR-424*, with all miRNAs being expressed at detectable levels. Expression of all miRNA members showed statistically significant (p ≦ 0.05) decreased expression in KB-CP.5 cells compared to the parental KB-3-1 cells. miR-16-1* showed the greatest loss in expression in KB-CP.5 cells, with miR-15a and -15b the smallest change in expression. miR-155 also showed a decrease in expression in KB-CP.5 cells (p ≦ 0.05) comparable to miR-15a but less than that of family members miR-15b/16/16-1*/424*. This indicates that miR-15a/15b/16/16-1*/424* and miR-155 have a lower expression in KB-3-1 cells consistent with their increased resistance to cisplatin.
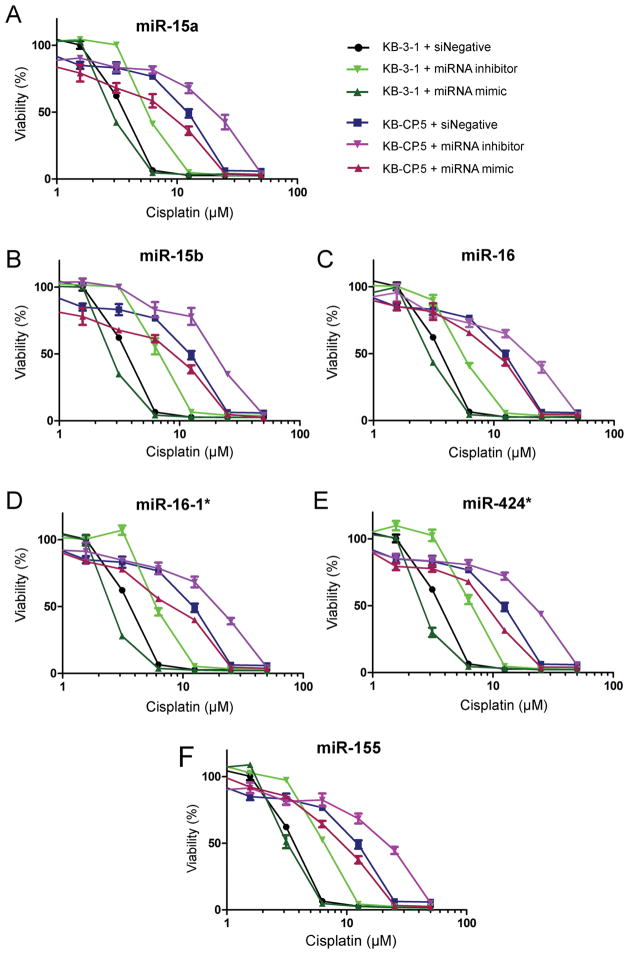
Figure 3. Response of KB cell lines to miRNA mimic/inhibitor data for the miR-15 family.
Cells were treated with a miRNA mimic or an miRNA inhibitor for each family member (miR-15a/15b/16/16-1*/424*) (10nM) and challenged with cisplatin (50 μM to 0 μM). miR-15a/15b/16/16-1*/424* all showed an increase in resistance when the miRNA was inhibited and an increase in sensitivity to cisplatin when the miRNA was increased.
miR-15a/15b/16/16-1*/424* target WEE1 and CHEK1 in KB-3-1 and KB-CP.5 cell lines
To determine whether manipulation of miRNA affects WEE1 and CHK1 levels, cells were treated with a miRNA mimic or miRNA inhibitor (siRNA directed against a miRNA precursor) for individual members of the miR-15a/15b/16/16-1*/424* family, and cell lysates were obtained at 48 h. Chk1 and Wee1 expression in response to an increase (mimic) or decrease (inhibitor) in miRNA were examined by Western blot (Fig. 2B–F). All miRNA family members elicited a decrease in WEE1 and CHK1 levels in response to miRNA mimics in both cell lines (Fig. 2C, E). KB-3-1 cells transfected with miRNA inhibitor showed an increase of CHK1 in all samples (Fig. 2D,F). miR-15a, miR-15b and miR-16 inhibitors transfected into KB-3-1 also increased expression of WEE1 (Fig. 2D). KB-CP.5 cells showed increased expression of WEE1 in response to miR-15a/15b/16/16-1* inhibitors, but did not show a marked response to miR-424* (Fig. 2F). All miRNA inhibitors increased the expression of CHK1 in KB-CP.5 cells (Fig. 2F).
Figure 2. Expression of miR-15 family members and their effect on expression of WEE1 and CHK1 kinases.
A, Expression of miR-15 family members and miR-155 in KB-3-1 and KB-CP.5, with RT-PCR. Samples were normalized to U6. Using ANOVA statistical analysis, columns comparing expression of one miRNA between KB-3-1 to KB-CP.5 have a p-value ≤ 0.05. Columns comparing different miRNAs to each other in the same cell line have a p-value of 0.0001. Western blots showing expression of WEE1 and CHK1 in response to transfection with miR-15a/15b/16/16-1*/424* miRNA mimics and miRNA. KB-3-1 cells transfected with mimics (B) and inhibitors (C), and KB-CP.5 cells transfected with mimics (D) and inhibitors (E) were collected and protein extracted after 72 h.
miRNA inhibitors confer a marked increase in cisplatin resistance
Loss-of-function/gain-of-function experiments were completed by transfecting sensitive and resistant cells with siNegative (control), miRNA mimics, or miRNA inhibitors. After 24 h all samples were challenged with cisplatin (dose gradient from 100 to 0 μM). miRNA loss of function by treatment with inhibitor against each miRNA for the miR-15a/15b/16/16-1*/424* family increased cellular resistance to cisplatin in KB-3-1 cells (Fig. 3, Supplementary Table S2). This was also observed for KB-CP.5 cells, although the low expression of miR-15a/15b/16/16-1*/424* in KB-CP.5 cells (Fig. 2A) means only minimal additional silencing of WEE1 and CHK1 is possible. Increasing the expression of miR-15a/15b/16/16-1*/424* decreases the expression of WEE1 and CHK1 and results in sensitivity to cisplatin. Cell survival curves demonstrate that all miRNA mimics for the miR-15a/15b/16/16-1*/424* family increase sensitivity to cisplatin for all data points. Transfection of miR-16-1* (Fig. 3D) and miR-424* (Fig. 3E) mimics conferred the greatest degree of sensitization. The effect of miRNA mimics and inhibitors was confirmed by RT-PCR (Supplementary Fig. S2), and no off-target effects were observed for other family members (Supplementary Fig. S3). Generally, miRNA inhibitors (siRNAs) lowered miRNA expression to undetectable levels, whereas the fold-increase in miRNA levels after transfection with miRNA mimic was not as strong. This difference accounts for the lesser effect of mimics compared with inhibitors.
The MTT assay acts as a measure of cellular proliferation, but does not give direct insight into whether cell death or arrest is at play in cells. This is partly because the MTT assay, as well as other analogous assays, provide insight into total cell number. The effect of this is that if cellular proliferation is arrested (which cisplatin is known to do), total cell count would be diminished without necessarily eliciting a high rate of cellular apoptosis. We adopted two experimental approaches to demonstrate that cisplatin’s effects on cell growth was related to loss of cell viability (apoptosis) and involved cell cycle arrest, as has been frequently characterized for cisplatin. For both approaches, we examined miR-16 as an exemplar of the family members identified in this report.
First, we assessed cellular apoptosis induced by cisplatin with either miR-16 inhibitor or mimic (with appropriate controls) to demonstrate whether apoptosis occurs. Cell death was assessed using the Annexin V apoptosis assay, which detects both phosphatidylserine levels on the external leaflet of the cell membrane (apoptotic cells, detected by Annexin V), and cell membrane leakage (permeabilization) indicated by propidium iodide entry (dead cells). We found that, compared with siNeg + cisplatin, both KB-3-1 and KB-CP.5 cell lines were sensitized to cisplatin in the presence of miR-16 mimic (increased apoptosis and death), and apoptosis and death were lowered when the cells were transfected with miR-16 inhibitor (Table 4, Supplementary Figure S5). For example, KB-3-1 cells treated with 5 mM cisplatin (siNeg as control) for 72 h were 81.4 ± 0.3 % viable (8.0 ± 0.5 % apoptotic, 9.9 ± 2.4 % dead). Co-treatment of cells with cisplatin and miR-16 mimic (down-regulation of Wee1/Chk1) resulted in strong sensitization of cells (19.2 ± 4.2 % apoptotic, 60.4 ± 7.3 % dead). Conversely, co-treatment of cells with cisplatin and miR-16 inhibitor (up-regulation of Wee1/Chk1) resulted in inhibition of cell death (0.7 ± 0.5 % apoptotic, 2.8 ± 1.2 % dead). Similar results were observed in KB-CP.5 cells treated with a higher concentration of cisplatin (25 mM, Table 4). The mimic or inhibitor alone did not have an effect on cell growth compared with negative control cells.
Table 4.
Cell viability (Annexin V/propidium iodide) and cell cycle analysis of KB-3-1 and KB-CP.5 cells transfected with either siNeg, miR-16 mimic or miR-16 inhibitor alone or in the presence of cisplatin (5 μM for KB-3-1 or 25 μM for KB-CP.5)
| KB-3-1 | KB-CP.5 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Viable (%) | Apoptotic (%) | Necrosis (%) | G1 (%) | G2 (%) | S (%) | Viable (%) | Apoptotic (%) | Necrosis (%) | G1 (%) | G2 (%) | S (%) | |
| siNeg | 99.3 ± 0.3 | 0.4 ± 0.2 | 0.18 ± 0.2 | 65.8 ± 3.4 | 5.5 ± 1.9 | 28.7 ± 5.3 | 99.0 ± 0.8 | 0.4 ±0.3 | 0.1 ± 0.1 | 54.0 ± 0.8 | 18.1 ± 0.9 | 28.0 ± 0.1 |
| siNeg + cisplatin | 81.4 ±0.3 | 8.0 ± 0.5 | 9.9 ± 2.4 | 0.66 ± 0.5 | 16.7 ± 3.4 | 82.6 ± 2.9 | 82.3 ± 1.5 | 5.7 ± 1.3 | 6.8 ± 0.3 | 1.7 ± 1.2 | 68.2 ± 3.4 | 30.1 ± 2.8 |
| miR-16 mimic | 99.2 ± 0.4 | 0.2 ± 0.1 | 0.1 ± 0.1 | 55.15 ± 0.5 | 7.5 ± 0.4 | 37.3 ± 0.3 | 98.6 ± 0.5 | 0.8 ± 0.6 | 0.2 ± 0.1 | 51.0 ± 0.5 | 17.1 ± 0.4 | 31.9 ± 0.2 |
| miR-16 mimic + cisplatin | 14.8 ± 3.2 | 19.2 ± 4.2 | 60.4 ± 7.3 | 1.93 ± 0.5 | 19.4 ± 0.5 | 78.7 ± 0.5 | 63.1 ± 2.3 | 15.6 ± 0.6 | 16.7 ± 2.2 | 0.3 ± 0.3 | 57.01 ± 0.3 | 42.7 ± 0.3 |
| miR-16 inhibitor | 99.3 ± 0.3 | 0.4 ± 0.3 | 0.2 ± 0.2 | 50.5 ± 0.2 | 16.7 ± 0.3 | 32.7 ± 0.3 | 98.6 ± 0.5 | 1.2 ± 0.7 | 0.3 ± 0.1 | 52.4 ± 0.6 | 18.5 ± 0.2 | 29.1 ± 0.7 |
| miR-16 inhibitor + cisplatin | 96.3 ± 0.9 | 0.7 ± 0.5 | 2.8 ± 1.2 | 0.5 ± 0.02 | 93.3 ± 0.3 | 6.2 ± 0.2 | 88.8 ± 1.3 | 3.2 ± 1.0 | 5.1 ± 0.7 | 1.6 ± 0.8 | 77.8 ±5.3 | 20.5 ± 4.7 |
KB-3-1 and KB-CP.5 cells were transfected with siNegative control, miR-16 mimic, or miR-16 inhibitor. After 24 hrs cells were treated with cisplatin, and harvested in PBS 72 hrs post cisplatin treatment. Cells were processed for apoptosis or cell cycle analysis as described in Materials & Methods. All data is in triplicate and presented as a percent mean +/− SD
Next, we assessed the impact of miR-16 inhibitor or mimic (with appropriate controls) on arrest of cells in the presence of cisplatin (Table 4, Supplementary Figure S6). We found that in line with the effects on Wee1 and Chk1 and cell viability (described above), miR-16 inhibitor in the presence of cisplatin elevated the proportion of cells in G2 phase (compared to cisplatin + siNeg). For example, KB-3-1 cells treated with cisplatin showed slow transit S phase (as described by Eastman (2), 82.6 ± 2.9 %) and some accumulation in the G2 phase (16.7 ± 3.4 %) compared with control cells (G2 = 5.5 ± 1.9 %). Presence of the miR-16 inhibitor resulted in dramatic arrest in the G2 phase (93.3 ± 0.3 %), and loss of S phase accumulation. KB-CP.5 cells treated with cisplatin alone showed normal S phase levels, and greater accumulation in G2 phase, when compared with parental cells, consistent with resistance and elevated checkpoint kinase expression (Fig. 1B). In CP.5 cells, miR-16 inhibitor resulted in greater G2 accumulation than cisplatin alone. Alternatively, the miR-16 mimic lowered the extent of G2 arrest to a lesser (but significant) extent (compared to cisplatin + siNeg) (Table 4). The mimic or inhibitor alone did not have an effect on cell cycle distribution.
Interference of miR-155 causes an increase in cisplatin resistance by silencing WEE1
Gain-of-function transfection experiments with miR-155 mimic in KB-3-1 cells did not sensitize cells to cisplatin. In KB-CP.5 cells there was a 20% decrease in percent growth when compared to siNegative treated cells (Fig. 3F). Western blots demonstrate that increasing the level of miR-155 in cell lines with miR-155 mimic silences WEE1 to undetectable protein levels (Fig. 2F). Loss-of-function experiments with miR-155 inhibitors increased resistance significantly (Fig. 3F) and restored expression of WEE1 (Fig. 2B). KB-3-1 cells transfected with miR-155 inhibitor were more resistant to cisplatin. This increased resistance was also seen in KB-CP.5 cells transfected with miR-155 inhibitor. These results indicate that the decreased expression of miR-155 in cisplatin-resistant cells contributes to increasing the amount of WEE1, thereby increasing resistance, but the interference of Wee1 alone by miR-155 is not enough to reverse the CP-r phenotype.
Discussion
We report here the results of two high-throughput screens, with the principal finding that a decrease in the miR-15abc/16/16abc/195/424/497 family mediates resistance to cisplatin in our model, and identified their gene targets, the kinases WEE1 and CHK1. This finding was achieved by developing two complementary high-throughput screens designed to identify kinases whose loss sensitized cells to cisplatin, and miRNA that, when increased by mimicking, achieved the same effect.
875 miRNA were screened for their ability to increase sensitivity to low doses of cisplatin when mimicked in cisplatin-resistant cells (KB-CP.5). Twelve miRNAs were shared amongst the top hits at both doses and 5 were from the same family. The miRNAs selected for further validation were predicted to target the kinase positive hits from the parallel screen, and all miRNAs were confirmed to influence cisplatin resistance by sensitizing cells to cisplatin when mimicked. We also examined miR-155, which was not identified in our screen but has been shown to target WEE1 (31) in our loss-of-function/gain-of-function validation studies. miR-155 did cause an increase in cisplatin resistance when its expression is decreased. However, increasing the abundance of miR-155 does very little to increase sensitivity to cisplatin, accounting for its lack of detection in our screen. An additional family member, miR-497, lay just outside the selection criteria, providing further support for the importance of this miRNA family in cisplatin sensitivity. Furthermore, miR-503, with 6 nt of seed sequence conserved with the 7-nt seed sequence of the miR-15abc/16/16abc/195/424/497/1907 family was also predicted to target CHK1.
The miR-15abc/16/16abc/195/424/497/1907 family are grouped together based on their common seed sequence, and have been identified as tumor suppressor miRNAs with decreased expression, mutations to the coding region, or absence in chronic lymphocytic leukemia, gastric, lung, and prostate cancers (22, 24, 33–36). This family has also been shown to influence apoptotic pathways and response to chemotherapy by targeting BCL2 in gastric cancer (36). However, this is the first report linking the regulation of WEE1 and CHK1 cell cycle kinases by the mi-R15abc/16/16abc/195/424/497 family in response to treatment with cisplatin. This leads us to postulate that increasing the expression of miR-15 family members could reverse resistance to chemotherapy in multiple types of cancers.
While several screens of varying scale have now been performed in the presence of cisplatin, none have examined the sensitization of CP-r cells (37). We believe that potential strategies for resolving cisplatin-based insensitivity is a critical unmet need in the clinic. Arora et al. recently reported a siRNA screen against kinases that played a role in the sensitivity of parental (nonresistant) SKOV cells to cisplatin, identifying ATR and CHK1, but not WEE1 (38). ATR, WEE1, and CHK1 are kinases that affect the G2/M checkpoint in the cell cycle, where cells arrest after exposure to cisplatin (2). Furthermore, CP-r cells develop prolonged G2 arrest, allowing a greater capacity for nuclear repair (11), supported by our finding that KB CP-r cells show increased expression of ATR, WEE1, and CHK1. While G2 arrest occurs in response to cisplatin, cell death occurs when the cells undergo an abortive attempt at mitosis after G2 arrest (2). Therefore, abrogating G2 by forcing the cell into mitosis before DNA repair has been adequately accomplished increases the efficacy of cisplatin. Decreasing the function or presence of WEE1 and CHK1, for example with small molecule inhibitors, accomplishes this needed abrogation (2, 12, 39). Here we show that this is also true by increasing miR levels with a mimic.
In response to DNA damage, sensor multiprotein complexes identify DNA lesions, following which ATR and ATM (cell cycle transducer proteins) are activated and signal the downstream checkpoint kinases (12). One of these checkpoint kinases Chk1, a serine/threonine kinase, is a housekeeper present in cells with a normally functioning cell cycle. When signaled by ATR and ATM in response to DNA damage, CHK1 is activated by S345 phosphorylation and prevents CDC2 from activating, thus preventing entry into mitosis (12). CHK1 also phosphorylates p53 and in conjunction with ATM/ATR stabilizes p53 allowing for its activation and accumulation which results in a reversible cell cycle arrest, without requiring de novo protein synthesis (40, 41). CHK1 is critical in modulating the G2/M checkpoint in the cell cycle when cells respond to DNA damaging agents (12) and is therefore one of the most important kinase targets related to the treatment of cancer (12, 42). WEE1 is a tyrosine kinase with a primary function of inactivating CDK1 (encoded by the CDC2 gene) by phosphorylating it, leading to G2 checkpoint arrest and therefore preventing entry into mitosis (14, 15). By regulating CDC2 activity, WEE1 determines the length of time spent in G2/M arrest (43).
Abrogation of the G2/M checkpoint by targeting CHK1 and WEE1 with kinase inhibitors has been shown to sensitize cancer cells to DNA-damaging drugs (14, 42). Kinase inhibitors against CHK1 and WEE1 are already in clinical trials but cells can develop resistance to the kinase inhibitors. These results highlight a potential strategy for a single therapeutic intervention targeted towards the multiple kinases that mediate cisplatin sensitivity.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute. We thank George Leiman for editorial assistance.
Footnotes
Disclosure of Potential Conflicts of Interest
We declare that we have no conflicts of interest related to the publication of this manuscript.
Authors’ Contributions
Conception and design: L.M. Pouliot, M.D. Hall, M.M. Gottesman
Development of methodology: L.M. Pouliot, M.D. Hall, S.E. Martin
Analysis and interpretation of data: L.M. Pouliot, Y.-C. Chen, J. Bai, R. Guha, S.E. Martin, M.D. Hall
Writing, review, and/or revision of the manuscript: L.M. Pouliot, M.M. Gottesman, M.D. Hall, S.E. Martin
References
- 1.Chu G. Cellular responses to cisplatin. The roles of DNA-binding proteins and DNA repair. J Biol Chem. 1994;269:787–90. [PubMed] [Google Scholar]
- 2.Eastman A. The Mechanism of Action of Cisplatin: From Adducts to Apoptosis. In: Lippert B, editor. Cisplatin: Chemistry and Biochemistry of a Leading Anticancer Drug. Zurich, Switzerland: Verlag Helvetica Chimica Acta; 1999. pp. 111–34. [Google Scholar]
- 3.Gottesman MM, Hall MD, Liang XJ, Shen DW. Resistance to Cisplatin Results from Multiple Mechanisms in Cancer Cells. In: Bonetti A, Muggia FM, Leone R, Howell SB, editors. Platinum and Other Heavy Metal Compounds in Cancer Chemotherapy. New York: Humana Press; 2009. pp. 83–8. [Google Scholar]
- 4.Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol Toxicol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- 5.Hall MD, Handley MD, Gottesman MM. Is resistance useless? Multidrug resistance and collateral sensitivity. Trends Pharmacol Sci. 2009;30:546–56. doi: 10.1016/j.tips.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen DW, Pouliot LM, Hall MD, Gottesman MM. Cisplatin Resistance: A Cellular Self-Defense Mechanism Resulting from Multiple Epigenetic and Genetic Changes. Pharmacol Rev. 2011;64:706–21. doi: 10.1124/pr.111.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bucher N, Britten CD. G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br J Cancer. 2008;98:523–8. doi: 10.1038/sj.bjc.6604208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Fraser M, Moll UM, Basak A, Tsang BK. Akt-mediated cisplatin resistance in ovarian cancer: modulation of p53 action on caspase-dependent mitochondrial death pathway. Cancer Res. 2006;66:3126–36. doi: 10.1158/0008-5472.CAN-05-0425. [DOI] [PubMed] [Google Scholar]
- 9.Basu A, Krishnamurthy S. Cellular responses to Cisplatin-induced DNA damage. J Nucleic Acids. 2010;2010:201367. doi: 10.4061/2010/201367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leijen S, Beijnen JH, Schellens JH. Abrogation of the G2 checkpoint by inhibition of Wee-1 kinase results in sensitization of p53-deficient tumor cells to DNA-damaging agents. Curr Clin Pharmacol. 2010;5:186–91. doi: 10.2174/157488410791498824. [DOI] [PubMed] [Google Scholar]
- 11.Sorenson CM, Eastman A. Mechanism of cis-diamminedichloroplatinum(II)-induced cytotoxicity: role of G2 arrest and DNA double-strand breaks. Cancer Res. 1988;48:4484–8. [PubMed] [Google Scholar]
- 12.Dai Y, Grant S. Methods to study cancer therapeutic drugs that target cell cycle checkpoints. Methods Mol Biol. 2011;782:257–304. doi: 10.1007/978-1-61779-273-1_19. [DOI] [PubMed] [Google Scholar]
- 13.Gagnon V, Van Themsche C, Turner S, Leblanc V, Asselin E. Akt and XIAP regulate the sensitivity of human uterine cancer cells to cisplatin, doxorubicin and taxol. Apoptosis. 2008;13:259–71. doi: 10.1007/s10495-007-0165-6. [DOI] [PubMed] [Google Scholar]
- 14.Hirai H, Arai T, Okada M, Nishibata T, Kobayashi M, Sakai N, et al. MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol Ther. 2010;9:514–22. doi: 10.4161/cbt.9.7.11115. [DOI] [PubMed] [Google Scholar]
- 15.Hirai H, Iwasawa Y, Okada M, Arai T, Nishibata T, Kobayashi M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009;8:2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- 16.Liu LZ, Zhou XD, Qian G, Shi X, Fang J, Jiang BH. AKT1 amplification regulates cisplatin resistance in human lung cancer cells through the mammalian target of rapamycin/p70S6K1 pathway. Cancer Res. 2007;67:6325–32. doi: 10.1158/0008-5472.CAN-06-4261. [DOI] [PubMed] [Google Scholar]
- 17.Shah MA, Schwartz GK. Cell cycle-mediated drug resistance: an emerging concept in cancer therapy. Clin Cancer Res. 2001;7:2168–81. [PubMed] [Google Scholar]
- 18.Shi Y, Frankel A, Radvanyi LG, Penn LZ, Miller RG, Mills GB. Rapamycin enhances apoptosis and increases sensitivity to cisplatin in vitro. Cancer Res. 1995;55:1982–8. [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 21.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cittelly DM, Das PM, Spoelstra NS, Edgerton SM, Richer JK, Thor AD, et al. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer. 2010;9:317. doi: 10.1186/1476-4598-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17:523–31. doi: 10.1038/cgt.2010.18. [DOI] [PubMed] [Google Scholar]
- 26.Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;111:478–86. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Liang XJ, Shen DW, Garfield S, Gottesman MM. Mislocalization of membrane proteins associated with multidrug resistance in cisplatin-resistant cancer cell lines. Cancer Res. 2003;63:5909–16. [PubMed] [Google Scholar]
- 28.Shen D, Pastan I, Gottesman MM. Cross-resistance to methotrexate and metals in human cisplatin-resistant cell lines results from a pleiotropic defect in accumulation of these compounds associated with reduced plasma membrane binding proteins. Cancer Res. 1998;58:268–75. [PubMed] [Google Scholar]
- 29.Shen DW, Liang XJ, Gawinowicz MA, Gottesman MM. Identification of cytoskeletal [14C]carboplatin-binding proteins reveals reduced expression and disorganization of actin and filamin in cisplatin-resistant cell lines. Mol Pharmacol. 2004;66:789–93. doi: 10.1124/mol.66.4.. [DOI] [PubMed] [Google Scholar]
- 30.Shen DW, Su A, Liang XJ, Pai-Panandiker A, Gottesman MM. Reduced expression of small GTPases and hypermethylation of the folate binding protein gene in cisplatin-resistant cells. Br J Cancer. 2004;91:270–6. doi: 10.1038/sj.bjc.6601956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tili E, Michaille JJ, Wernicke D, Alder H, Costinean S, Volinia S, et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A. 2011;108:4908–13. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2 ΔΔCT Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Pekarsky Y, Zanesi N, Croce CM. Molecular basis of CLL. Semin Cancer Biol. 2010;20:370–6. doi: 10.1016/j.semcancer.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, et al. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–9. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 35.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 36.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–9. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 37.Nijwening JH, Kuiken HJ, Beijersbergen RL. Screening for modulators of cisplatin sensitivity: unbiased screens reveal common themes. Cell Cycle. 2011;10:380–6. doi: 10.4161/cc.10.3.14642. [DOI] [PubMed] [Google Scholar]
- 38.Arora S, Bisanz KM, Peralta LA, Basu GD, Choudhary A, Tibes R, et al. RNAi screening of the kinome identifies modulators of cisplatin response in ovarian cancer cells. Gynecol Oncol. 2010;118:220–7. doi: 10.1016/j.ygyno.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Montano R, Chung I, Garner KM, Parry D, Eastman A. Preclinical Development of the Novel Chk1 Inhibitor SCH900776 in Combination with DNA-Damaging Agents and Antimetabolites. Mol Cancer Ther. 2012;11:427–38. doi: 10.1158/1535-7163.MCT-11-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tse AN, Carvajal R, Schwartz GK. Targeting checkpoint kinase 1 in cancer therapeutics. Clin Cancer Res. 2007;13:1955–60. doi: 10.1158/1078-0432.CCR-06-2793. [DOI] [PubMed] [Google Scholar]
- 41.Ashwell S, Zabludoff S. DNA damage detection and repair pathways--recent advances with inhibitors of checkpoint kinases in cancer therapy. Clin Cancer Res. 2008;14:4032–7. doi: 10.1158/1078-0432.CCR-07-5138. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kellogg DR. Wee1-dependent mechanisms required for coordination of cell growth and cell division. J Cell Sci. 2003;116:4883–90. doi: 10.1242/jcs.00908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.