SUMMARY
In plants, where cells cannot migrate, asymmetric cell divisions (ACDs) must be confined to the appropriate spatial context. We investigate tissue-generating asymmetric divisions in a stem cell daughter within the Arabidopsis root. Spatial restriction of these divisions requires physical binding of the stem cell regulator SCARECROW (SCR) by the RETINOBLASTOMA-RELATED (RBR) protein. In the stem cell niche, SCR activity is counteracted by phosphorylation of RBR through a cyclinD6;1-CDK complex. This cyclin is itself under transcriptional control of SCR and its partner SHORT ROOT (SHR), creating a robust bistable circuit with either high or low SHR-SCR complex activity. Auxin biases this circuit by promoting CYCD6;1 transcription. Mathematical modeling shows that ACDs are only switched on after integration of radial and longitudinal information, determined by SHR and auxin distribution, respectively. Coupling of cell-cycle progression to protein degradation resets the circuit, resulting in a “flip flop” that constrains asymmetric cell division to the stem cell region.
INTRODUCTION
In Arabidopsis, several factors mediating asymmetric cell divisions (ACDs) have been identified, but little is known on how control of their activity produces precise spatial patterning, which is key to the development of the body plan (Abrash and Bergmann, 2009). Endodermis and cortex tissues are generated in the Arabidopsis root meristem by two successive ACDs. The cortex/endodermis initial (CEI) is a stem cell that self-renews and generates a cortex/endodermis initial daughter (CEID) cell. The CEID undergoes a single periclinal asymmetric division, and the progeny generates endodermis and cortex tissues (Figure 1A). The GRAS family transcription factors SHORT ROOT (SHR) and SCARECROW (SCR) play a prominent role in the CEI and CEID ACDs acting as a heterodimer and are required for the specification and maintenance of the root stem cell niche (Cui et al., 2007; Di Laurenzio et al., 1996; Helariutta et al., 2000; Sabatini et al., 2003). SHR moves from internal tissues to the endodermis (Helariutta et al., 2000). There, it gains efficient nuclear localization, and further movement is restricted by SCR (Heidstra et al., 2004; Cui et al., 2007; Welch et al., 2007). In addition, ACDs of several root stem cells require the RETINOBLASTOMA-RELATED (RBR) protein. RBR interacts genetically with SCR, but the molecular mechanism by which it restricts ACDs to the stem cell niche has not yet been identified (Wildwater et al., 2005). The Arabidopsis CYCLIND6;1 gene (CYCD6;1), which potentially mediates RBR phosphorylation, is a direct transcriptional target of SCR, suggesting a possible connection between SCR and RBR activity (Sozzani et al., 2010).
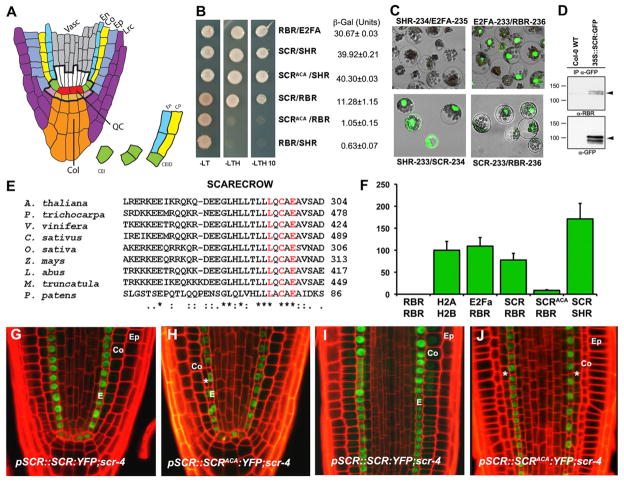
Figure 1. A Conserved LxCxE Motif in SCR Mediates Direct Binding to RBR.
(A) Root stem cell niche organization and cell transitions and divisions defining ground tissue lineages.
(B) Yeast two-hybrid analyses showing and quantifying SCR-RBR interaction. RBR-E2FA and SCR-SHR combinations are positive controls, and RBR-SHR is the negative control.
(C) SCR-RBR binding by BiFC in Arabidopsis mesophyll protoplasts. RBR-E2FA and SCR-SHR are positive controls.
(D) Coimmunoprecipitation of RBR with α-GFP antibody in WT and 35S::SCR:GFP root extracts. Black arrow marks endogenous RBR in top panel and SCR-GFP in lower panel.
(E) Protein sequence alignment of SCR orthologs in seed plants and P. patens moss showing conservation of the LxCxE motif.
(F) In vivo interaction strengths from split Renilla luciferase assay in mesophyll protoplasts. RLUs were normalized to H2A-H2B interaction strength. Arrow bar represents SEM.
(G–J) Confocal laser scanning microscope (CLSM) of longitudinal root sections of 5 dpg. scr4-1 plants complemented with WT SCR (G and I) and SCRAxCxA (H and J). Ep, epidermis; Co, cortex; E, endodermis;*, extra ground tissue layer. See also Figure S1.
In animals and plants, RB proteins control G1- to S-phase progression in the cell cycle. In animals, CyclinD/CDK complexes phosphorylate RB and inhibit RB binding to E2F/DP transcription factor complexes that modulate cell-cycle progression (Temple-ton et al., 1991; Krek et al., 1994; reviewed in Harbour and Dean, 2000). In plants, RBR is phosphorylated in a cell-cycle-specific manner by several Cyclin/CDK complexes (Boniotti and Gutierrez, 2001; Nakagami et al., 2002; Takahashi et al., 2010). In animals, RBs have also been implicated in cellular differentiation through modulation of the activity of tissue-specific transcription factors (Chen et al., 1996; Chen et al., 2007; Berman et al., 2008; Nalam et al., 2009; Calo et al., 2010). The plant RBR protein shares conserved residues with other plant and animal RBs, primarily in the motifs that define interactions with E2F transcription factors and those indispensable for binding diverse proteins containing the conserved Leu-x-Cys-x-Glu (LxCxE motif; Lee et al., 1998; Lendvai et al., 2007; reviewed in Dick, 2007).
Local reduction of RBR in the root meristem expands the stem cell pool without altering cell-cycle rates, suggesting that RBR regulates stem cell transitions by promoting differentiation of stem cell daughters (Wildwater et al., 2005). In addition, RBR is required for the maintenance of stem cells in the shoot and for differentiation of precursor cells for stomata (Borghi et al., 2010).
Here, we show that RBR binds the ACD regulator SCR through its LxCxE motif. We demonstrate that this interaction, together with the activity of the RBR regulator CYCD6;1, defines the precise position of ACDs in the stem cell area through two nested feedforward loops. The resulting network creates a robust bistable switch that is attenuated by a stem-cell-associated auxin maximum and by mitotic progression, thereby linking the auxin gradient along the longitudinal axis with the SHR distribution pattern in the radial axis.
RESULTS
RBR Physically Interacts with SCR through the Conserved LxCxE Motif
We first investigated the molecular basis for previously observed genetic interactions between SCR and RBR. Yeast two hybrid assays indicated that RBR and SCR directly interact in vitro, although less strongly than the SCR-SHR and RBR-E2FA combinations used as positive controls (Figure 1B). Direct interaction between RBR and SCR was also observed in Arabidopsis mesophyll protoplasts by using bimolecular fluorescence complementation (BiFC) assays (Figure 1C). By coimmunoprecipitation assays from root extracts, we also observed that SCR and RBR form a complex in planta (Figure 1D).
An LxCxE motif N-terminal to the GRAS domain in the SCR protein is highly conserved in SCR orthologs from seed plants to the moss Physcomitrella patens (Figure 1E). To determine the relevance of the LxCxE motif for the SCR-RBR interaction, we generated a variant of the SCR complementary DNA (cDNA) in which the LxCxE motif was converted into AxCxA (SCRAxCxA) and tested its capacity to bind RBR. The SCRAxCxA variant interacted with SHR with the same efficiency as wild-type (WT) SCR, but the interaction with RBR was disrupted (Figure 1B). Split-luciferase experiments (Fujikawa and Kato, 2007; Chen et al., 2008) produced SCR-SHR and RBR-E2Fa interactions equally strong as an H2A-H2B interaction (Figure 1F). RBR-SCR interaction was weaker, and RBR-SCRAxCxA was reduced to 11% of the RBR-SCR interaction (Figure 1F). We concluded that SCR and RBR interact directly and that this interaction depends on the LxCxE motif in SCR.
Disruption of the SCR-RBR Interaction Promotes CEID-like ACDs
To test the relevance of the SCR-RBR interaction in planta, we generated protein fusions to yellow fluorescent protein (YFP) by using either the SCR wild-type cDNA or the SCRAxCxA cDNA under the transcriptional control of the SCR promoter. Both constructs, pSCR::SCR:YFP and pSCR::SCRAxCxA:YFP, were transformed independently into the scr4-1 mutant background.
Seedlings of multiple stable transformants, homozygous for the pSCR::SCR:YFP and pSCR::SCRAxCxA:YFP transgenes, complemented the macroscopic defects in the scr4-1 mutant and restored cotyledon and primary root size, indicating that SCR and SCRAxCxA are both functional (data not shown). scr4-1;pSCR::SCR:YFP roots fully complemented the scr4-1 phenotype and displayed the characteristic SCR expression pattern in the quiescent center (QC), ground tissue initials and mature endodermal layer (Figure 1G; compare with Figures S1A and S1B available online). Roots of scr4-1;pSCR::SCRAxCxA: YFP (hereafter referred to as scr4-1;SCRAxCxA) seedlings similarly complemented the scr4-1 phenotype. However, meristematic endodermis cells expressing the SCRAxCxA fusion performed extra periclinal divisions capable of generating a complete extra ground tissue layer (Figures 1G–1J, asterisks) from late embryo stage onward (Figures S1E and S1F, asterisk).
We investigated the identity of the extra layer observed in scr4-1;SCRAxCxA roots by using markers to distinguish cortex from endodermis tissue. Three markers demonstrated that disruption of the interaction between SCR and RBR led to an extra ACD that originates in the inner ground tissue cell and again separates cortex and endodermis identity (Figures S1C, S1D, S1G, and S1H). The extra ACD in the scr4-1;SCRAxCxA line indicates that RBR interaction with WT SCR counteracts recurrent ACD-promoting activity.
In the WT, SCR promotes ACD in the CEI and CEID cells together with its heterodimeric interaction partner SHR. When we transformed the scr4-1;shr double mutant with pSCR:: SCR:YFP and pSCR::SCRAxCxA:YFP constructs, both lines displayed the shr phenotype lacking ACDs (“M” in Figures S1I and S1J). These results indicate that the extra ACD observed in the scr4-1;SCRAxCxA roots, like the ACD in WT, is SHR dependent.
RBR Negatively Regulates SCR-SHR Transcriptional Targets
Well-characterized direct targets of SCR and SHR are the MAGPIE (MGP) and NUTCRAKER (NUC) genes (Cui et al., 2007). A genetic link between RBR and SCR-SHR transcriptional activity was suggested by Wildwater et al. (2005), who showed that the expression domain of NUC (At5g44160) expands as a consequence of RBR silencing. After RBR transactivation induced by dexamethasone (Dex, as previously described in Wildwater et al., 2005), pMGP::MGP:GFP expression gradually decreased in roots harboring the RBR-transactivation construct (Figures 2A–2C). This reduction in expression was evident well before the full differentiation of the meristem invoked by prolonged RBR overexpression.
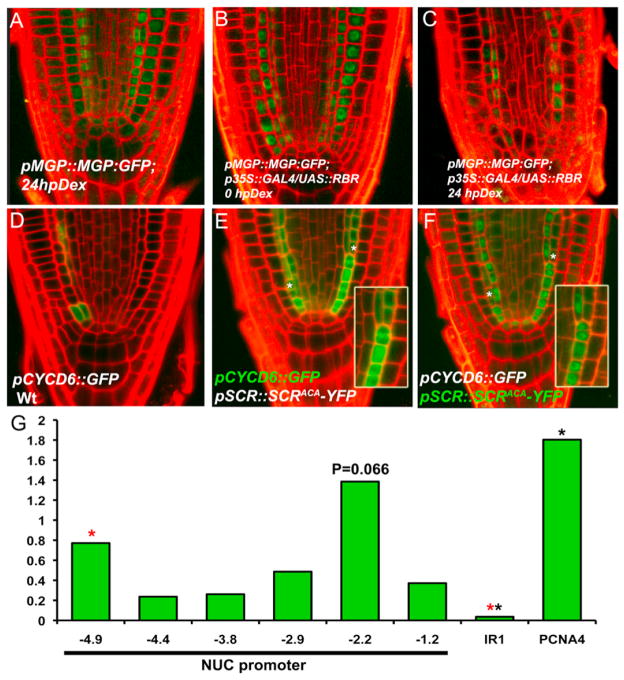
Figure 2. RBR-SCR Interaction Affects the Expression of SCR-SHR Direct Targets.
(A–C) pMGP::MGP:GFP protein fusion expression in WT (A) and inducible RBR overexpressor line before (B) and after transfer for 24 hr (C) to Dex-amethazone (hpDex).
(D–F) pCYCD6;1::GFP fusion expression in WT (D) and in scr4-1;SCRAxCxA (E); note the correlation between expansion of pCYCD6;1::GFP expression domain and appearance of extra ACDs, marked with asterisks; asymmetric segregation of pSCR::SCRAxCxA:YFP in the same plant (F).
(G) ChIP-qPCR using RBR antibody and primers to specific regions of NUC, PCNA, and IR1 promoters. Black asterisks, Student’s t test (p < 0.05); fragment −2.2 showed variable enrichment in biological replicates.
SHR and SCR directly activate the D-type cyclin CYCD6;1, whose expression is enriched in the CEI/CEID cells and whose forced expression in the ground tissue produced extra divisions, similar to those in scr4-1;SCRAxCxA roots (Sozzani et al., 2010). CYCD6;1 is a potential regulator of RBR phosphorylation. If RBR phosphorylation by CYCD6;1 inhibited RBR action on SCR, this would lead to a feedforward loop (model 1, Figure 3G). We predicted that the scr4-1;SCRAxCxA mutation, which prevents RBR inhibition of SCR activity, leads to CYCD6;1 expression outside of the CEI and CEID. Indeed, whereas the pCYCD6;1::GFP expression domain in the scr4-1;SCR background, as in the WT context, was predominantly confined to the ground tissue initials (Figure 2D), it expanded in the endodermal layer of scr4-1;SCRAxCxA seedlings in perfect correlation with the extra ACDs (Figures 2E and 2F).
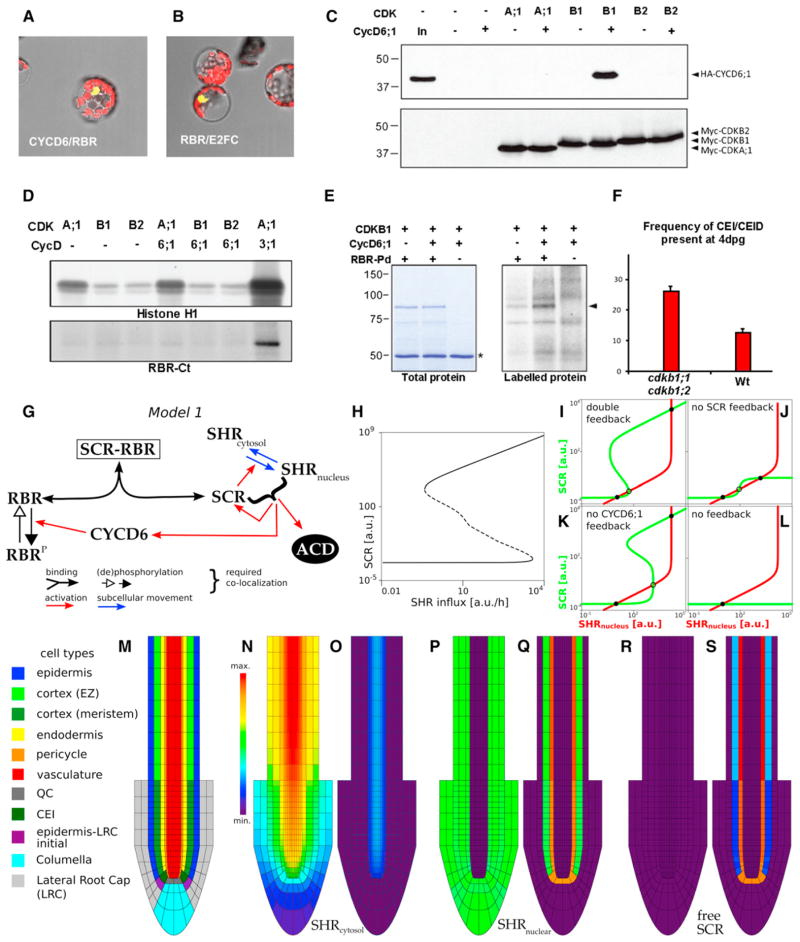
Figure 3. Specific Interaction between CDKB1 and CYCD6;1 Forms a Protein Complex that Phosphorylates RBR.
(A and B) BiFC assays for in vivo interaction between CYCD6 and RBR (A) with E2FC/RBR as positive control (B).
(C) Myc-CDK and HA-CYCD6;1 proteins expressed in Arabidopsis protoplasts. Protein gel blots of complexes immunoprecipitated with anti-c-Myc antibodies using anti-HA and anti-c-Myc antibodies to detect CYCD6;1 binding and CDK expression, respectively.
(D) Myc-CDK expressed in Arabidopsis protoplasts either alone or together with HA-CYCD6;1 or HA-CYCD3;1 and precipitated with anti-c-Myc antibody. Kinase activity of immunocomplexes in the presence of [γ-32P]ATP using histone H1 and GST-RBR-Ct proteins as substrates and detected by autoradiography of protein gels.
(E) Kinase activity of CDKB1 and CDKB1-CYCD6;1 expressed in protoplasts in the presence of [γ-32P]ATP using GST-RBR pocket domain (GST-RBR-Pd) as substrate, detected by autoradiography. Asterisk marks IgG heavy chain.
(F) Frequency of undivided CEI and CEIDs in roots of WT and ckdb1;1 cdkb1;2 seedlings 4 dpg. Differences are significant (t test < 0.05); error bars indicate SEM.
(G) Schematic of model 1.
(H) Bifurcation diagram with equilibrium levels of free SCR as a function of SHR influx into the cell. Dashed line indicates unstable equilibria. Both a high and a low stable SCR level exist over a wide range of influx.
(I–L) Phase plane analysis of model 1 using quasi-steady-state assumptions (see Figure S2). (I) Model 1 presents two stable equilibria (solid black circles, one of high SCR and nuclear SHR levels and the other of low SCR and nuclear SHR levels) separated by an unstable equilibrium (open circle). (J and K) When the activation mediated by CYCD6;1 or the feedback of SCR and nuclear SHR on SCR transcription is turned off, the model continues presenting bistability.
(L) Without either of these feedbacks, no bistability occurs for any parameter setting.
(M) Root layout for all spatial simulations.
(N–S) Cytosolic SHR (N and O), nuclear SHR (P and Q), and free SCR (R and S) for weak (N, P, and R) and strong (O, Q, and S) activation mediated by CYCD6;1 (see “Modeling Procedures,” Figure S2, and Tables S1 and S2). Molecular weight standards are indicated in kilodaltons.
SHR and SCR bind to the NUC promoter (Cui et al., 2007). We hypothesized that binding of SCR to RBR takes place in the context of target gene promoters. To test this, we performed replicated chromatin immunoprecipitation (ChIP) experiments followed by quantitative PCR (ChIP-qPCR) on the NUC promoter. We found that RBR consistently associated with specific NUC promoter fragments located at −4.9 Kb (Figure 2G, red asterisk; Student’s t test, p < 0.05) and at −2.2 Kb upstream from the ATG (Figure 3A; p = 0.066) but that it was only weakly enriched in line with its indirect binding to DNA.
We conclude that RBR negatively regulates SHR-SCR transcriptional targets such as NUC and MGP, which in turn promote ACD (Welch et al., 2007). In addition, RBR constrains the expression of another SHR-SCR target, CYCD6;1, which may itself regulate RBR activity.
Specific Binding of CYCLIND6;1 to CDKB1 Yields an Active Complex that Phosphorylates RBR
BiFC assays revealed that CYCD6;1 interacted with RBR in Arabidopsis protoplasts (Figure 3A) in a manner similar to the positive control RBR-E2FC (Figure 3B). Previous large-scale BiFC interaction mapping of Arabidopsis cell-cycle regulators indicated association between CYCD6;1 and A-, B-, and D-type CDK subunits (Boruc et al., 2010). We transiently coex-pressed epitope-tagged CYCD6;1 and CDKs in Arabidopsis protoplasts and observed specific interaction between CDKB1 and CYCD6;1 proteins (Figure 3C). Immunoprecipitation-protein kinase assay against histone H1 and a glutathione S-transferase (GST) fusion to the C-terminal polypeptide encompassing the last 235 amino acids (GST-RBR-Ct) of Arabidopsis RBR revealed that a CDKA;1-CYCD3;1 complex phosphorylated both histone H1 and GST-RBR-Ct but detected no significant increase in histone H1 or GST-RBR-Ct phosphorylation levels upon CYCD6;1 binding to CDKB1 (Figure 3D). On the other hand, immuno-precipitation-kinase assays using a GST fusion to the 608-amino-acid-long complete pocket domain (see Experimental Procedures for a detailed description of RBR fragments used) revealed that CYCD6;1 binding increased basal activity of CDKB1 (Figure 3E). Our data indicate that an active CDKB1-CYCD6;1 kinase complex targets one or more phosphorylation sites within the RBR pocket domain.
The CDKB1 kinases to which CYCD6;1 specifically binds are encoded by two genes, CDKB1;1 and CDKB1;2. Although CDKB1;1 is expressed in a range of tissues (Segers et al., 1996), a transcriptional fusion to the β-glucuronidase gene (pCDKB1;1:GUS) and a translational fusion to GFP (pCDKB1; 1::CDKB1;1:GFP) revealed high levels of CDKB1;1 expression and protein accumulation in CEI cells as they undergo ACD (Figures S2C and S2D). To detect the protein fusion, reduction of proteasome activity with MG132 treatment was necessary, suggesting additional posttranslational regulation. Double-mutant cdkb1;1 cdkb1;2 seedlings revealed a higher frequency of undivided CEIDs as compared to WT (Figures 3F and S2B), indicating that ACDs are delayed in cdkb1 double mutants. Consistent with the specific CDKB1-CYCD6;1 interaction, this phenotype is similar to that reported for the cycd6;1 mutant (Sozzani et al., 2010). Collectively, our results suggest that spatiotemporal fine tuning of RBR phosphorylation in the ground tissue occurs through tight regulation of the CYCD6;1-CDKB1;1 protein pair.
The SHR/SCR/RBR/CYCD6;1 Network Generates a Bistable Radial Patterning Switch
We addressed whether the nested feedback circuit among SHR, SCR, RBR, and CYCD6;1 could result in the precise spatial confinement of ACDs. We generated coupled ordinary differential equations (ODEs) to describe the wiring of the network (Figure 3G; Supplemental Information for details) and focused on analyzing equilibrium levels of nuclear SHR and free SCR (unbound to RBR), as ACDs occur when both levels are high and SCR-RBR levels are low. Analysis and numerical simulations of the model showed that the network ensures highly robust bistability in which nuclear SHR and free SCR levels can flip between a low and a high state. Figure 3H depicts the equilibria in terms of free SCR levels. Bistability can be found over a very wide range of SHR influx levels. For low values of SHR influx, there is only one stable state, illustrating that sufficient SHR influx is needed to gain the potential of the ACD cell state, whereas very high SHR influx causes the low equilibrium to disappear, leading to the flip to the ACD cell state. Conversely, once the ACD cell state is triggered, only very low SHR levels can bring the cell back into the normal state.
This hysteresis effect is due to the fact that SCR sequestering by RBR prevents the positive feedback loops from taking effect, which is necessary to increase SCR concentrations. This dynamic behavior is observed over a broad parameter range, indicating intrinsic robustness within the network wiring. To address to which extent the bistability is due to the SCR feedback on its own transcription or to the CYCD6-mediated feedback loop that promotes free SCR, we mathematically analyzed the cases in which one or both of the feedback loops are removed (Figures 3I–3L and S2E–S2H). The qualitative behavior of bistability was preserved when any one of the two feedbacks was removed (Figures 3J, 3K, S2F, and S2G). However, if both are turned off, the system failed to present bistability for any possible parameter setting (Figures 3L and S2H). We therefore conclude that it is the combined (and redundant) action of two feedback loops that determines the huge potential for bistability underlying the behavior of ACD specification.
To explore how these interactions are influenced by the cellular environment in the root, we inserted the network in every cell of an in silico root (Figure 3M). Because SHR production is limited to the vasculature while degraded everywhere, a radial gradient in SHR levels was formed. Interestingly, within the spatial simulations, the SHR influx-dependent bistability leads to two possible scenarios. One possibility is that the highest SHR levels, found close to the vasculature, are not sufficient to trigger the ACD state (Figures 3N, 3P, and 3R). In that case, SHR keeps its graded radial distribution (Figure 3N), which is never observed in vivo. Alternatively, the positive feedback loops are sufficiently strong that all cells close to the vasculature are flipped to the ACD cell state (Figures 3O, 3Q, and 3S). In the latter case, due to the efficient sequestering of SHR in the endodermis, CEI/CEID, and QC, cells farther from the stele do not receive enough SHR to switch to the high equilibrium. This result was consistent for different parameter settings and spatial implementations (data not shown). The modeling result that the complete endodermal cell file triggers the ACD state is not observed in vivo. Hence, the network explains the radial position of ACDs but does not contain a mechanism to bias this to the CEI/CEID cells.
Auxin Promotes SHR/SCR-Dependent CYCD6;1 Transcription to Trigger ACDs in the Longitudinal Axis
The plant growth factor auxin is a key factor in positioning the niche, and its polar transport is required for longitudinal positioning of CEI/CEID markers (Sabatini et al., 1999). Therefore, auxin might mediate restriction of CYCD6;1 expression and the associated ACD in the longitudinal axis. When we increased auxin concentration with IAA (the major active auxin) and NPA (a polar auxin transport inhibitor), expression of pCYCD6;1::GFP expanded, coinciding with extra periclinal divisions that often led to an extra ground tissue layer in primary roots (Figure 4A). After the same treatment, Col0;pSHR::SHR:GFP roots revealed the SHR protein fusion in the inner cell layer, indicating that auxin induced ACDs (Figure 4B). In scr4-1;SCRAxCxA;pCYCD6;1::GFP roots, where RBR cannot repress SCR activity, IAA/NPA treatment induced successive ectopic ACDs in the transit amplifying zone (Figure 4C), correlating with strong shootward-extended expression of pCYCD6;1::GFP (Figure 4D).
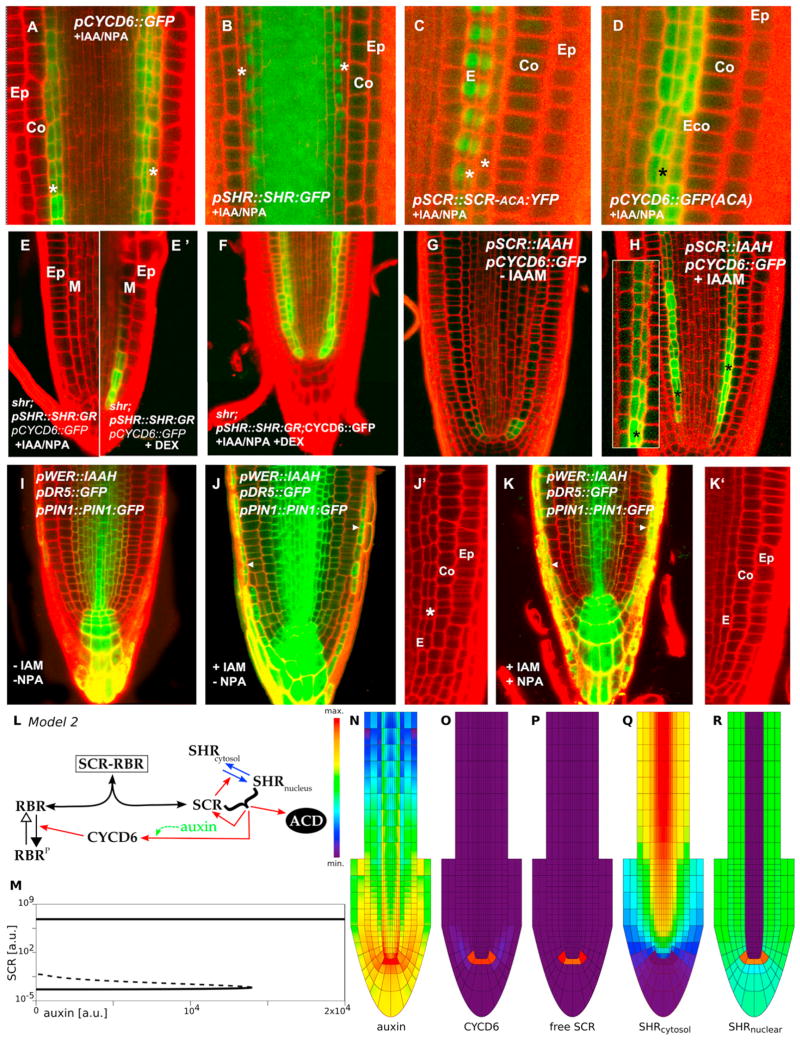
Figure 4. Network Model within Tissue Context Presents Bistable Switch Where Auxin-Dependent CYCD6;1 Activation by SHR-SCR Limits ACD to the CEI.
(A–K) IAA+NPA treatments in Col-0;pCYCD6;1::GFP (A), Col-0;pSHR::SHR:GFP (B), and scr4-1;ACA; pCYCD6;1::GFP (C) and (D). shr;pSHR::SHR:GR; pCYCD6;1::GFP seedlings treated with IAA+NPA (E), same seedling after SHR induction by Dex treatment showing pCYCD6;1::GFP expression (E′), and seedling treated with both IAA+NPA and Dex for 24 hr (F). Effect of local increase of auxin levels in endodermis revealed in the pSCR::IAAH; pCYCD6;1::GFP line in absence (G) and presence (H) of the substrate IAM. Phenotype and expression patterns of Col-0;pWER::IAAH;pPIN1::PIN1:GFP;pDR5::GFP roots of 5 dpg seedlings grown in MS media and transferred for 2 days to media in absence (I) and presence of the substrate IAM (J and J′) and in the presence of IAM and NPA (K and K′). Arrowheads in (J) and (K) point to increased pDR5::GFP expression in the LRC, indicating auxin synthesis. Ep, epidermis; Co, cortex; E, endodermis; Eco, extra cortex layer; M, mixed identity ground tissue layer; *, extra ground tissue layer.
(L) Schematic of model 2. The green arrow indicates the extension of the model, which takes the effect of auxin into account.
(M) Bifurcation diagram showing equilibrium levels of free SCR as a function of auxin levels. A switch-like behavior occurs when auxin levels are increased, but the system does not “turn off” when the levels are subsequently decreased.
(N–R) The spatial simulations show the triggering of the ACD cell state in the QC and CEI/CEID cells. (N) auxin; (O) CYCD6;1; (P) free SCR; (Q) cytosolic SHR; and (R) nuclear SHR levels. (See also “Modeling Procedures,” Figure S3, and Tables S2 and S3.)
IAA/NPA treatment induced pCYCD6;1::GFP expression in the shr;pSHR::SHR:GR mutant background only after DEX-induced nuclear localization of SHR (Figures 4E and 4E′). When shr;pSHR::SHR:GR seedlings were grown continuously in both DEX and IAA/NPA, pCYCD6;1::GFP expression expanded, and extra ACDs occurred (Figure 4F). We concluded that auxin-mediated induction of the CYCD6;1 promoter requires SHR and SCR activity.
To increase auxin levels only in the endodermis by bacterial biosynthetic enzyme IAAH, we introgressed the pSCR::IAAH transgene in the Col0;pCYCD6;1::GFP background. Roots of seedlings grown in IAM-containing medium, but not control roots (Figure 4G), expanded pCYCD6;1::GFP expression, which is correlated with extra ACDs (Figure 4H). Because it remained possible that auxin synthesized in the endodermis fluxed around to activate an ACD stimulating factor elsewhere, we drove IAAH from the epidermal/lateral root cap WEREWOLF promoter (pWER::IAAH). When grown on IAM-containing medium, extra ACDs occurred in the endodermis, and pDR5::GFP and pPIN1:: PIN1:GFP signals increased in the ground tissue and the vascular tissue, indicating that auxin synthesized in the outer layers flows to the inner tissues and triggers extra ACDs (Figures 4I, 4J, and 4J′). However, on IAM- and NPA-containing medium, reflux of auxin from the WER domain was blocked, as revealed by higher levels of pDR5::GFP in the LRC and low expression of both markers in the vascular and ground tissues; under these conditions, no extra ACDs were observed (Figures 4K and 4K′). Laser ablation experiments, which alter auxin flow in predictable manners (van den Berg et al., 1997; Xu et al., 2006), independently supported that a local increase of auxin levels in the endodermis is critical to trigger new ACDs but needs the presence of SHR and SCR (Figures S4A–S4H).
An Auxin-Attenuated Switch Promotes ACD at the Stem Cell Niche
To explore the spatial-dynamical consequences of auxin feedback on CYCD6 transcription, we added this interaction to our ODE network, now representing model 2 (Figure 4L, green arrow). For an isolated cell, in a nonspatial setting, we found a switch-like response in free SCR levels linked to the auxin concentration (Figure 4M). Next, we generated a spatial model by concomitantly describing auxin dynamics based on experimentally observed PIN distributions (Grieneisen et al., 2007; Laskowski et al., 2008) and with information on the detailed organization of the stem cell niche and root cap (Figure S3, details in Supplemental Information). The modeled auxin dynamics in this context predicted that, through the auxin-attenuated network within the root context, CYCD6;1, nuclear SHR, and SCR become localized to the QC and CEI (Figures 4N–4R).
Although, compared to model 1, model 2 clearly presents an improvement in capturing the regulation of the ACDs, it fails to explain the confinement of SHR to the endodermis. This is because, in model 2, only the cells that enter the ACD state can block the SHR throughput from the vasculature to the external cell files. Thus, SHR moves from endodermal cells that are not in the ACD state into the next cell layer (Figure 4Q), as in scr mutants (Heidstra et al., 2004; Sena et al., 2004).
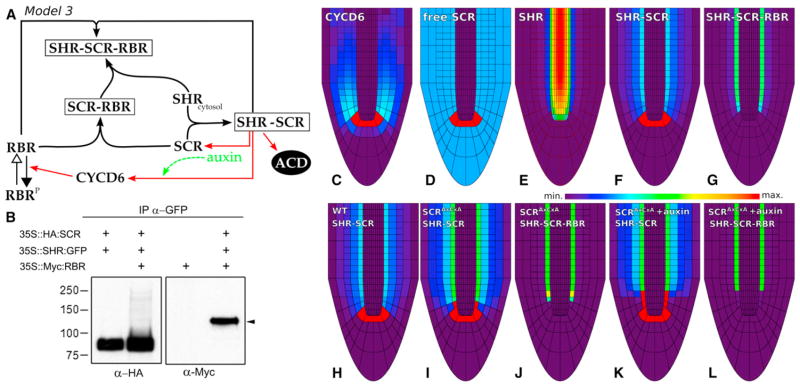
The lack of SHR confinement contradicted experimental data, which suggested that RBR binding to SCR does not prevent SCR from interacting with SHR but instead inhibits activation of downstream targets by binding the SHR-SCR complex. To test whether a ternary complex can indeed be formed, we coex-pressed SHR-GFP fusion protein with epitope-tagged SCR and RBR in Arabidopsis protoplasts and purified complexes on a GFP affinity matrix. Immunoblot analysis of affinity-purified complexes confirmed the previously observed SCR-SHR heterodimer and indicated that SCR-SHR interaction is not compromised by the RBR protein (Figure 5B). Western analysis also revealed copurification of the RBR protein, suggesting that the three proteins can form a complex (Figure 5B). We therefore extended model 2 to explicitly take into account not only the SCR-RBR complex but also the SHR-SCR and the SHR-SCR-RBR ternary complex (Figure 5A). The third model presents, in a parsimonious way, our current understanding of the regulatory circuit and the distribution of key factors underlying ACDs (Figures 5C–5G and S4I–S4M).
Figure 5. Auxin Concentration Influences CYCD6;1 Expression and Modulates Ground Tissue ACDs.
(A) Schematic of model 3.
(B) Complex formation between SHR-GFP, HA-SCR, and Myc-RBR proteins expressed in Arabidopsis protoplasts and purified by binding to GFP affinity beads.
(C–G) Spatial simulations show the triggering of the ACD cell state in the QC and CEI/CEID cells and the confinement of SHR in the endodermis. Profiles within the root tip of (C) CYCD6;1; (D) free SCR; (E) SHR; (F) SHR-SCR; and (G) SHR-SCR-RBR levels. (See also “Modeling Procedures,” Figure S4, and Movie S1.)
(H–J) A 75% reduction of the binding of SCR to RBR, as to mimic the SCR (see also Movie S2).
(K and L) Treatment of the SCR with auxin (see also Movie S2). Color bar represents relative concentration levels. See Supplemental Information, Figure S4, and Table S4 for full details.
Model 3 revealed that, whereas RBR complexes are absent in the ACD region, as basically all RBR is in its phosphorylated form, high levels of the ternary complex in the endodermis prevent both ACDs and SHR transport into the cortex (Figures 5G and S4M). Implementing the SCRAxCxA modification, which decreases the binding of RBR to SCR, resulted in high levels of the SHR-SCR complex not only the CEI but also in the first endodermal cell and CEID, which is in agreement with the observed extended CEI activity (Figures 5H–5J). After simulating auxin immersion (Grieneisen et al., 2007), high levels of the SHR-SCR complex were even further propagated spatially (Figure 5K), together with a further reduction of the inhibitory complex (Figure 5L). This confirms the quantitative impact of auxin levels on the process of ACD specification through the network and mirrors the experimental outcomes of auxin treatment promoting extra ACDs. However, although model 3 explains how auxin and SHR influx trigger the “on” state of the bistable SHR-SCR-RBR circuit, as well as the radial confinement of SHR, it does not reveal how the system is reset. Because the network functions through a double feedback loop (one of which is auxin dependent), once the system is flipped to the “on” state to undergo ACDs, even a dramatic decrease in auxin levels will not be able to turn it back “off” (Figures S4N and S4O).
Protein Degradation Can Reset the Bistable ACD Switch
The solution to this apparent failure was suggested by the ACD itself. The triggering of an ACD is a cell-cycle event, and regulatory proteins are often degraded during specific cell-cycle phases. Therefore, we analyzed the levels of functional RBR, SHR, and SCR protein fusions expressed from their own promoters during the cell cycle. Prior to mitosis, endodermis-expressed RBR:YFP, SHR:YFP, and SCR:YFP protein fusions were primarily localized to the nucleus. However, elongated cells prior to cell plate formation displayed a weaker and cytoplasmic signal (Figures S5A–S5C′), and postdivision nuclear levels were also decreased.
To record multiple cells in specific cell-cycle phase, we standardized conditions for in planta hydroxyurea (HU)-induced cell-cycle synchronization (Figures S5D–S5K′; Cools et al., 2010). pSHR::SHR:YFP decreased in abundance in individual endodermal cells followed between 19 and 22 hr HU, when the 35S::H2B:RFP protein fusion resolved into a mitotic figure, and such decrease was also observed for pSCR::SCR:YFP pRBR::RBR:CFP fusions (Figures S5L–S5S).
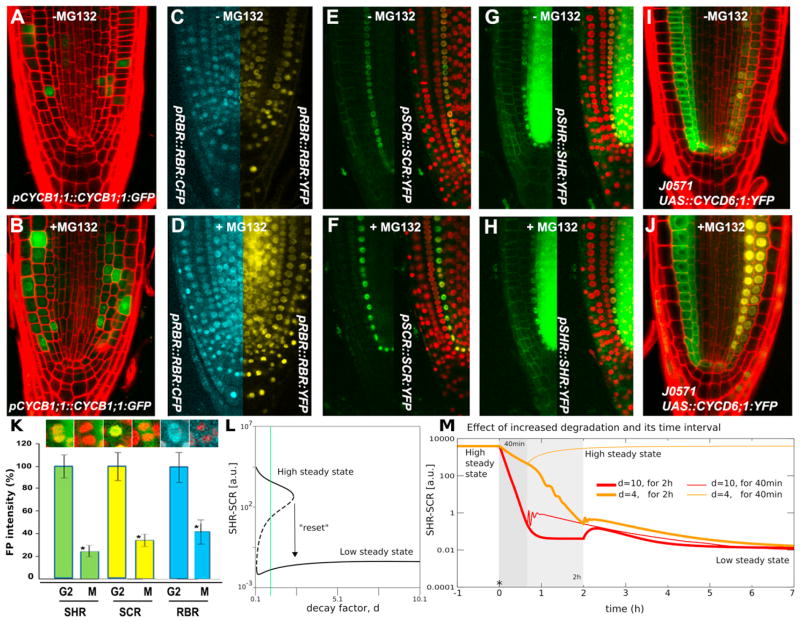
To investigate further whether specific protein degradation machinery mediated the decrease in SHR, SCR, and RBR concentration, we utilized the 26S proteasome inhibitor MG132. In the presence of MG132, the fluorescence of a CYCB1;1:GFP control line increased (Figures 6A and 6B; n ≥ 7). Roots expressing RBR and SCR protein fusions also revealed increased signal after MG132 treatment (Figures 6C–6F). No increase of pSHR::SHR:YFP signal was evident after MG132 treatment (Figure 6H) when compared to the control (Figure 6G). These data suggest that SCR and RBR are subject to 26S-proteasome-mediated degradation, whereas the decrease in SHR concentration during cell division appears to be mediated by other factors.
Figure 6. Protein Degradation Turns “Off” the Switch.
(A–K) Expression patterns prior to and after MG132 treatment for pCYCB1;1:: CYCB1;1:GFP (A and B), pRBR::RBR:CFP/pRBR::RBR:YFP (C and D), pSCR::SCR:YFP/p35S::H2B:RFP (E and F), pSHR::SHR:YFP/p35S::H2B:RFP (G and H), and J0571/pUAS::CYCD6;1:YFP (I and J). Quantification of total fluorescence intensity before and after mitosis for pSHR::SHR:YFP/p35S::H2B:RFP, pSCR::SCR:YFP/p35S::H2B:RFP, and pRBR::RBR:CFP/p35S::H2B:RFP plants (K); Student’s t test was used to assess the statistical significance of the distributions (*p < 0.05). Arrow bar represents SEM.
(L) Bifurcation diagram showing equilibrium SHR-SCR levels as a function of the level of enhanced protein degradation, d.
(M) Time dynamics for intermediate (40 min, thin lines) and long (2 hr, thick lines) time span of enhanced protein degradation. Red, d = 10; orange, d = 4. See also Figure S5.
Some plant D Cyclins are targets for degradation by the 26S proteasome (Genschik et al., 1998; Lechner et al., 2002; Plan-chais et al., 2004; Sanz et al., 2011). Indeed, when CYCD6;1: YFP was expressed in cortex and endodermal layers along the root meristem, fluorescence was detected in the nucleus of some endodermis cells and in the cytoplasm of others. In contrast, after MG132 application, strong nuclear-localized YFP signal appeared throughout both cortex and endodermis layers (Figures 6I and 6J). We concluded that proteasome activity destabilizes the CYCD6;1:YFP protein. We validated the dynamics of protein degradation during divisions in vivo by recording the behavior of SCR and SHR proteins in endo-dermis cells by using the double marked lines p35S:: H2B:RFP;pSCR::SCR:YFP and p35S::H2B:RFP;pSHR::SHR:GFP and observed rapid protein degradation (Figure 6K and Movies S3 and S4).
We analyzed whether the observed cell-cycle-mediated degradation was sufficient to reset the ACD switch in model 3. Figures 6L and S5X show bifurcation diagrams for the equilibrium level of SHR-SCR as a function of the increased level of protein degradation. Without elevated protein degradation, there were three equilibria, two of which were stable. This implies that the first cell within the endodermal cell file could indeed be either in the “on” state or in the “off” state, but, having inherited its state from the CEID, it would be expected to be in the “on” state. If, however, cell division was accompanied by increased protein degradation, it would be sufficient to flop cells from the “on” state back down to a lower state, switching the system off (see Figures 6L and 6M).
The observed rapid protein degradation corresponded to the quantitative predictions of our model. The importance of the right balance between the level of accelerated protein degradation and the time interval during which accelerated degradation takes place is further illustrated in Figure 6M, in which shorter time windows need higher degradation rates for turning the ACD state “off” without relapse to the “on” state. This behavior of our model predicted that protease inhibitor treatment could itself trigger ACDs. MG132 application indeed consistently triggered ectopic periclinal cell divisions associated with CycD6 expression that were asymmetric based on the persistence of SCR expression only in inner cells (Figures S5T–S5V).
DISCUSSION
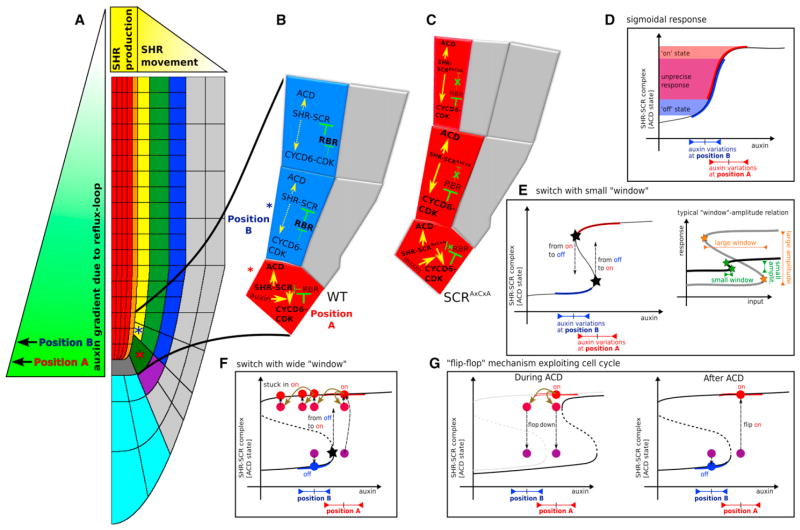
We describe a nested feedback circuit in Arabidopsis that ensures the accurate spatiotemporal control of asymmetric cell divisions. Modeling studies reveal that this circuit integrates information of two developmental axes, auxin-mediated tissue polarity, and the radial SHR expression domain and translates this into a precisely located asymmetric cell division state, which is schematized by the simplified model in Figures 7A–7C.
Figure 7. “Flip-Flop” Mechanism Exploiting Cell Cycle.
(A–C) Schematic of root with SHR and auxin gradient. Red asterisk indicates CEI/CEID cell in ACD “on” state, receiving higher auxin levels (position A); blue asterisk indicates endodermal/cortex cell in CEI “off” state, receiving lower auxin levels (position B).
(D) Due to natural auxin variations at position A and B, a sigmoidal response would lead to an imprecise outcome.
(E) (left) With a bistable switch, a small window of bistability would be required to change the ACD cell state (right). Such a small window of bistability, combined with highly distinct cell differentiation states, lacks robustness.
(F) A robust wide window of bistability leads to sustained ACDs in the endodermal cells.
(G) Enhanced protein breakdown during cell cycle shifts the window of bistability, causing ACD cell states in the daughter cells to turn down (flop); a rapid collapse in sustenance of the ACD cell state (left). Afterward, a rapid recovery of the ACD cell state (flip on) can take place in CEI/CEID cells due to high auxin levels, but not in endodermal and cortex cells, which turn off their ACD cell state (right).
Our results suggest a specific role for the CYCD6-CDKB1 complex in which it phosphorylates RBR to influence SCR transcriptional activity. Based on our genetic and biochemical data, RBR phosphorylation is carried out by a complex of CYCD6;1 with either CDKB1;1 or CDKB1;2. The cell-type-specific transcriptional regulation of some of the Arabidopsis CYCDs and studies reporting 26S-proteasome-dependent CYCD3;1 and CYCD2;1 degradation (Planchais et al., 2004; Sanz et al., 2011) suggest that tight spatiotemporal regulation of CYCDs at both transcriptional and posttranslational levels may be common among the highly diversified plant D cyclins. The influence of CYCD6;1-CDKB1-mediated RBR phosphorylation on the SCR-SHR activity needs to be resolved at the mechanistic level but points to a role for RBR and cyclin-CDK complexes in modulating the transcriptional capacity of factors controlling cell transitions in a specific cell lineage. Our findings are in line with recent in vivo studies in animals in which RBs are shown to promote cellular differentiation by interaction with tissue-specific transcription factors (Calo et al., 2010) but provide, in addition, a mechanism by which this role is orchestrated in space and time.
Our data demonstrate that the auxin maximum spatially correlates with and positively influences CYCD6;1 transcription in the CEI/CEID cells, in which high levels of CYCD6;1 (along with CDKB1) phosphorylate RBR. An auxin-responsive element (ARE) in the promoter region of the CYCD6;1 gene suggests that auxin-responsive factors (ARFs) could read out auxin concentrations on the CYCD6;1 promoter. This would serve to potentiate SHR-SCR complex activity on the CYCD6;1 promoter because the auxin input is unable to act in the absence of SHR. This scenario is consistent with the additive stimulation of ACDs observed after auxin increase and simultaneous uncoupling of SCR action from RBR inhibition by the SCRAxCxA allele.
We show that the mechanism of correct ACD localization depends on a radial SHR gradient and a longitudinal auxin gradient (Figure 7A). The feedback of the network on SHR transport guarantees that there is only a single cell file with ACD potential, and auxin triggers the correct cells within that file. A strong bistability due to a nested positive feedback loop, combined with resetting of the high state through protein degradation, underlies the “readout” of the axial information. Other scenarios could program this readout as well but do not lead to such a robust cell fate specification.
A first alternative scenario could rely on a steep sigmoidal response curve. However, this would not allow for the sudden switch in the cellular response that is critical for daughter cells to present very different behaviors after cell division. Moreover, a sigmoidal response curve would make root development very sensitive for auxin levels within the CEI/CEID region, which naturally present many fluctuations during development (due to gravitropism, variable shoot-derived auxin influx, etc.). As indicated in Figure 7D, in the sigmoidal scenario, natural fluctuations in auxin levels in the CEI/CEID and in endodermal cells could present them with comparable SHR-SCR levels.
A second alternative scenario could be that of a quantitatively different bistable switch. In this scenario, bistability is possible only within a small window of auxin levels, such that high auxin levels trigger the ACD state, whereas slightly lower levels cause the dynamics to fall back to the low SHR-SCR state, thereby removing the requirement of increased protein turnover rates (Figure 7E). This scenario, however, imposes strict requirements on the network dynamics. First, both bifurcations have to be very carefully tuned because the auxin concentration within the cell undergoing the ACD should be high enough to promote the “on” state coming from the “off” state. Concomitantly, the concentration within one of the daughter cells should be low enough to promote the “off” state coming from the “on” state. These requirements impose strong constraints on the kinetics of the network. For a dynamical system to present two states that differ substantially, it tends to be accompanied by a broad window for bistability as well (Figure 7E, inset). A network presenting both very different characteristic states and a narrow window of bistability would require high cooperativity, which makes it very sensitive to parameter changes.
In short, robustness can most readily be achieved through a broad window for bistability (Figures 7F and 7G), which, however, comes with a price—an extra requirement for the resetting of the cells.
Cell-cycle factors also produce a bistable behavior, which is fundamental to establish the reentry in cell cycle after serum deprivation (Yao et al., 2008). In our case, the circuit not only provides bistability but also the spatial and temporal constraints that attenuate it to make it specific to the stem cell area. Given the advantages of an attenuated bistable switch over other mechanisms, it will be interesting to find out whether this regulatory cassette forms a recurring basic regulatory circuit among others (Alon, 2007). We propose the term “flip-flop circuit” for this and similar genetic networks.
There remain at least two issues to be addressed to further match our model’s predictions and experimental observations. First, our model predicts that high levels of SHR-SCR activity should also form within the QC. As this is not the case, additional network components within the QC may suppress the high activity levels of SHR-SCR complex at that location. The QC contains a modified transcriptional program (Sarkar et al., 2007), and it remains to be elucidated how this program represses the ACD-promoting activity of the SCR complex. Second, targets of SHR and SCR such as NUC and MGP are themselves required for SHR movement (Welch et al., 2007). This indicates that more intricate feedback loops may be present in the circuit described here, whose analysis will refine our understanding of its function.
Finally, it has recently been shown how plant transcription factors can trigger cytoskeletal rearrangements for division plane reorientation (Dhonukshe et al., 2012). It will be interesting to explore whether the network described here connects to similar mechanistic factors that trigger division plane rotation during asymmetric cell division.
EXPERIMENTAL PROCEDURES
Microscopic Analysis
Arabidopsis thaliana plants were grown as described in Sabatini et al. (2003) (see Extended Experimental Procedures). IAA/NPA treatments were done for 24 to 48 hr in liquid media similarly to Pérez-Torres et al. (2008). For CEI frequency measurements, plants were grown and roots were prepared for confocal microscopy as described (Nieuwland et al., 2009). To test 26S-proteasome-dependent protein degradation, 4 days postgermination (dpg) seedlings were transferred to liquid MS media supplemented with 50 μM MG132 (Sigma-Aldrich). Similar conditions were used to enable visualization of CDKB1;1::GFP protein fusion. For confocal microscopy, roots were mounted in 10 μM propidium iodide. Root tissue sections were performed as in Willemsen et al. (1998) and were visualized by using Nomarski optics. Casparian strip staining with berberine hemisulphate (Sigma) is described in Lux et al. (2005).
Yeast Two-Hybrid Assay
Yeast two-hybrid (Y2H) interactions were studied by using the ProQuest Two-Hybrid System (Invitrogen Life Technologies). Coding sequences of RBR, SCRAxCxA, and E2FA were amplified and fused to both pDEST32 BD and pDEST22 AD vectors. Compatible Y2H constructs for SCR and SHR were previously generated, and Y2H analyses were performed as described (Welch et al., 2007). To quantify interaction strengths, three experimental and technical replicates of β-galactosidase assays, with CPRG as substrate, were performed.
Bimolecular Fluorescence Complementation Assays
For BiFC analysis, we subcloned RBR and E2Fa cDNAs in vectors pARC233, pARC234, pARC235, and pARC236 by Gateway LR reactions to generate C- and N-terminal fusions to the two YFP fragments. SHR and SCR were generated as previously described (Welch et al., 2007). YFP fluorescence was recorded by using a Leica SP2 CLSM. Col-0 mesophyll protoplasts were transfected according to Yoo et al. (2007). Results are from three biological replicates with three technical replicates each.
For split Renilla luciferase analysis, cDNAs were cloned by Gateway LR recombination into pDuExB and pDuExP vectors (Fujikawa and Kato, 2007). A minimum of two biological replicates of 50,000 mesophyll protoplasts were transfected in triplicate with 2 μg of each DNA and 1 μg of transfection control. Proteins were extracted and analyzed for firefly and renilla luciferase activity with the Dual Luciferase Reporter Assay System (Promega) in a Glomax 96 microplate luminometer (Promega). Relative luciferase activity was calculated as described (Ozawa et al., 2001). Results were expressed as percentage of average H2A-H2B interaction.
Chromatin Immunoprecipitation
ChIP was carried out on root material of 5-day-old Col-0 seedlings. IP was performed in the absence (negative control) and presence of antibody specific for AtRBR1 protein as described in Horváth et al. (2006). NUC promoter qPCR primers were designed to amplify fragments between 100 and 200 bp spanning the 6 kb promoter region of the gene and were designed to be in ascending order upstream away from the NUC ATG (see Table S5). Enrichment was calculated by comparison of samples after IP with and without antibody. We used as negative control a random intergenic region (IR) (between At3g03660–70) and as positive control, the promoter region of PCNA1 (At1g07370) (Kosugi and Ohashi, 2002). Each primer pair was tested with a minimum of two biological replicates, with three technical replicates each. Student’s t tests were performed to analyze statistical significance.
Kinase Assays
For protein kinase assays, CDK-cyclin complexes were immunoprecipitated with anti-c-Myc antibody from lysate of transfected Arabidopsis protoplasts. Immunocomplexes were washed as described in Extended Experimental Procedures and were washed once with kinase buffer containing 25 mM Tris-HCl (pH 7.8), 15 mM MgCl2, and 1 mM DTT. Phosphorylation assays were performed in 20 μl kinase buffer supplemented with 0.25 mg ml−1 histone H1 or 0.06 mg ml−1 GST-RBR-C-terminal protein and 5 μCi of [γ-32P]ATP (Perkin Elmer) for 40 min at room temperature and then terminated by the addition of Laemmli sample buffer. Reaction mixtures were separated by SDS-PAGE and stained with CBB, and substrate phosphorylation was revealed by autoradiography. For details in other methods, see Extended Experimental Procedures.
Regulatory Network Modeling
Regulatory networks were formulated as coupled ordinary differential equations or were embedded within a spatial context of the root (see Grieneisen et al., 2007), where simulations of auxin dynamics were performed by concurrently solving for diffusion, permeability, and decay of auxin by using an Alternating Direction Implicit (ADI) method (Peaceman and Rachford, 1955). For details on formal descriptions of regulatory networks, see Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Dominique Bergmann for critically reviewing this manuscript; Fred Sack, Lieven de Veylder, Naohiro Kato, Dorus Gadella, Joachim Goedhart, Tom Beeckman, and Tom Bennett for materials; and Anahí Pérez-Torres, Luis Herrera-Estrella, and Juan Carlos del Pozo for technical advice. B.S. was supported by an ERC Advanced Investigator Fellowship and by ALW-ERAPG grant 855.50.017. A.C.-R. was supported by EMBO-ALTF 1114-2006 and CONACYT 000000000092916 grants. S.D.-T. was funded by Ministerio de Educacion y Ciencia, Spain, and by Marie Curie IEF (IEF-2008-237643). V.A.G. was supported by the Dorothy Hodgkin Fellowship. V.A.G. and A.F.M.M. were supported by the UK Biological and Biotechnology Research Council (BBSRC) via grant BB/J004553/1 to the John Innes Centre. I.B. was sponsored by an NWO VIDI grant. P.N.B. was funded by NIH grant R01-GM043778. L.B. was supported by Swedish Research Council and by Carl Tryggers Stiftelse grants. Work in James A.H. Murray laboratory was supported by the BBSRC grant BB/G00482X and by the ERASysBio+ initiative under the EU FP7 ERA-NET Plus scheme.
Footnotes
Supplemental Information includes Extended Experimental Procedures, seven figures, five tables, and four movies and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.07.017.
References
- Abrash EB, Bergmann DC. Asymmetric cell divisions: a view from plant development. Dev Cell. 2009;16:783–796. doi: 10.1016/j.devcel.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Berman SD, Yuan TL, Miller ES, Lee EY, Caron A, Lees JA. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol Cancer Res. 2008;6:1440–1451. doi: 10.1158/1541-7786.MCR-08-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti MB, Gutierrez C. A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 2001;28:341–350. doi: 10.1046/j.1365-313x.2001.01160.x. [DOI] [PubMed] [Google Scholar]
- Borghi L, Gutzat R, Fütterer J, Laizet Y, Hennig L, Gruissem W. Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell. 2010;22:1792–1811. doi: 10.1105/tpc.110.074591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E. Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell. 2010;22:1264–1280. doi: 10.1105/tpc.109.073635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466:1110–1114. doi: 10.1038/nature09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- Chen D, Opavsky R, Pacal M, Tanimoto N, Wenzel P, Seeliger MW, Leone G, Bremner R. Rb-mediated neuronal differentiation through cell-cycle-independent regulation of E2f3a. PLoS Biol. 2007;5:e179. doi: 10.1371/journal.pbio.0050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools T, Iantcheva A, Maes S, Van den Daele H, De Veylder L. A replication stress-induced synchronization method for Arabidopsis thaliana root meristems. Plant J. 2010;64:705–714. doi: 10.1111/j.1365-313X.2010.04361.x. [DOI] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Weits DA, Cruz-Ramirez A, Deinum EE, Tindemans SH, Kakar K, Prasad K, Mähönen AP, Ambrose C, Sasabe M, et al. A PLETHORA-auxin transcription module controls cell division plane rotation through MAP65 and CLASP. Cell. 2012;149:383–396. doi: 10.1016/j.cell.2012.02.051. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Dick FA. Structure-function analysis of the retinoblastoma tumor suppressor protein - is the whole a sum of its parts? Cell Div. 2007;2:26. doi: 10.1186/1747-1028-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa Y, Kato N. Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J. 2007;52:185–195. doi: 10.1111/j.1365-313X.2007.03214.x. [DOI] [PubMed] [Google Scholar]
- Fülöp K, Tarayre S, Kelemen Z, Horváth G, Kevei Z, Nikovics K, Bakó L, Brown S, Kondorosi A, Kondorosi E. Arabidopsis anaphase-promoting complexes: multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle. 2005;4:1084–1092. [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J. Cell cycle-dependent proteolysis in plants. Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor mg132. Plant Cell. 1998;10:2063–2076. doi: 10.1105/tpc.10.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B. Mosaic analysis using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- Horváth BM, Magyar Z, Zhang Y, Hamburger AW, Bakó L, Visser RG, Bachem CW, Bögre L. EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 2006;25:4909–4920. doi: 10.1038/sj.emboj.7601362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 2002;29:45–59. doi: 10.1046/j.1365-313x.2002.01196.x. [DOI] [PubMed] [Google Scholar]
- Krek W, Ewen ME, Shirodkar S, Arany Z, Kaelin WG, Jr, Livingston DM. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AFM, Scheres B. Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 2008;6:e307. doi: 10.1371/journal.pbio.0060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E, Xie D, Grava S, Pigaglio E, Planchais S, Murray JAH, Parmentier Y, Mutterer J, Dubreucq B, Shen WH, Genschik P. The AtRbx1 protein is part of plant SCF complexes, and its down-regulation causes severe growth and developmental defects. J Biol Chem. 2002;277:50069–50080. doi: 10.1074/jbc.M204254200. [DOI] [PubMed] [Google Scholar]
- Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- Lendvai A, Pettkó-Szandtner A, Csordás-Tóth E, Miskolczi P, Horváth GV, Györgyey J, Dudits D. Dicot and monocot plants differ in retinoblastoma-related protein subfamilies. J Exp Bot. 2007;58:1663–1675. doi: 10.1093/jxb/erm022. [DOI] [PubMed] [Google Scholar]
- Lux A, Morita S, Abe J, Ito K. An improved method for clearing and staining free-hand sections and whole-mount samples. Ann Bot (Lond) 2005;96:989–996. doi: 10.1093/aob/mci266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A. Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell. 2002;14:1847–1857. doi: 10.1105/tpc.002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam RL, Andreu-Vieyra C, Braun RE, Akiyama H, Matzuk MM. Retinoblastoma protein plays multiple essential roles in the terminal differentiation of Sertoli cells. Mol Endocrinol. 2009;23:1900–1913. doi: 10.1210/me.2009-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland J, Maughan S, Dewitte W, Scofield S, Sanz L, Murray JAH. The D-type cyclin CYCD4;1 modulates lateral root density in Arabidopsisby affecting the basal meristem region. Proc Natl Acad Sci USA. 2009;106:22528–22533. doi: 10.1073/pnas.0906354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Kaihara A, Sato M, Tachihara K, Umezawa Y. Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Anal Chem. 2001;73:2516–2521. doi: 10.1021/ac0013296. [DOI] [PubMed] [Google Scholar]
- Peaceman DW, Rachford HH., Jr The numerical solution of parabolic and elliptic differential equations. J Soc Indust Appl Math. 1955;3:28–41. [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell. 2008;20:3258–3272. doi: 10.1105/tpc.108.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchais S, Samland AK, Murray JA. Differential stability of Arabidopsis D-type cyclins: CYCD3;1 is a highly unstable protein degraded by a proteasome-dependent mechanism. Plant J. 2004;38:616–625. doi: 10.1111/j.0960-7412.2004.02071.x. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, Scheres B. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Dewitte W, Forzani C, Patell F, Nieuwland J, Wen B, Quelhas P, De Jager S, Titmus C, Campilho A, et al. The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell. 2011;23:641–660. doi: 10.1105/tpc.110.080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Segers G, Gadisseur I, Bergounioux C, de Almeida Engler J, Jacqmard A, Van Montagu M, Inzé D. The Arabidopsis cyclin-dependent kinase gene cdc2bAt is preferentially expressed during S and G2 phases of the cell cycle. Plant J. 1996;10:601–612. doi: 10.1046/j.1365-313x.1996.10040601.x. [DOI] [PubMed] [Google Scholar]
- Sena G, Jung JW, Benfey PN. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development. 2004;131:2817–2826. doi: 10.1242/dev.01144. [DOI] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–132. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi I, Kojima S, Sakaguchi N, Umeda-Hara C, Umeda M. Two Arabidopsis cyclin A3s possess G1 cyclin-like features. Plant Cell Rep. 2010;29:307–315. doi: 10.1007/s00299-010-0817-9. [DOI] [PubMed] [Google Scholar]
- Templeton DJ, Park SH, Lanier L, Weinberg RA. Nonfunctional mutants of the retinoblastoma protein are characterized by defects in phosphorylation, viral oncoprotein association, and nuclear tethering. Proc Natl Acad Sci USA. 1991;88:3033–3037. doi: 10.1073/pnas.88.8.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007;21:2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Willemsen V, Wolkenfelt H, de Vrieze G, Weisbeek P, Scheres B. The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development. 1998;125:521–531. doi: 10.1242/dev.125.3.521. [DOI] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B. A molecular framework for plant regeneration. Science. 2006;311:385–388. doi: 10.1126/science.1121790. [DOI] [PubMed] [Google Scholar]
- Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.