Summary
To ensure equal chromosome segregation during mitosis, the macromolecular kinetochore must remain attached to depolymerizing microtubules, which drive chromosome movements. How kinetochores associate with depolymerizing microtubules, which undergo dramatic structural changes forming curved protofilaments, has yet to be defined in vertebrates. Here, we demonstrate that the conserved kinetochore-localized Ska1 complex tracks with depolymerizing microtubule ends and associates with both the microtubule lattice and curved protofilaments. In contrast, the Ndc80 complex, a central player in the kinetochore-microtubule interface, binds only to the straight microtubule lattice and lacks tracking activity. We demonstrate that the Ska1 complex imparts its tracking capability to the Ndc80 complex. Finally, we present a structure of the Ska1 microtubule binding domain that reveals its interaction with microtubules and its regulation by Aurora B. This work defines an integrated kinetochore-microtubule interface formed by the Ska1 and Ndc80 complexes that associates with depolymerizing microtubules, potentially by interacting with curved microtubule protofilaments.
Keywords: Mitosis, Kinetochore, Microtubule, Winged helix domain
Introduction
In eukaryotes, the physical distribution of chromosomes to the daughter cells during mitosis requires a force-generating connection between chromosomal DNA and spindle microtubules. The multi-protein kinetochore complex, comprised of more than 100 different proteins in human cells, assembles upon centromeric DNA to mediate microtubule interactions (Cheeseman and Desai, 2008). Although several molecular motors localize to kinetochores, work over the past two decades has suggested that the primary driver of chromosome movement is microtubule dynamics (McIntosh et al., 2010). During microtubule polymerization, GTP-bound tubulin dimers are incorporated into the microtubule lattice. Upon assembly, GTP is rapidly hydrolyzed to GDP (Desai and Mitchison, 1997). GDP-tubulin has a curved conformation relative to GTP-tubulin, but is forced into a straight conformation in the context of the microtubule lattice (Wang and Nogales, 2005). Thus, microtubules store potential energy that is released during microtubule depolymerization when GDP-tubulin subunits exposed at the microtubule end undergo a conformational change resulting in the peeling away of individual microtubule protofilaments. These bending protofilaments have been demonstrated to generate force with a theoretical maximum of 65 pN per microtubule (Grishchuk et al., 2005). However, to harness this force and to move processively, kinetochores must remain associated with these dramatically changing structures.
A key challenge has been to define the specific molecules that mediate kinetochore-microtubule interactions, and to determine the mechanisms by which these proteins remain associated with depolymerizing microtubules. The central player at the kinetochore-microtubule interface is the conserved Ndc80 complex, a component of the larger KNL1/Mis12 Complex/Ndc80 Complex (KMN) network (Cheeseman et al., 2006). Loss of Ndc80 function results in catastrophic defects in kinetochore-microtubule attachments (DeLuca et al., 2002). The Ndc80 complex has been shown to track with polymerizing and depolymerizing microtubules when artificially oligomerized on the surface of a microsphere (Powers et al., 2009). However, monomeric Ndc80 complex lacks this activity. In addition to the Ndc80 complex, the fungal Dam1 complex plays a critical role in chromosome segregation in budding yeast and is capable of forming striking ring-like structures around microtubules (Miranda et al., 2005; Westermann et al., 2006), although the existence of this ring in cells has been debated. In budding yeast, the Ndc80 and Dam1 complexes work together to mediate connections to microtubules, with the Dam1 complex facilitating processive interactions of the Ndc80 complex with microtubules (Lampert et al., 2010; Tien et al., 2010). However, the Dam1 complex is absent from metazoa, including humans, and the nature of an integrated kinetochore-microtubule interface capable of processively associating with depolymerizing microtubules has yet to be defined in vertebrates.
We and others have previously implicated the human Ska1 complex as an important kinetochore component involved in the formation of kinetochore-microtubule interactions (Daum et al., 2009; Gaitanos et al., 2009; Hanisch et al., 2006; Raaijmakers et al., 2009; Theis et al., 2009; Welburn et al., 2009). Depletion of the Ska1 complex from human cells results in a checkpoint-dependent mitotic arrest with mis-aligned chromosomes. However, the mechanisms by which the Ska1 complex acts to mediate kinetochore-microtubule interactions were unclear. Here, we took combined biochemical, biophysical, structural, and cell biological approaches to analyze the functions and activities of the Ska1 complex for its interactions with microtubules. Based on our data, we propose a model in which the distinct biochemical activities of the Ska1 and Ndc80 complexes form an integrated interface to couple chromosome movement to microtubule depolymerization.
Results
The C-terminus of Ska1 contains the conserved microtubule-binding domain of the Ska1 complex
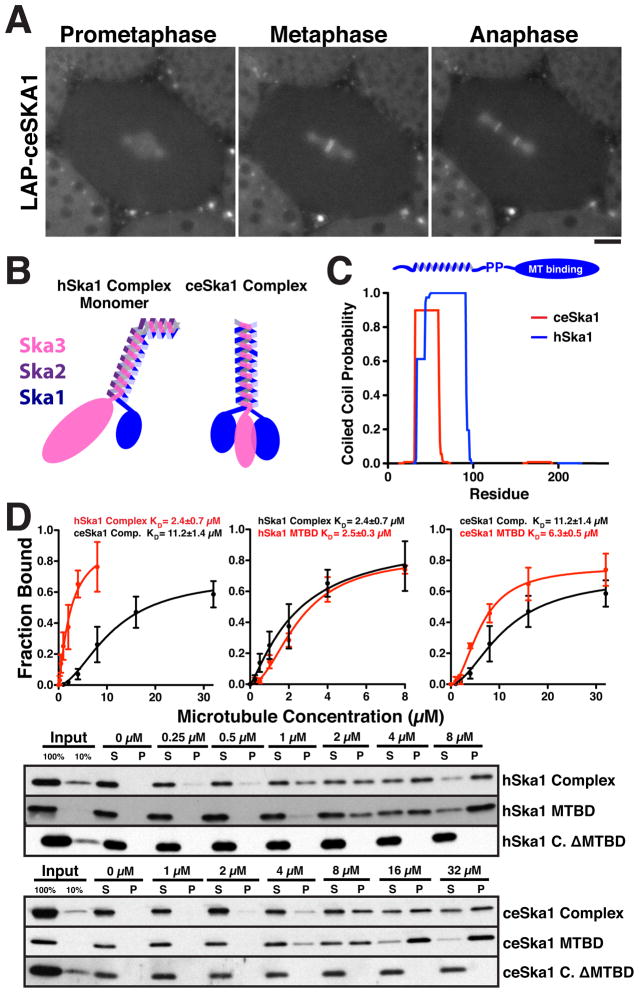
The Ska1 complex is an integral component of the kinetochore-microtubule interface in human cells, but to date the Ska1 complex has not been described outside of vertebrates. To define the conserved biochemical properties of the Ska1 complex, we sought to identify divergent homologues of Ska1. Homology-based searches identified C. elegans SKA-1 (Y106G6H.15; Fig. S1A). Based on RNAi experiments, SKA-1 is not essential for embryonic viability in C. elegans (Arshad Desai, personal communication). Similar to human Ska1 (Welburn et al., 2009), GFP-SKA-1 localized to the mitotic spindle and kinetochores from prometaphase through anaphase (Fig. 1A; Movie S1). One-step immunoprecipitations of GFP-SKA-1 from C. elegans adults identified a complex of SKA-1 and F54E7.8 (Fig. S1B), which displays limited sequence similarity to Ska3 (data not shown). We did not identify a counterpart to Ska2. Sedimentation velocity analytical ultracentrifugation (SVAUC) of the reconstituted C. elegans Ska1 complex demonstrated that it is composed of two copies of SKA-1 and a single copy of SKA-3 (Fig. 1B; Fig. S1D, E). In contrast, our previous biochemical analysis (Welburn et al., 2009) and recent structural data (Jeyaprakash et al., 2012) suggests that the human Ska1 complex is composed of two copies each of Ska1, Ska2, and Ska3 (Fig. 1B).
Figure 1. The C-terminus of Ska1 is the conserved microtubule-binding domain of the Ska1 complex.
(A) GFP-ceSKA1 localizes to the mitotic spindle and kinetochores throughout mitosis. Fluorescence images from different mitotic stages of a one-cell C. elegans embryo stably expressing GFP-ceSKA-1. Scale bar, 5 μm. (B) Models for the molecular composition of the human and C. elegans Ska1 complexes based on (Jeyaprakash et al., 2012; Welburn et al., 2009) and Fig. S1. (C) Graph showing the predicted probability for formation of a coiled-coil structure for hSka1 and ceSKA-1. The N-terminus of Ska1 contains a conserved coiled-coil region. (D) Quantification (Mean ± SD, n = 3, Dissociation constants ± SE) (top) and Western blots (bottom) showing microtubule co-sedimentation for the human and C. elegans Ska1 complexes, the full Ska1 complexes lacking the C-terminal microtubule binding domain (MTBD) in Ska1, and the Ska1 microtubule binding domain alone (100 nM, 75 mM KCl). Western blots were probed with antibodies against hSka1 or the full ceSKA1 complex. The C-terminus of Ska1 protein is necessary and sufficient for Ska1 complex microtubule binding. (See also Figure S1)
The identification of the divergent C. elegans Ska1 complex revealed that the C-terminal half of Ska1 is the most highly conserved (Fig. S1A), suggesting that this may represent the microtubule-binding domain, whereas the N-terminus contains a conserved coiled-coil (Fig. 1C). In microtubule co-sedimentation assays, the reconstituted C. elegans and human Ska1 complexes both bound to taxol stabilized microtubules with apparent dissociation constants of 11.3 μM and 2.4 μM, respectively (Fig. 1D; Fig. S1E). In addition, the C-terminus of both human Ska1 (residues 132–255, KD = 2.5 μM) and C. elegans SKA-1 (residues 118–243, KD = 6.3 μM) bound to microtubules directly (Fig. 1D, S1E), whereas complexes lacking the Ska1 C-terminus failed to bind to microtubules (Fig. 1D), but did not affect complex formation (Fig. S2A, B). Importantly, all antibodies used in this study have similar standard curves in Western blots, allowing us to reliably determine relative affinities (Fig. S1F). These results demonstrate that the C-terminal half of Ska1 is the conserved microtubule-binding domain of the Ska1 complex.
Ska1 microtubule binding is essential for mitotic progression and the formation of robust kinetochore-microtubule attachments
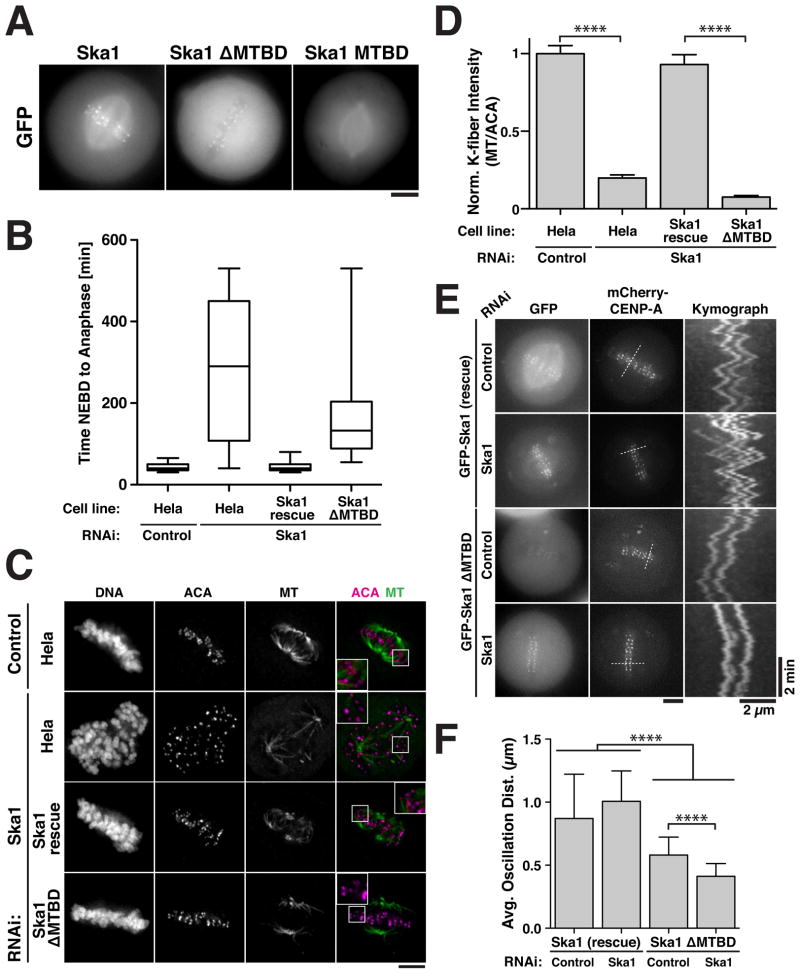
We next sought to determine the contribution of the Ska1 microtubule-binding activity to chromosome segregation. Full-length GFP-Ska1 localizes to kinetochores and the mitotic spindle (Fig. 2A). In contrast, the hSka1 N-terminus lacking the microtubule binding domain (Ska1 ΔMTBD; residues 1–131) localized to kinetochores, but not spindle microtubules (Fig. 2A). GFP-Ska1 containing only the microtubule-binding domain (residues 132–255) localized to the mitotic spindle, but not kinetochores (Fig. 2A). Depletion of any Ska1 complex subunit leads to the loss of function and delocalization of the entire complex, resulting in a mitotic arrest with scattered chromosomes and the loss of kinetochore-microtubule attachments (Daum et al., 2009; Gaitanos et al., 2009; Hanisch et al., 2006; Raaijmakers et al., 2009; Theis et al., 2009; Welburn et al., 2009). Full length GFP-Ska1 efficiently rescued depletion of endogenous Ska1 (Fig. 2B–F). In contrast, RNAi-based replacement of endogenous Ska1 with GFP-Ska1 ΔMTBD resulted in a significant increase in the time from nuclear envelope breakdown (NEBD) to anaphase onset to 174 min (Fig. 2B) compared to 44 minutes for controls or cells in which full length GFP-Ska1 replaces the endogenous protein (Fig. 2B). Following depletion of endogenous Ska1, the majority of cells expressing GFP-Ska1 ΔMTBD contained aligned chromosomes, demonstrating that the Ska1 microtubule binding activity is not required for initial chromosome alignment (Fig. 2C). However, cells expressing GFP-Ska1 ΔMTBD displayed strongly reduced cold stable kinetochore fibers (K-fibers; Fig. 2C and 2D), which monitor the stability of kinetochore-microtubule interactions. In addition, based on time-lapse analysis, we found that sister chromatid oscillations were significantly impaired in cells lacking the Ska1 microtubule binding domain (Fig. 2E and 2F, Movie S2). We also observed modest defects in mitotic timing, cold stable kinetochore-fibers, and sister chromatid oscillations for GFP-Ska1 ΔMTBD in the presence of the endogenous proteins (Fig. 2) consistent with dominant negative effects due to the dimerization of the Ska1 complex. These results demonstrate that the microtubule binding activity of Ska1 is necessary for the formation of robust kinetochore-microtubule interactions and proper kinetochore movement, which in turn are required for timely mitotic progression.
Figure 2. The microtubule-binding activity of the Ska1 complex is required for mitotic progression and kinetochore-microtubule attachments.
(A) Fluorescent images showing the localization of GFP-tagged hSka1 constructs into HeLa cells. Full length GFP-Ska1 localizes to kinetochores and the mitotic spindle. GFP-tagged hSka1 lacking the MTBD (residues 1–131) localizes only to kinetochores, whereas the MTBD (residues 132–255) localizes exclusively to the mitotic spindle. Scale bar, 5 μm. (B) HeLa cell lines stably expressing GFP-tagged and RNAi resistant full-length Ska1 or Ska1 ΔMTDB were depleted for endogenous Ska1 by RNAi. Cells were imaged by time-lapse microscopy 36 h after siRNA transfection and the time from nuclear envelope breakdown (NEBD) to anaphase was quantified (Box plot showing range, median, lower and upper quartile, n = 42–58 cells per condition). (C) Immunofluorescence of HeLa cells stably expressing LAP-Ska1 or LAP-Ska1 ΔMTDB depleted for endogenous Ska1 following cold treatment for 10 min. Cells were immuno-stained against centromeres (ACA, magenta), microtubules (DM1α, green), and DNA. (D) Quantification of kinetochore-microtubule intensity shown in (C) (Mean ± SEM, 18–22 cells per conditions, 10 kinetochores per cell, P < 0.0001, t-test). Kinetochores were chosen at random and adjacent microtubule intensity was quantified and normalized using the ACA signal. (E) Live cell fluorescence images and kymograph showing the oscillation of sister kinetochores in HeLa cells stably expressing LAP-Ska1 or LAP-Ska1 ΔMTDB and mCherry CENP-A depleted for endogenous Ska1. (F) Quantification of sister chromatid oscillations from (E) measuring the distance that a kinetochore pair moved prior to reversing direction (Mean ± SD, LAP-Ska1 Control n = 7 cells, 129 oscillations, Ska1 RNAi n = 6 cells, 119 Oscillations; LAP-Ska1 ΔMTDB Control n = 11 cells, 178 oscillations, Ska1 RNAi n = 7 cells, 199 Oscillations; P < 0.0001, t-test).
The Ska1 complex diffuses on microtubules as complex containing multiple microtubule binding sites
To analyze the microtubule-binding activity of individual molecules of the human and C. elegans Ska1 complexes, we next visualized the Ska1 complex containing GFP-Ska1 using total internal reflection fluorescence microscopy (TIR-FM; Fig. 3A). Human GFP-Ska1 complex readily diffused on microtubules (Fig. 3B; Movie S3A), similar to the human Ndc80-GFP complex (Fig. 3B; Movie S3B). We note that for the biochemical experiments in this paper, we used well-behaved truncated versions of Ndc80 and Nuf2 that include all microtubule-binding and adjacent regions and behaves identically to full-length Ndc80 complex (termed Ndc80 “Broccoli”; see Fig. S2D, E). Based on the intensity distributions and the lack of bleaching events in our TIRF analysis, hNdc80 “Broccoli”-GFP is mostly monomeric on microtubules (Fig. 3C; also see Powers et al., 2009). In contrast, the GFP-hSka1 complex intensity distribution displayed two distinct peaks; particles with a mean brightness similar to the Ndc80-GFP complex, and particles that have a two-fold increased intensity (Fig. 3C), suggesting that GFP-hSka1 complex is present on microtubules as monomers and dimers. Indeed, time-lapse analysis of GFP-hSka1 complex binding events revealed bleaching steps with the intensity value of a single GFP fluorophore (Fig. 3D), confirming the presence of two GFP-Ska1 molecules. This is consistent with our previous biochemical analysis (Welburn et al., 2009) and recent structural work (Jeyaprakash et al., 2012) indicating that the hSka1 complex dimerizes to generate a complex with two microtubule binding sites. In contrast, in the absence of Ska3, which is required to dimerize the Ska1 complex (Welburn et al., 2009), the GFP-hSka1/2 complex behaved as a monomer (Fig. S3A, B, Movie S3C). GFP-hSka1 complex, GFP-hSka1/2, and Ndc80 “Broccoli”-GFP diffused on microtubule with diffusion coefficients of 0.09 μm2/s, 0.18 μm2/s, and 0.03 μm2/s respectively (Fig. 3E, S3C), consistent with previous observations made for the Ndc80 complex (Powers et al., 2009).
Figure 3. The Ska1 complex diffuses on microtubules.
(A) Schematic of the TIRF setup used to visualize microtubules and individual microtubule binding proteins. (B) Representative kymographs with microtubule position along the horizontal axis and time along vertical axis showing one-dimensional diffusion of GFP-hSka1 complex (left, 100 pM) and Ndc80 “Broccoli”-GFP (right, 50 pM) on taxol-stabilized microtubules labeled with HiLyte 647. (C) Intensity distribution of the binding events for 100 pM GFP-hSka1 complex (Mean ± SD, red, n = 20676) and 50 pM Ndc80 “Broccoli”-GFP (Mean ± SD, blue, n = 13411). Intensities were determined by fitting a 2D Gaussian distribution to each identified spot, ignoring first and last frames of tracks. (D) Photo-bleaching analysis of Ska1 complex particles. Left, kymograph and corresponding fluorescent intensity for one photobleaching event. Right, graph showing the average intensity before and after photobleaching events (Mean ± SD, n = 32). (E) Mean square displacement (MSD, Mean ± SEM) plotted against time for 100 pM GFP-hSka1 complex (red, n = 187) and 50 pM Ndc80 “Broccoli”-GFP (blue, n = 258). Diffusion coefficients were calculated from the slope of the linear fit of the MSD. (F) Distribution of the residence times of GFP-hSka1 complex (100 pM, n = 502) and hNdc80 “Broccoli”-GFP (50 pM, n = 359) binding events to taxol stabilized microtubules (Rate constants ± SE). (See also Figure S3)
Under the conditions tested, the residence time distribution of GFP-hSka1 complex displays bi-phasic dissociation kinetics (Fig. 3E) - a rapid phase with a rate constant of kfast = 3.4 ± 0.3 s−1 and a slow phase with a rate constant of kslow = 0.15 ± 0.02 s−1. The disassociation rate of the rapid phase is similar to the behavior of monomeric GFP-hSka1/2 (koff = 4.9 ± 0.4 s−1) (Fig. S3D, E), suggesting that the rapid phase represents hSka1 complex monomers. Indeed, analysis of short-lived (<0.6 s) hSka1 complex binding events indicated that these were primarily monomeric based on their intensity distribution (Fig. S3F). Ndc80 “Brocolli”-GFP has a dissociation constant of koff = 0.53 ± 0.03 s−1 (Fig. 3F). All measured dissociation rate constants were faster than the bleaching rate constant determined for immobilized Ndc80-GFP under identical imaging conditions (kbleach = 0.11 ± 0.01 s−1; data not shown).
Similar to the hSka1 complex, the C. elegans GFP-ceSKA1 complex also associates with microtubules as a mixture of dimers and monomers. However, in contrast to hSka1 complex, the sfGFP-ceSKA1 complex was primarily monomeric under the tested conditions (Fig. S3G–J, Movie S3D). Manual inspection of the less abundant long-lived binding events (white arrow, Fig. S3G) revealed that their intensity was approximately 2-fold higher compared to monomeric ceNdc80-sfGFP (Fig. S3I and J, Movie S3E), consistent with the presence of two ceSKA1 subunits. We note that sfGFP-ceSKA1 was used in these assays at 40-fold higher concentrations (4 nM) relative to GFP-hSka1 complex (100 pM). We conclude that even at concentrations of 4 nM, the ceSKA1 complex primarily contains one ceSKA1 subunit, indicating that it dissociates under these conditions.
The human (D = 0.09 μm2/s) and C. elegans (D = 0.11 μm2/s) Ska1 complex both diffused on microtubules with similar diffusion coefficients (Fig. 3E, S3K), whereas the C. elegans Ndc80 complex (D = 0.08 μm2/s) diffuses more rapidly than its human counterpart (D = 0.03 μm2/s) (Fig. 3E, S3K). Furthermore, dimeric sfGFP-ceSKA1 binding events dissociate more rapidly (koff = 0.42 s−1) from microtubules than GFP-hSka1 complex dimers (kslow = 0.15 s−1) (Fig. 3F, S3L), consistent with the lower affinity of the ceSKA1 complex in microtubule pelleting assays. In contrast the ceNdc80 “Broccoli” has a lower dissociating constant (koff = 0.34 s−1) than hNdc80 “Broccoli” (koff = 0.53 s−1).
Thus, even at low concentrations (100 pM) the human Ska1 complex can bind to microtubules as a dimer containing two Ska1 subunits. These observations do not exclude the formation of higher order oligomers at higher protein concentrations as we have previously reported (Welburn et al., 2009). In total, these results demonstrate that diffusion on the microtubule lattice is a conserved property of the Ska1 complex.
The Ska1 complex tracks with depolymerizing microtubule ends
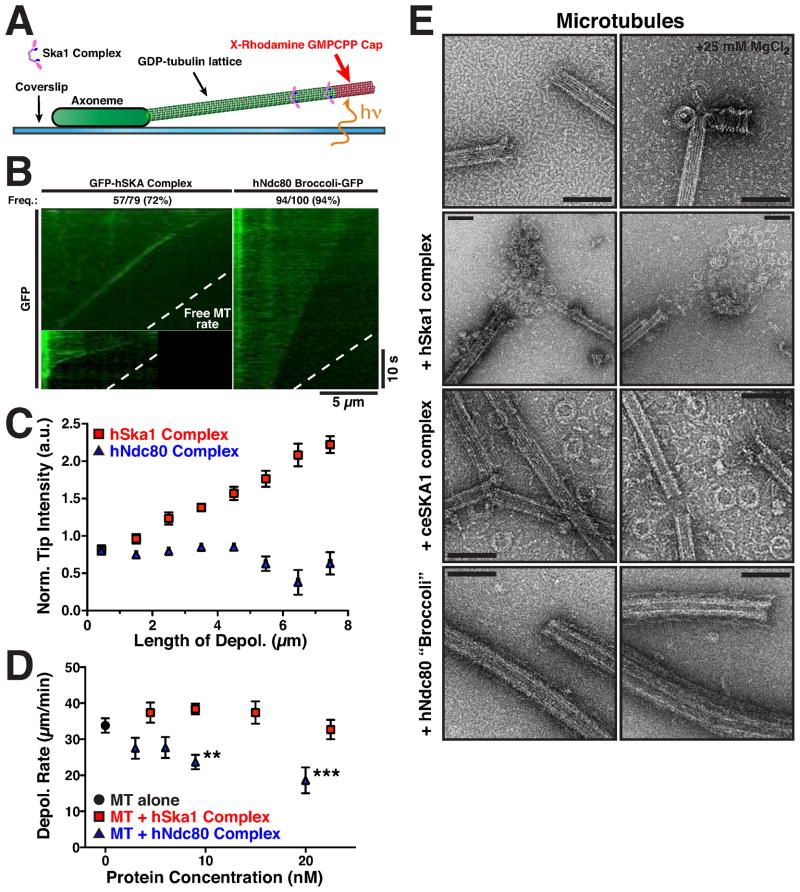
Previous work demonstrated that the Ska1 complex can couple movement of 0.5 μm microspheres to microtubule depolymerization (Welburn et al., 2009), but the mechanism by which this occurs was unknown. Previous work has shown that some microtubule associated proteins facilitate bead “rolling” when conjugated to the bead surface (Grishchuk et al., 2008), an activity that is unlikely to be physiologically relevant at kinetochores. To test whether the Ska1 complex itself can remain associated with depolymerizing microtubule ends, we visualized GFP-hSka1 complex bound to microtubules that were induced to depolymerize by ablation of a stabilizing GMPCPP-cap (Fig. 4A). We found that GFP-hSka1 complex accumulated at microtubule ends during the majority of depolymerization events observed (70%; n = 79). In most cases, the hSka1 complex tracked processively with the depolymerizing end until it reached the microtubule seed (Fig. 4B; Movie S4). The hSka1 complex present at the microtubule end consistently increased intensity throughout the time course of microtubule depolymerization (Fig. 4C, Movie S4) suggesting that an accumulation of GFP-Ska1 complexes from the microtubule lattice is occurring at the depolymerizing end. In contrast, hNdc80 “Broccoli”-GFP diffusely decorated microtubules and failed to track with depolymerizing microtubule ends (Fig. 4B, Movie S5). We also did not observe binding of the ceSKA1 complex to microtubules under these experimental conditions, most likely due to dissociation of the ceSKA1 complex at the low (nM) protein concentrations required for this experiment (see above).
Figure 4. The Ska1 complex tracks with depolymerizing microtubule ends and induces the formation of curved microtubule structures.
(A) Schematic of the experimental setup used to visualize the GFP-hSka1 complex binding to depolymerizing microtubules. Microtubules were induced to depolymerize by ablating a stabilizing GMPCPP cap with excitation light. (B) Representative kymographs showing the binding of GFP-hSka1 complex (left, 9 nM) and Ndc80 “Broccoli”-GFP (right, 9 nM) to depolymerizing microtubules (Dashed line indicates median protein-free microtubule depolymerization rate). Tracking events were identified as kymographs with substantial periods of an increased fluorescence signal at the microtubule end relative to the lattice. (C) Graph showing the normalized intensity of the GFP-hSka1 (n = 47-108) complex or GFP-Ndc80 complex (Mean ± SEM, n = 4–72) at the depolymerizing microtubule end over time. The Ska1 complex accumulates constantly at the depolymerizing end. (D) Depolymerization rates of microtubules in the presence and absence of a range of GFP-hSka1 complex and Ndc80 “Broccoli”-GFP concentrations (Mean ± SEM, n = 18–87). The presence of of 9 nM (P < 0.01) and 20 nM (P < 0.001) Ndc80 “Broccoli”-GFP significantly reduces the rate of microtubule depolymerization (Kruskal-Wallis test with Dunn’s post test), which is unaffected by similar concentrations of the Ska1 complex. Depolymerization rates were determined from the slopes of the corresponding kymographs. (E) TEM images of the dynamic microtubules in the absence and presence of 25 MgCl2, or the Ska1 and Ndc80 complexes (Scale bars, 100 nm). The Ska1, but not the Ndc80 complex, induces the formation of protofilament rings.
In these depolymerization assays, we found that addition of ≥9 nM Ndc80 “Broccoli”-GFP significantly reduced the depolymerization rate of microtubules from 33.8 μm/min to 23.7 μm/min (Fig. 4D), whereas addition of the Ska1 complex did not retard microtubule depolymerization. Together with the lack of tracking activity for the Ndc80 complex, this suggests that the interactions of the Ska1 and Ndc80 complexes with depolymerizing microtubules are fundamentally different. In total, these results demonstrate that the hSka1 complex can processively track with depolymerizing microtubules.
The Ska1 complex induces the formation of curved microtubule structures
The experiments described above demonstrate that the Ska1 complex can robustly track with depolymerizing microtubules (Fig. 4B). To structurally define the interaction between the Ska1 complex and depolymerizing microtubules, we incubated dynamic microtubules in the presence of Ska1 complex. Transmission electron microscopy (TEM) analysis demonstrated that addition of equimolar amounts of either the human or C. elegans Ska1 complex to microtubules resulted in the formation of numerous protofilament rings or spiral structures, often in direct proximity to microtubule ends (Fig. 4E). Spiral-like structures can be induced by addition of 25 mM MgCl2, although in this case the spirals are typically directly connected to the microtubule end (Fig. 4F; also see Mandelkow et al., 1991). In contrast, such structures were rarely observed for microtubules alone or microtubules incubated in the presence of the Ndc80 complex (Fig. 4E). These results demonstrate that the Ska1 complex has the conserved ability to induce and stabilize the formation of protofilament rings or spiral-like tubulin structures.
The Ska1 complex directly associates with curved microtubule structures
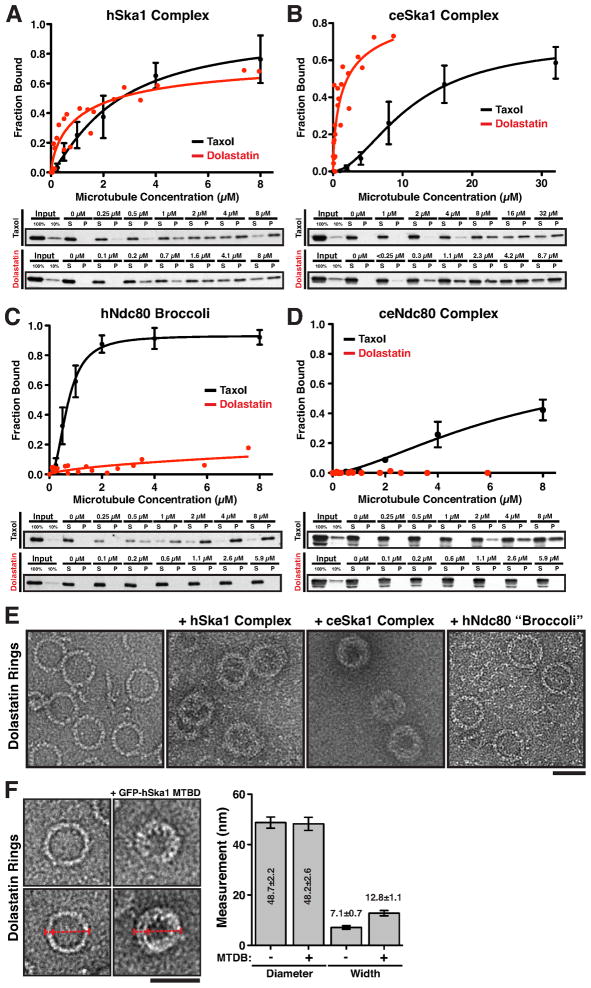
The Ska1 complex induces the formation of ring-like protofilament structures when incubated with dynamic microtubules (Fig. 4E). Previous work on microtubule tracking proteins has focused on their association with the straight microtubule lattice proximal to the depolymerizing microtubule end. However, the depolymerizing end, which consists of bent microtubule protofilaments (Mandelkow et al., 1991), also represents a potential site of interaction to remain associated with depolymerizing microtubules. To assess the relative affinities of the Ska1 complex binding to straight and curved microtubule structures, we compared the binding of the human and C. elegans Ska1 complexes to straight taxol-stabilized microtubules and dolastatin-10-induced tubulin rings, which mimic curved protofilament structures (Bai et al., 1999; Boukari et al., 2007). Under equivalent conditions, the hSka1 complex bound with similar affinities to both taxol-stabilized microtubules and dolastatin rings (Fig. 5A), whereas the ceSka1 complex bound significantly better to dolastatin-10 induced microtubule rings (Fig. 5B). In contrast, both the human and C. elegans Ndc80 complexes bound preferentially to straight microtubules (Fig. 5C, 5D; also see Alushin et al., 2010).
Figure 5. The Ska1 complex, but not the Ndc80 complex, associates with depolymerizing microtubule end mimics.
(A–D) (Top) Quantification of microtubule co-sedimentation assays (Mean ± SD, n = 3) and (Bottom) representative Western blots of microtubule co-sedimentation assays of (A) hSka1 complex (100 nM, 75 mM KCl), (B) ceSKA1 complex (100 nM, 75 mM KCl), (C) hNdc80 “Broccoli” (100 nM, 75 mM KCl), and (D) ceNdc80 complex (100 nM, 75 mM KCl) binding to taxol stabilized microtubules (black) and dolestatin-10 induced protofilament rings (red). (E) Transmission electron microscopy (TEM) images of dolestatin-10 induced protofilament rings alone or decorated with the ceSKA1 complex or hSka1 complex, and in the presence of hNdc80 “Broccoli” (Scale bars, 40 nm). (F) Left, TEM images showing individual dolestatin-10 rings in the presence or absence of the GFP-hSka1 microtubule-binding domain (MTBD; Scale bar, 50 nm). Right, graph showing the outer diameter and width of the dolestatin-10 rings in each condition (Mean ± SD, n = 58 per condition). In the presence of Ska1 MTBD, the width of the ring is increased, but not the outer diameter indicating that Ska1 binds to the inner surface.
We next imaged the Ska1 complex bound to dolastatin-induced rings by TEM. In the presence of the Ska1 complex, we observed binding to the inner face of the dolastatin rings (Fig. 5E), which corresponds to the outer surface of a microtubule protofilament. Similarly, in the presence of GFP-hSka1 MTBD, the outer diameter of the dolastatin-10 induced rings is unchanged, but thickness the rings increased by 5 nm (Fig. 5F) consistent with GFP-hSka1 MTBD binding to the inner surface. In contrast, the dolastatin rings remained largely undecorated in the presence of the Ndc80 complex (Fig. 5E). In total, these results demonstrate that the Ska1 complex, but not the Ndc80 complex, directly associates with curved microtubule structures.
The Ska1 Complex and Ndc80 complex bind to microtubules synergistically
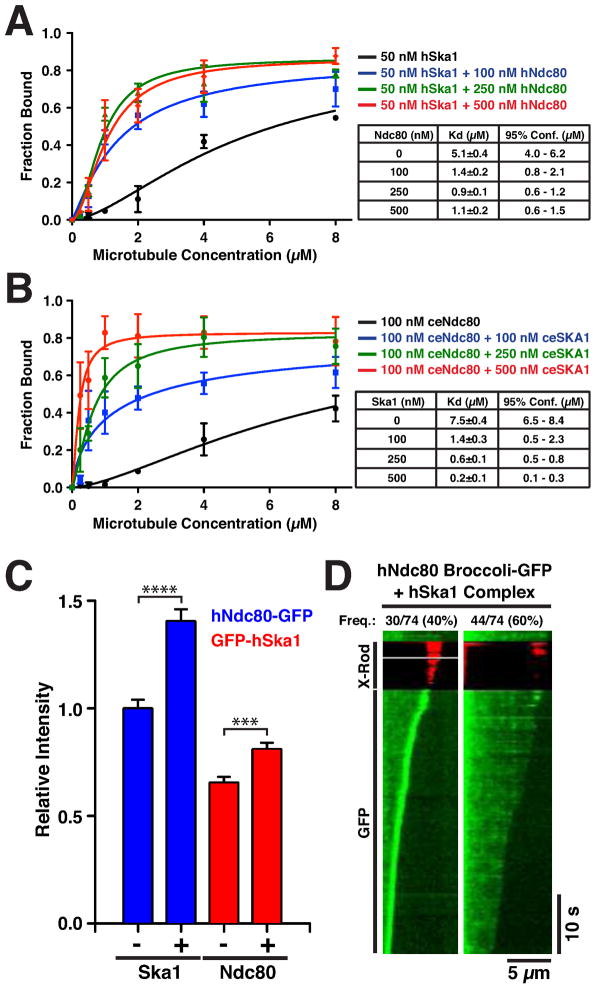
Previous work demonstrated that the Ndc80 complex is required for Ska1 complex localization to kinetochores (Chan et al., 2012; Raaijmakers et al., 2009; Welburn et al., 2009; Zhang et al., 2012). As the Ndc80 and Ska1 complexes are both implicated in the formation of stable kinetochore-microtubule interactions, we next sought to define how these activities are coordinated. To test the relationship between the Ndc80 and Ska1 complexes for microtubule binding, we conducted microtubule co-sedimentation assays using both the hSka1 complex and hNdc80 “Broccoli”. These experiments demonstrated that the hNdc80 complex increases the affinity of the hSka1 complex for microtubules by up to 8-fold in a dose-dependent manner (Fig. 6A, S4A), suggesting that there is a direct interaction between the hNdc80 and hSka1 complexes in the context of microtubules. Similarly, the C. elegans Ndc80 complex increased the affinity of the C. elegans Ska1 complex for microtubules and vice versa (Fig. 6B, S4B–C). In contrast, we were unable to detect a direct interaction between the Ndc80 and Ska1 complexes in solution (data not shown), suggesting that the synergy requires the microtubule lattice. The affinity of the human Ska1 MTBD for taxol stabilized microtubules was unaffected by the presence of Ndc80 complex, indicating that the interaction is mediated by the non-microtubule binding portion of the Ska1 complex (Fig. S4D). These results demonstrate that there is an evolutionarily conserved interaction between the Ska1 complex and the Ndc80 complex in the context of microtubules.
Figure 6. The Ska1 complex confers microtubule tracking activity to the Ndc80 complex.
(A) The Ska1 complex and Ndc80 complex bind to microtubules synergistically. Quantification of microtubule co-sedimentation assays for hSka1 complex binding to taxol-stabilized microtubules alone and in the presence of a range of concentration of hNdc80 “Broccoli” (Mean ± SD, n = 3). (B) Quantification of microtubule pelleting assays of C. elegans Ndc80 complex binding to taxol-stabilized microtubules alone and in the presence of a range of ceSKA1 complex (n = 3). (C) Quantification of the fluorescence intensity (Mean ± SEM) on microtubules of hNdc80 “Broccoli”-GFP (blue) in the absence (n = 95) and presence of hSka1 (n = 136; P < 0.0001, t-test), and GFP-hSka1 (red) in the absence (n = 73) and presence of hNdc80 “Broccoli” (n = 81; P < 0.001, t-test). (D) Representative kymographs (microtubule position along horizontal axis, time along vertical axis) of time-lapse movies showing hNdc80 “Broccoli”-GFP (9 nM) binding to depolymerizing microtubules in the presence of unlabeled hSka1 complex (9 nM, n = 74). Tracking events were identified as kymographs with substantial periods of an increased fluorescence signal at the microtubule end relative to the remaining lattice. When both Ndc80 and Ska1 are present, the GMPCPP-caps are often converted into cargo and become transported by the shortening microtubules. (See also Figure S4)
The Ska1 complex promotes the association of the Ndc80 complex with depolymerizing microtubule ends
To test the synergy between the Ska1 and Ndc80 complexes in the context of dynamic microtubules, we visualized hNdc80 “Broccoli”-GFP on depolymerizing microtubules in the presence and absence of unlabeled hSka1 complex. The presence of the hSka1 complex increased the overall signal intensity of the hNdc80 complex on microtubules and vice versa, confirming that the Ska1 and Ndc80 complexes bind microtubules synergistically (Fig. 6C). Although hNdc80 “Broccoli”-GFP does not track with depolymerizing microtubule ends on its own (Fig. 4B, Movie S5), in 40% of the depolymerization events observed in the presence of unlabeled hSka1 complex, hNdc80 “Broccoli”-GFP formed bright complexes that tracked the depolymerizing microtubule (Fig. 6D, Movie S6). In the remaining 60% of events, no tracking was observed (Fig. 6D). The tracking assemblies of Ndc80 complex in the presence of the Ska1 complex are substantially brighter than those observed for the Ska1 complex alone. These bright assemblies most likely represent remnants of the stabilizing GMPCPP-cap, decorated with hNdc80 “Broccoli”-GFP, that are transported along the depolymerizing microtubule in a Ska1-dependent manner (Movie S6). In contrast, if only the Ska1 or Ndc80 complex was present, the GMPCPP-caps dissociated from the depolymerizing microtubule (Movie S4, S5). GFP-hSka1 complex also formed bright tracking assemblies in the presence of unlabeled hNdc80 “Broccoli” (Fig. S4G), indicating that the identity of the GFP-labeled protein does not influence the formation of these assemblies, but that the presence of both complexes is necessary. The presence of both hSka1 complex and hNdc80 “Broccoli” significantly reduced the rate of microtubule depolymerization regardless of which complex is labeled with GFP, demonstrating that the integrated assembly resists microtubule depolymerization (Fig. S4G). These results demonstrate that the Ndc80 and Ska1 complexes display a synergistic interaction at the depolymerizing microtubule end and their combined action can sustain movement of cargo along the depolymerizing microtubule.
The Ska1 microtubule binding domain contains conserved basic surfaces that are regulated by Aurora B phosphorylation
To define the mechanism by which Ska1 binds to microtubules, we next determined the high resolution structure of the C. elegans Ska1 microtubule binding domain using Nuclear Magnetic Resonance (NMR; Fig. S5A, B; PDB accession # 2LYC). Our structural analysis demonstrates that the Ska1 microtubule binding domain is a variation of the winged helix domain (Fig. 7A, B, S5C). The winged helix domain is a ~100 amino acid domain that has previously been implicated primarily as a DNA binding motif, but which can also function as a protein-protein interaction domain (Gajiwala and Burley, 2000; Haering et al., 2004). In addition to the three helix-bundle and anti-parallel beta-sheets found in canonical winged-helix domains, the Ska1 microtubule binding domain contains an extended helix (αB) as part of three helix bundle and an additional helix (αC) inserted between alphaB and βA. Furthemore, αB is preceded by a loop (loopA) and a short helix (αA) (Fig. 7B). E130 in αA and R229 form a highly conserved salt bridge positioning αA and loopA relative to the three-helix bundle (Fig. S5D, S1A green stars).
Figure 7. The Ska1 microtubule binding domain is a winged-helix domain, regulated by Aurora B kinase.
(A) Cartoon representation of an ensemble of NMR-structures of the C. elegans Ska1 MTBD. (B) Topology diagram of the C. elegans Ska1 MTBD, showing the elements of the canonical winged-helix domain (grey). (C) Surface representation of the C. elegans Ska1 MTBD, showing conservation scores calculated using the Consurf Server (Ashkenazy et al., 2010). (D) Surface representation of the C. elegans Ska1 MTBD, showing electrostatic surface potential calculated using the Adaptive Poisson-Boltzman Solver software (Baker et al., 2001). (E) Surface representation of the C. elegans Ska1 MTBD, indicating conserved arginine residues (blue) and residues corresponding to Aurora B consensus sites in hSka1 (red). (F) (Top) Quantification of microtubule co-sedimentation assays (Mean ± SD, n = 3, 100 nM Ska1 complex, 75 mM KCl) and (Bottom) representative Western blots of microtubule co-sedimentation assays of hSka1 complex (black), hSka1 complex R155A, R236A, R245A (blue), and hSka1 complex S185D, S242D (red). Mutation of conserved arginine residues to alanine or Aurora B consensus sites is aspartate abolishes microtubule binding of the Ska1 complex. (G) Model showing the coordination of Ska1 and Ndc80 microtubule binding. The Ndc80 complex senses the polarity of the microtubule and integrates the Ska1 complex into the kinetochore. The Ska1 complex binds to the curved microtubule protofilaments at the depolymerizing microtubule ends, facilitating processive attachment to depolymerizing microtubules. (See also Figure S5).
Further analysis of the structure revealed that three highly conserved arginine residues (R141, R225, R234, Fig. S1A blue stars) located in loopA, αE, and βB are part of a conserved basic patch on the surface on the Ska1 microtubule binding domain (Fig. 7C–E). As binding of the Ska1 complex to microtubules requires the acidic C-terminal tails of tubulin (Welburn et al., 2009), we reasoned that this basic patch might represent the microtubule binding surface. Consistent with this, mutation of the homologous arginine residues in human Ska1 (R155, R236, R245) to alanine eliminated microtubule binding activity in vitro (Fig. 7F), and resulted in a mitotic delay and a reduction of kinetochore-fibers in human cells, similar to cells expressing hSka1 ΔMTBD (Fig. S5F–I).
Recent work has identified multiple phosphorylation sites in the Ska1 complex that are regulated by Aurora B (Chan et al., 2012). These sites were proposed to regulate the recruitment of the Ska1 complex to kinetochores, but not its microtubule binding activity. Interestingly, all four Aurora B phosphorylation sites identified in human Ska1 (Chan et al., 2012) are located in the region that we have defined here as the microtubule binding domain. Mutation of two of these Aurora B phosphorylation sites (S185, S242 in hSka1 MTDB) to aspartate to mimic constitutive phosphorylation dramatically reduced the microtubule binding activity of the Ska1 complex in vitro (Fig. 7F) and resulted in a mitotic delay and reduced kinetochore-fibers in vivo (Fig. S5F–I). However, these defects were slightly less severe than the arginine mutant or the deletion of the MTBD. Interestingly, S185 and S242 are located in distinct regions within the microtubule binding domain. The residue corresponding to S242 (R231 in C. elegans MTBD) resides in the direct vicinity of the basic patch identified above, suggesting that the introduction of negative charge by S242 phosphorylation might disrupt the interaction of the positively charged patch with microtubules (Fig. 7E). In contrast, the residue corresponding to S185 (Q171 in C. elegans MTBD) is located in a loop between αB and αC on the opposite face of the microtubule binding domain (Fig. 7E). This suggests that two distinct surfaces play a role in microtubule binding, potentially binding to adjacent tubulin monomers. Individual mutation of S185 or S242 also reduced microtubule binding activity, although to a lesser extent than the double mutant (data not shown). Importantly, mutation of the conserved arginine residues or Aurora B phosphorylation sites did not affect the folding of the microtubule binding domain (Fig. S5E). In total, these results demonstrate that the Ska1 complex uses a winged helix domain to bind to microtubules and that point mutations that disrupt microtubule binding phenocopy deletion of the Ska1 MTBD. Thus, microtubule binding is a crucial mitotic function of the Ska1 complex.
Discussion
Chromosome movement driven by microtubule dynamics is essential for proper cell division. Here, we defined conserved biochemical activities of the Ska1 complex, a key player in coupling chromosome movement to microtubule dynamics. Our work provides mechanistic and structural insights into the interaction of the Ska1 complex with depolymerizing microtubules and demonstrates that its activity is integrated with the Ndc80 complex to generate an assembly that can remain processively associated with depolymerizing microtubules.
The Ska1 complex tracks depolymerizing microtubules using a unique mechanism
Since it was first demonstrated that microtubule depolymerization is sufficient to move chromosomes (Coue et al., 1991), a key goal has been to identify the molecular players involved in this process. The Dam1 complex was the first kinetochore component that was shown to track with depolymerizing microtubules (Westermann et al., 2006), but the Dam1 complex is only present in fungi and is not essential for chromosome segregation in fission yeast (Sanchez-Perez et al., 2005). The striking formation of ring-like Dam1 structures around microtubules has led to forced walk or ring-coupler models to explain how the kinetochore maintains attachment to depolymerizing microtubules (Efremov et al., 2007; Miranda et al., 2005; Westermann et al., 2006). The Ndc80 complex is conserved throughout eukaryotes and is essential for chromosome segregation in all organisms where it has been studied (Cheeseman and Desai, 2008). Recently, Powers and colleagues proposed that the Ndc80 complex remains attached to depolymerizing microtubules by biased diffusion (Powers et al., 2009). However, the Ndc80 complex can only track with depolymerizing microtubules when it is artificially induced to oligomerize (Powers et al., 2009). The Dam1 and Ndc80 complexes from budding yeast have also been shown to synergize in forming load-bearing attachments to depolymerizing microtubules (Lampert et al., 2010; Tien et al., 2010).
The models previously described for the Ndc80 and Dam1 complexes were under the implicit assumption that each complex interacts with the straight microtubule lattice (Efremov et al., 2007; Hill, 1985). Here, we demonstrated that the human Ska1 complex autonomously tracks with depolymerizing microtubules. We propose that the Ska1 complex remains associated with depolymerizing microtubules by not only associating with and diffusing along the straight microtubule lattice, but also by interacting with the curved microtubule structures at depolymerizing microtubule ends. Consistent with this, both the human and C. elegans Ska1 complexes associate with dolastatin-10-induced rings, which mimic the curved structures formed at depolymerizing microtubule ends, indicating that binding of curved microtubule structures is a key conserved property of the Ska1 complex. We note that the Dam1 complex has been shown to form rings around microtubules, but this behavior is fundamentally distinct from the Ska1 complex in which dolastatin induces the formation of a protofilament ring to which the Ska1 complex binds. Interestingly, the Ndc80 complex neither associates with dolastatin-10-induced rings nor tracks with depolymerizing microtubules under the conditions tested, indicating that its association with microtubules is inherently different from the Ska1 complex. This is further supported by the observation that the presence of the Ska1 complex induces the formation of protofilament rings when added to dynamic microtubule polymers, whereas the Ndc80 complex does not.
We also demonstrated that the Ska1 complex associates with microtubules using a variation of a winged-helix domain, while the Ndc80 complex utilizes a calponin-homology domain (Ciferri et al., 2008). Previous work has demonstrated that the “toe-print” of the Ndc80 calponin homology domain contains residues from two consecutive tubulin monomers (Alushin et al., 2010). Thus, protofilament bending could distort this binding site and thereby reduce the affinity of the Ndc80 complex for curved microtubules. In contrast, the Ska1 complex maintains, or in case of the C. elegans homologue even increases, its binding affinity when associating with curved protofilaments. The long-axis of the Ska1 MTBD is approximately the same length as the longest axis of the tubulin monomer (~5 nm). Thus, the Ska1 MTBD domain could interact with consecutive tubulin monomers and the binding surfaces, such as the C-terminal charged tail, could be ideally positioned when the protofilaments assume a curved conformation. Alternatively, the Ska1 MTBD could have a binding site on the tubulin monomer that is unaffected by the conformation of the protofilament. In case of the C. elegans homologue, additional interactions may be formed with dolastatin-10 induced protofilament rings.
In total, we propose that the Ska1 complex can associate with the bending protofilaments of depolymerizing microtubules, thereby assuring that the kinetochore remains attached to the microtubule when it encounters a depolymerizing microtubule end. The Ndc80 complex preferentially binds to the straight microtubule lattice, thus stabilizing it relative to the bent conformation and counteracting microtubule depolymerization (Fig. 7G). To mediate kinetochore-microtubule attachments these distinct microtubule-binding activities must be coordinated to facilitate robust kinetochore-microtubule attachments.
The Ska1 and Ndc80 complexes form an integrated microtubule binding assembly
We demonstrated that the Ska1 and Ndc80 complexes bind to microtubules synergistically, a property that is conserved from humans to nematodes, indicating that the Ska1 complex and Ndc80 complexes interact directly in the context of microtubules. Our analysis suggests that this is a specific effect, as we were unable to detect a synergy between the Ska1 microtubule-binding domain and the Ndc80 complex (Fig. S4D), or between the human Ska1 complex and the C. elegans Ndc80 complex (data not shown). Furthermore, under the conditions tested, the Ndc80 complex alone does not have the capacity to remain associated with depolymerizing microtubules. In contrast, in the presence of the Ska1 complex, large Ndc80 assemblies are processively transported along depolymerizing microtubule. The rate of microtubule depolymerization in the presence of these assemblies is strongly reduced relative to free microtubules, indicating that these assemblies resist microtubule depolymerization. Therefore, we propose that Ska1 and Ndc80 complexes interact to form a unique coupler that can remain associated with depolymerizing microtubules and harness their energy to drive chromosome movement (Fig. 7G). The Ndc80 complex can sense microtubule orientation and integrates the Ska1 complex into the macromolecular kinetochore, whereas the activity of the Ska1 complex is necessary to remain attached to depolymerizing microtubules by associating with both curved protofilaments and the straight microtubule wall. Although the Ndc80 and Ska1 complexes can coordinate to track microtubules, future studies will be required to study this behavior in the presence of a load, such as would be present at kinetochores in cells.
Our cell biological analysis suggests that the Ska1 and Ndc80 complexes make distinct contributions to kinetochore microtubule attachments in vivo. In the absence of Ska1 complex microtubule binding activity, the Ska1 complex localizes to kinetochores and together with the Ndc80 complex is sufficient to direct chromosome alignment and end-on kinetochore microtubule attachments (Fig. 2). Indeed, when Ska1 complex function is eliminated entirely, cells are unable to form kinetochore-microtubule attachments (Gaitanos et al., 2009; Raaijmakers et al., 2009; Welburn et al., 2009), although others have instead suggested that the Ska1 complex is involved in spindle assembly checkpoint silencing (Daum et al., 2009). Importantly, in the absence of Ska1 microtubule binding activity kinetochore fibers and kinetochore oscillations are both strongly reduced, suggesting that the Ndc80 complex microtubule binding activity is insufficient to carry out these functions. Instead, the unique microtubule binding activity of the Ska1 complex is required to maintain kinetochore fibers and to power kinetochore oscillations, indicating that is required to allow the kinetochore to remain associated with depolymerizing microtubules.
Experimental Procedures
Cell culture and RNAi
Stable cell lines derived from HeLa cells were generated as previously described (Shah et al., 2004). Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin/streptomycin, and L-glutamine at 37°C in a humidified atmosphere with 5% CO2. RNAi experiments were conducted using Lipofectamine RNAiMAX (Invitrogen) using the manufacturers instructions. siRNAs against Ska1 were described previously (Welburn et al., 2009).
Immunofluorescence and Microscopy
Immunofluorescence in human cells was conducted as described previously (Welburn et al., 2009). Microtubules were visualized using the DM1α (Sigma) at 1:2000. To detect kinetochores, anti-centromere antibodies (Antibodies, Inc., Davis, CA) were used at 1:100. To visualize GFP-tagged Ska1, the intrinsic GFP fluorescence was used. Cy2-, Cy3-, and Cy5-conjugated secondary antibodies (Jackson Laboratories) were used at 1:100. DNA was visualized using 10 μg/ml Hoechst. For additional information, see the Supplemental Methods.
Total internal reflection fluorescence microscopy
TIR-FM movies were acquired with a Nikon Ti-E inverted microscope with perfect focus, equipped with an Andor Revolution 500 XD laser system (488 nm 50 mW, 640 nm 100 mW), a Ti-TIRF-E motorized illumination unit (Nikon), an Apo 100x/1.49 TIRF objective (Nikon), 1.5x auxiliary magnification, and an Andor iXon3 897 EMCCD-camera using the Andor iQ2 software. 600 images were acquired under continuous illumination with 45 ms exposure times (20 fps). A single image of the microtubules was taken before and after each movie. All imaging was carried out at 30°C. The movies were analyzed with a custom algorithm and manually screened for false positives. For a detailed protocol see supplemental experimental procedures.
Microtubule co-sedimentation assays
Microtubule pelleting assays were conducted as described previously (Cheeseman et al., 2006) using taxol stabilized microtubules or dolastatin-10 induced rings. Dolastatin-10 induced rings were generated by incubating 17.3 μM tubulin-dimer in BRB80 with 6% DMSO and 40 μM dolastatin-10 (provided by the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute) for 1 hr at room temperature. Rings were purified by pelleting through a cushion of BRB80 with 40% glycerol for 10 min at 300000xg at room temperature. Rings were resuspended in BRB80 with 6% DMSO and 40 μM dolastatin-10. The amount of rings in pellet fractions was quantified by Coommassie staining relative to a standard curve of tubulin on the same gel. Antibodies to detect the Ndc80 complex and the ceSKA1 complex were raised against Ndc80 “Bonsai” and full ceSKA1 complex and affinity purified, as described previously (Desai et al., 2003). The hSka1 antibody was described previously (Welburn et al., 2009).
Dynamic microtubule tracking assays
Tracking assays were conducted as described previously (Grishchuk and Ataullakhanov, 2010; Grishchuk et al., 2005). Briefly, microtubules were grown from coverslip-attached Chlamydomonas axonemes in the presence to bovine tubulin and GTP. Stabilizing caps where then assembled by elongating microtubules in the presence GMPCPP, and with Rhodamine-labeled tubulin. After washing chambers with these coverslip-anchored segmented microtubules, GFP-labeled proteins were perfused in motility buffer (BRB80, 4 mM MgCl2, 5 mg/ml BSA, 0.4 mg/ml Casein, 10 mM DTT). Microtubule depolymerization was induced by a brief exposure to a bright green light. Images were acquired and processed using Metamorph, using a Zeiss Axiophot 2, with a 100x 1.4NA objectice, and a Andor iXon3 camera, under continuous illumination at 32°C. Tip intensity was determined with line scans along kymographs, which were background corrected and normalized by the lattice intensity.
Electron Microscopy
Dynamic microtubules were assembled by incubating 45 μM tubulin (Cytoskeleton, Inc.) in 50 mM PIPES pH 6.8, 4 mM MgCl2, 12% (V/V) DMSO, and 2 mM GTP at 34 C for 1 hour. Microtubules were diluted in BRB80 to 4.5 μM in the presence of 2 μM hSka1 complex, ceSKA1 complex, or hNdc80 “Broccoli”, immediately added to glow discharged electron microscopy grids and stained with 1% uranyl acetate. Dolastatin-10 induced rings were assembled as described above and incubated at 1 μM with a 1.5-fold excess of hSka1 complex, ceSKA1 complex, or hNdc80 “Broccoli”. Images were acquired using FEI Tecnai and FEI Spirit transmission electron microscopes equipped with Teitz (2k × 2k) and Gatan (4k × 4k) CCD cameras.
NMR Spectroscopy
All NMR samples were prepared in 20 mM potassium phosphate pH 6.5, 150 mM NaCl, 1.5 mM DTT. Spectra were recorded on Varian/Agilent Inova 600 MHz and 500 MHz and Bruker 700 MHz and 900 MHz spectrometers at 298 K. Backbone and side chain resonances were picked and assigned manually in CARA (Keller, 2004), NOEs were picked and assigned manually in CcpNmr (Vranken et al., 2005). Torsion angle constraints were derived from chemical shifts using the program TALOS+ (Shen et al., 2009). NOE intensities were integrated and converted into distance constraints using CcpNmr (Vranken et al., 2005). Hydrogen bond restraints for beta-sheets and helices were assigned when supported by NOEs and secondary structure prediction. 50/20 structures were created in a simulated annealing protocol using CYANA (Guntert et al., 1997). Structures were visualized with PyMOL and MOLMOL (Koradi et al., 1996). PROCHECK-NMR was used to assess the stereochemical quality of the structure (Laskowski et al., 1996). For a detailed description of the structure determination see the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The Ska1 complex tracks with depolymerizing microtubules.
The Ska1 complex associates with curved protofilament structures.
The Ndc80 and Ska1 complexes bind to and track microtubules synergistically.
The Ska1 complex microtubule binding domain has a winged-helix structure.
Acknowledgments
We thank members of the Cheeseman laboratory and Angelika Amon, Arshad Desai, and Peter Reddien for discussions and critical reading of the manuscript, Gary Brouhard and Stefan Westermann for advice on conducting the TIRF experiments, and the staff of W. M. Keck microscopy facility at Whitehead. We thank Arshad Desai for sharing unpublished experimental results, Mary Porter for a gift of axonemes, and Fazly Ataullakhanov and Vadim Mustyatsa for help with image analysis. This work was supported by awards to IMC from the Searle Scholars Program, the Human Frontiers Science Foundation, the Leukemia & Lymphoma Society, and a grant from the NIH/National Institute of General Medical Sciences (GM088313). Further support came from NIH grants to RAM (GM052468-17), HA (NIDDK-K01-DK085198) and GW (P01-GM-047467), and MIT Faculty Start-up Funds to MB. ELG is a Kimmel scholar and IMC is a Thomas D. and Virginia W. Cabot Career Development Professor of Biology.
Footnotes
Accession Numbers
Coordinates for the ceSKA1 winged-helix domain have deposited in the Protein Data Bank with the accession code 2LYC.
The Supplemental Information includes five figures, six movies, supplemental experimental procedures, and supplemental references and can be found with this article online at XYZ.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai R, Durso NA, Sackett DL, Hamel E. Interactions of the sponge-derived antimitotic tripeptide hemiasterlin with tubulin: comparison with dolastatin 10 and cryptophycin 1. Biochemistry. 1999;38:14302–14310. doi: 10.1021/bi991323e. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukari H, Sackett DL, Schuck P, Nossal RJ. Single-walled tubulin ring polymers. Biopolymers. 2007;86:424–436. doi: 10.1002/bip.20752. [DOI] [PubMed] [Google Scholar]
- Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. The Journal of cell biology. 2012;196:563–571. doi: 10.1083/jcb.201109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. The Journal of cell biology. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum J, Wren J, Daniel J, Sivakumar S, McAvoy J, Potapova T, Gorbsky G. Ska3 Is Required for Spindle Checkpoint Silencing and the Maintenance of Chromosome Cohesion in Mitosis. Curr Biol. 2009 doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. The Journal of cell biology. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes & development. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov A, Grishchuk EL, McIntosh JR, Ataullakhanov FI. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc Natl Acad Sci USA. 2007:19017–19022. doi: 10.1073/pnas.0709524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. The EMBO journal. 2009;28:1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Ataullakhanov FI. In Vitro Assays to Study the Tracking of Shortening Microtubule Ends and to Measure Associated Forces. Method Cell Biol. 2010;95:657–676. doi: 10.1016/S0091-679X(10)95033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, Mcintosh JR. Force production by disassembling microtubules. Nature. 2005:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. Journal of molecular biology. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J. Structure and stability of cohesin’s Smc1-kleisin interaction. Molecular Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Hanisch A, Silljé HHW, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci USA. 1985:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, Conti E. Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Molecular Cell. 2012;46:274–286. doi: 10.1016/j.molcel.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Keller R. The Computer Aided Resonance Assignment Tutorial. Goldau: CANTINA Verlag; 2004. [Google Scholar]
- Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Volkov V, Ataullakhanov FI, Grishchuk EL. Tubulin depolymerization may be an ancient biological motor. J Cell Sci. 2010;123:3425–3434. doi: 10.1242/jcs.067611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JJL, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci. 2009;122:2436–2445. doi: 10.1242/jcs.051912. [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. The EMBO journal. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Current biology: CB. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis M, Slabicki M, Junqueira M, Paszkowski-Rogacz M, Sontheimer J, Kittler R, Heninger A, Glatter T, Kruusmaa K, Poser I, et al. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J. 2009 doi: 10.1038/emboj.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JPI, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Wang H-W, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- Zhang G, Kelstrup CD, Hu XW, Kaas Hansen MJ, Singleton MR, Olsen JV, Nilsson J. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci. 2012;125:3243–3253. doi: 10.1242/jcs.104208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.