Abstract
Low-penetrance alleles associated with breast cancer risk have been identified in population-based studies. Most risk loci contain either no or multiple potential candidate genes. Rat mammary carcinoma susceptibility 1b (Mcs1b) is a quantitative trait locus (QTL) on RN02 that confers decreased susceptibility when Copenhagen (COP) resistant alleles are introgressed into a Wistar Furth (WF) susceptible genome. Five WF.COP congenic lines containing COP RN02 segments were compared. One line developed an average of 3.4 ± 2.0 and 5.5 ± 3.6 mammary carcinomas per rat ± SD when females were Mcs1b resistant homozygous and Mcs1b heterozygous, respectively. These phenotypes were significantly different from susceptible genotype littermates (7.8 ± 3.1 mean mammary carcinomas per rat ± SD, P = 0.0001 and P = 0.0413, respectively). All other congenic lines tested were susceptible. Thus, Mcs1b was narrowed to 1.8 Mb of RN02 between genetic markers ENSRNOSNP2740854 and g2UL2-27. Mammary-gland-graft carcinoma-susceptibility assays were used to determine that donor (P = 0.0019), but not recipient Mcs1b genotype (P = 0.9381), was associated with ectopic mammary carcinoma outcome. Rat Mcs1b contains sequence orthologous to human 5q11.2, a breast cancer susceptibility locus identified in multiple genome-wide association studies. Human/rat MAP3K1/Map3k1 and MIER3/Mier3 are within these orthologous segments. We identified Mier3 as a candidate Mcs1b gene based on 4.5-fold higher mammary gland levels of Mier3 transcripts in susceptible compared to Mcs1b resistant females. These data suggest that the human 5q11.2 breast cancer risk allele marked by rs889312 is mammary-gland autonomous, and MIER3 is a candidate breast cancer susceptibility gene.
Keywords: breast cancer susceptibility, rat Mcs1b, rat mammary cancer, complex disease genetics, comparative genetics
Introduction
Low-penetrance breast cancer susceptibility alleles have been identified using human genome-targeted and genome-wide association study (GWAS) designs [1–13]. Apart from identifying complex disease risk-associated genetic variation, human studies alone are limited in pinpointing candidate genes and functional characterization of susceptibility associated loci. Comparative genetics based on experimental organisms such as Rattus norvegicus (laboratory or Norway rat) recapitulate the complex genetics, and may complement and enhance human studies of breast cancer risk and prevention [14, 15].
The laboratory rat provides a good model of human breast cancer. Rat mammary carcinomas closely resemble human breast carcinomas in histopathology, hormonal responsiveness, and potential environmental etiologies [16–22]. Rats develop spontaneous, carcinogen, and oncogene induced mammary carcinomas [23]. Several rat strains representing different spectrums of genetic variation in susceptibility to mammary carcinogenesis exist [24–29]. Genotypic differences in susceptibility are not necessarily due to carcinogen-induced differences in metabolism or DNA damage [26, 30].
Multiple rat mammary carcinoma susceptibility (Mcs) quantitative trait loci (QTLs) have been identified [28, 29, 31]. Rat Mcs QTLs that are concordant to human breast cancer risk alleles may be used to identify genes and mechanisms controlling breast cancer susceptibility in women. Comparative genetic approaches have been used to identify MCS5A1 and MCS5A2, which are common noncoding breast cancer risk alleles on human Chr 9 [1]. Human MCS5A1 has been confirmed to associate with breast cancer risk in additional population-based studies [9]. Another rat QTL, Mcs6, was mapped to an orthologous region of human Chr 12, which potentially associates with breast cancer risk [32]. Rat genetics will continue to play an important role in elucidating breast cancer risk alleles, candidate genes, and molecular mechanisms.
Rat Mcs1b was initially mapped to a 16 cM region of rat Chr 2 [33]. Positional mapping was completed in WF.COP congenics by introgression of a segment of resistant Copenhagen (COP) Chr 2 into a susceptible Wistar-Furth (WF) genetic background. Mcs1b resistant alleles decrease mammary carcinoma multiplicity compared to susceptible WF alleles. We have mapped rat Mcs1b to a shorter genomic interval, and found that it contains the rat ortholog to a GWAS identified human breast cancer risk associated allele at human Chr 5q11.2 that is marked by SNP rs889312 [2]. This locus has been confirmed to associate with human breast cancer in different human populations and tumor sub-types [34–39]. We have further utilized rat Mcs1b congenic lines to investigate functional aspects of these alleles and identify Mier3 as a strong mammary cancer susceptibility gene.
Materials and Methods
Animals and Phenotyping
Congenic rat lines were maintained in an AAALAC-approved facility on a 12 h light/dark cycle and provided LabDiet 5001 Rodent Diet (PMI® Nutrition International) and water ad libitum. All animal protocols were approved by the University of Louisville Animal Care and Use Committee. Congenics are defined as genetic lines that carry defined COP alleles introgressed into the inbred WF/NHsd (Harlan) genome. Information for genetic markers defining ends of COP alleles carried by each congenic line T, N3, F3, W2, U2, and I4 is available at the UCSC Genome Browser (www.genome.ucsc.edu), Rat Genome Database (http://rgd.mcw.edu/), NLM NCBI, or Supplementary Table S1. At 50–55 days of age, 7,12-dimethylbenzanthracene (DMBA, 20 mg/mL sesame oil) was given by a single oral gavage (65 mg DMBA/kg body mass). Mammary carcinoma susceptibility phenotypes were determined by counting (multiplicity) mammary carcinomas ≥ 3×3 mm2 that developed 15 weeks after carcinogen [33].
Mammary Gland Grafting
Mammary gland grafting experiments used WF.COP line N3 females at N16F5, N16F6, and N16F7 generations to supply Mcs1b resistant alleles. At the N15 generation, line N3 was backcrossed to the inbred WF/NHsd strain and re-fixed at the N16 generation for the Mcs1bCOP allele it carries. Using congenics, opposed to F1 rats, provided rats homozygous at Mcs1b, which provided higher penetrance without influence from other COP resistance alleles. Both abdominal and adjacent inguinal mammary glands with lymph nodes removed were excised from 30–35 day old donor females. Mammary tissue was finely minced in a Petri dish on ice, divided into four equal volumes, and placed onto the interscapular white fat pad of 30–35 day old recipients (1 donor per 4 recipients). DMBA was administered as before. Interscapular fat pads were evaluated at 15 weeks following DMBA for graft site tumor development. Ectopic mammary tumors were histologically verified to be carcinomas. Interscapular fat pad tissue was whole mounted and stained with aluminum carmine to determine if an ectopic mammary gland had developed.
Resequencing
Total RNA samples, extracted using TriReagent (Molecular Research Center) and standard chloroform/isopropanol precipitation, were from WF/NHsd and WF.COP lines N3 and T. TURBO DNase (Life Technologies) was used to reduce DNA contamination and cDNA was made using Superscript III reverse transcriptase (Life Technologies). In some instances sequences were not attainable from cDNA. To obtain these sequences genomic DNA, extracted from frozen spleen or liver tissues using standard phenol-chloroform/isopropanol precipitation, was used. Ampliphied samples were cleaned with the QIAquick PCR Purification Kit (Qiagen) prior to using the BigDye Terminator v.3.1 Cycle Sequencing Kit (Life Technologies). Sequencing reactions were purified with Agencourt CleanSeq magnetic beads (Beckman Coulter) and analyzed by the University of Louisville Center for Genetics & Molecular Medicine DNA Core using an ABI PRISM 7700 Sequence Detection System (Life Technologies). Primer sequences for amplifying and sequencing Mcs1b ORFs and 3′-UTRs are in Supplementary Table S2. Nucleic acid sequences were submitted to NCBI/GenBank and assigned accession numbers JQ013728 thru JQ013737.
Quantitative PCR
Total RNA was isolated with TRI-Reagent (Molecular Research Center) from flash-frozen and homogenized tissues. To reduce possible solvent and DNA contamination RNA samples were further processed by a 1/10 v/v 3M sodium acetate and 2.5x v/v 100% ethanol wash on ice for 10 minutes followed by 80% ethanol wash followed by Turbo DNAse (Life Technologies). Total RNA quantity and quality were measured with a Nanodrop 1000 (Fisher Scientific) and a Bioanalyzer with RNA 6000 NanoChips (Agilent). Reverse transcription reactions (20 μl f.v.) that contained 1μg total RNA, 0.5x RNAsecure, 5μM random hexamers, 25ng/μL oligo(dT18), and 0.5 mM dNTPs were incubated 5 minutes at 65° C prior to adding 1× first strand buffer, 100mM DTT, and 1μL Superscript III (Life Technologies). Reactions were incubated 5 m at 25° C, 1 h at 50° C, and 15 m at 70° C. Quantitative PCR was performed as previously published [1] using TaqMan MGB probes (Life Technologies) and primers in Supplementary Table S3. Except, 60 nM each Rplp2 primer and 120 nM VIC-labeled Rplp2 probe were used as an endogenous control. Fluorescence values were measured using SDS v2.3 software (Life Technologies).
Plasmid Construction and MIER3 Expression
Homo sapiens MIER3 ORF (NCBI/GenBank ref|NM_152622.3) from pooled human breast total RNA (#AM6952, Life Technologies) was cloned into pEGFP-C1 vector at EcoRI and KpnI sites. Primer sequences for cDNA amplification were 5′-CGGAATTCTATGGCGGAGGCTTCTTTTGGAAGT and 5′-CGGGGTACCCTCAGAGTGTAGGGCCGCGTGC. MDA-MB-231 (#HTB-26) and T47D (#HTB-133) cell lines were purchased from American Type Culture Collection (ATCC) in May 2011, cultured, respectively, in DMEM with 10% FBS and RPMI 1640 with 10% FBS and 0.2 U/mL bovine insulin, and cyropreserved after one passage for future use. Cell authentication was guaranteed by ATCC and morphology was confirmed under a phase-contrast light microscope. Cells (6.25 × 105 MDA-MB-231/well or 2 × 105 T47D/well) in 6-well plates were transiently transfected with 2.5 μg of pEGFP-hMIER3 or pEGFP-C1 plasmids using Lipofectamine LTX/Plus (Life Technologies). Samples were visualized at 24 h using a phase-contrast or confocal (Olympus IX51 40x objective) microscope. Cells were grown on cover slips in chambers (Lab-Tek #177445), washed with PBS, fixed for ten minutes with 10% paraformaldehyde, and washed again with PBS prior to DAPI (2 ng/mL PBS) staining for five minutes. Cover slips were mounted on microscope slides using fluorescent mounting solution (DAKO). Fluorescent images were captured using cellSens Dimension software (Olympus).
Genomics and Statistical Analysis
Genome assemblies used were Homo sapiens version GRCh37/hg19 and Rattus norvegicus version 3.4/rn4. Mammary carcinoma multiplicity phenotypes were compared by nonparametric Mann Whitney tests. Results from mammary gland grafting experiments were analyzed using logistic regression. Donor and recipient genotypes were incorporated as dependent variables. In independent models, graft site tumor outcome and grafting ability were used as independent variables. Quantitative PCR (QPCR) data were analyzed using ANOVAs with log2 (Target quantity/Rplp2 quantity) as the dependent variable. Independent variables for comparing mammary gland transcript levels were Mcs1b genotype and DMBA exposure. Mcs1b genotype and tissue source were independent variables for mammary carcinoma and non-diseased mammary tissue QPCRs. Fisher’s PLSD tests were used to compare groups following a significant F-test (α ≤ 0.05). Statview software (SAS Institute) was used.
Results
Fine-Mapping Mcs1b using WF.COP Congenics
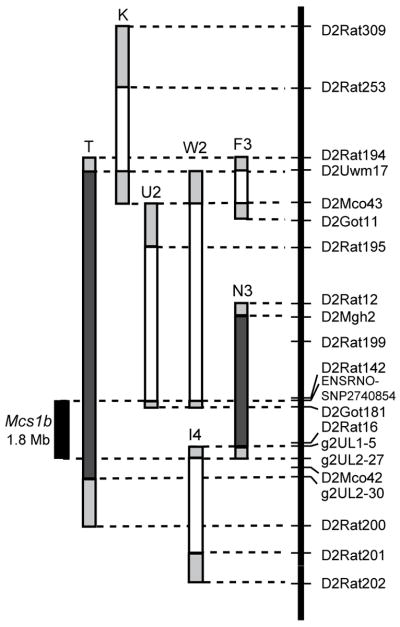
Subsequent comparative genetics work is reduced if QTLs are mapped to short syntenic intervals. Five congenic lines that contained different resistant COP rat Chr 2 segments of the Mcs1b candidate region from D2Uwm17:D2Rat200 (Chr2:32051320-48762858) on a susceptible WF genetic background were tested to narrow Mcs1b (Fig. 1). Mammary carcinoma susceptibility phenotypes were determined using tumor multiplicity at 15 weeks following DMBA induction of mammary carcinogenesis. A shorter segment of COP Chr 2 that was contained in congenic line N3 conferred a decreased Mcs1b phenotype similar to line T (Table 1). Females of line N3 that were Mcs1b resistant homozygous or heterozygous developed, respectively, 56% and 30% less mammary carcinomas per rat than Mcs1b susceptible homozygous (WF/WF) female littermates from line N3. Congenic lines F3, W2, U2, and I4 each contained different COP Chr 2 segments (Fig. 1). All these lines had Mcs phenotypes similar to littermates with susceptible WF genotypes (Table 1). Comparison of microsatellite DNA and published rat SNPs located in the 0.66 Mb of genomic sequence between the distal and proximal ends of lines N3 and I4 yielded no genetic variation between resistant COP and susceptible WF alleles (Supplementary Table S4). Therefore, we were unable to define a precise distal end to Mcs1b. When considered together, our congenic line data delimit Mcs1b to a ~1.8 Mb of rat Chr 2 that spans from SNP ENSRNOSNP2740854 to microsatellite g2UL2-27, which corresponds to rat Chr2:42364155-44195382 (Fig. 1).
Figure 1. Rat Chr 2 map of WF.COP lines delimiting Mcs1b to 1.8 Mb.
Markers used to genotype WF.COP congenics are listed in relative positions on the y-axis. Lines are labeled with letter-number combinations and designated with filled dark-gray bars to indicate Mcs1b resistant alleles. Lines that are drawn with unfilled bars represent COP intervals incapable of conferring decreased susceptibility or resistance to mammary carcinoma development. The filled light-gray bars at ends of each congenic segment are intervals of unknown genotype. Lines T and K were published previously [33].
Table 1.
Mammary carcinoma multiplicity phenotypes (mean mammary carcinomas per rat ± SD) by genotype for WF.COP Chr 2 congenic lines used to map Mcs1b to 1.8 Mb
| WF.COP Chr2 region | Line | COP/COP (COP/WF) | n | WF/WF | n | P value* |
|---|---|---|---|---|---|---|
| D2Uwm17/g2UL2-30 | T† | 3.5±2.2 | 21 | 8.3±3.3 | 19 | 0.0010 |
| D2Uwm17/D2Ulb4 | F3 | 9.6±4.1 | 32 | 8.8±3.5 | 32 | 0.8433 |
| D2Mgh2/g2UL1-5 | N3 | 3.4±2.0 | 25 | 7.8±3.1 | 25 | 0.0001 |
| N3 | (5.5±3.6) | 15 | 0.0413 | |||
| D2Ulb4/ENSRNOSNP2740854 | W2 | 6.0±1.8 | 18 | 5.9±3.2 | 9 | 0.8498 |
| D2Rat116/ENSRNOSNP2740854 | U2 | 5.7±3.9 | 6 | 6.3±3.3 | 12 | 0.8866 |
| g2UL2-27/D2Rat201 | I4 | 9.3±3.0 | 19 | 7.9±3.7 | 13 | 0.2470 |
Abbreviations: WF, Wistar Furth; COP, Copenhagen; Chr, chromosome
P values from Mann Whitney tests
Line T phenotype published previously by Haag et al. Cancer Research, 63:5808–5812, 2003
Mcs1b Mammary-Gland-Graft Carcinoma Susceptibility
To determine if the Mcs1b resistant allele acted to reduce mammary cancer susceptibility in a mammary gland autonomous manner, we subjected animals with ectopically transplanted mammary-gland tissue to DMBA-induced mammary carcinoma susceptibility assays. Females used in mammary grafting assays had a WF genetic background and either Mcs1b resistant or susceptible WF alleles. We expected these animals to have compatible immune systems; and thus, not reject reciprocal mammary gland grafts. To test this expectation, it was determined empirically that recipients did not reject mammary tissue grafts from donors of different Mcs1b genotypes. The respective total number of recipients (r) and ectopic mammary-graft positive recipient (+r) females in susceptible (S) and resistant (R) reciprocal donor:recipient transplant groups are reported in Table 2. There were no statistically significant associations between mammary tissue grafting ability and donor or recipient genotype.
Table 2.
Mammary gland graft-site and carcinoma outcome analyses.
| Raw data from mammary gland grafting assays | ||||
|---|---|---|---|---|
| Mcs1b Donor : Recipient Genotype | ||||
| S : S | S : R | R : S R : R | ||
| Total recipients (n) | 28 | 25 | 23 | 22 |
| MG-graft positive (n) | 27 | 23 | 18 | 22 |
| MG-graft AND tumor positive (n) | 15 | 13 | 4 | 5 |
| Logistic regression of graft site mammary gland outcome | |||
|---|---|---|---|
| Effect | Coefficient | P value | Odds Ratio (95% CI) |
| Donor | 0.99 | 0.1869 | 2.69 (0.62 – 11.68) |
| Recipient | −1.31 | 0.1160 | 0.27 (0.05 – 1.38) |
| Intercept | 2.70 | 0.0004 | |
| Logistic regression of mammary gland graft site tumor outcome | |||
|---|---|---|---|
| Effect | Coefficient | P value | Odds Ratio (95% CI) |
| Donor | 1.48 | 0.0019 | 4.40 (1.73 – 11.18) |
| Recipient | −0.04 | 0.9381 | 0.96 (0.39 – 2.36) |
| Intercept | −1.22 | 0.0045 | |
Abbreviations: S, susceptible genotype; R, Mcs1b resistant genotype; n, number of females; MG, mammary gland; CI, confidence interval
Data from ectopic mammary gland graft positive recipients were analyzed to test for associations of donor and recipient genotypes with carcinoma development at the ectopic mammary gland site. Donor mammary tissue from Mcs1b resistant (R) females, when grafted into interscapular white fat pads of either R or susceptible (S) recipients resulted in fewer females developing ectopic mammary carcinomas compared to recipients of either genotype that received mammary tissue from S genotype donors (Fig. 2). Donor, but not recipient genotype, was significantly associated with graft-site mammary carcinoma outcome (Table 2). These results indicated that Mcs1b conferred resistance is mammary gland autonomous. This also suggested that work to characterize this locus should initially focus on mammary tissue.
Figure 2. Rat Mcs1b is mammary gland autonomous.
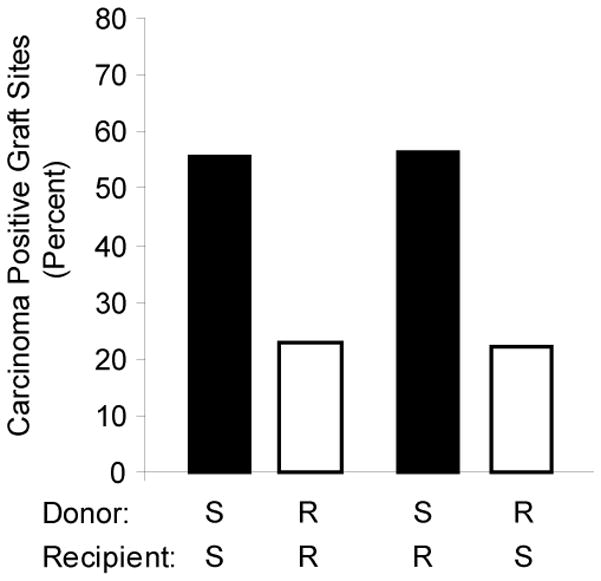
Percentage of mammary gland graft positive recipients that developed ectopic mammary gland carcinomas are shown for each susceptible (S) and Mcs1b resistant (R) donor:recipient group. Groups with a S donor are shown as filled bars, and groups with a R donor are shown as unfilled bars. The total number of mammary gland graft positive recipients that were evaluated for tumor outcome in each group were, respectively, 27, 22, 23, and 18 for S:S, R:R, S:R, and R:S.
Resequencing Mcs1b Potential Candidate Open Reading Frames
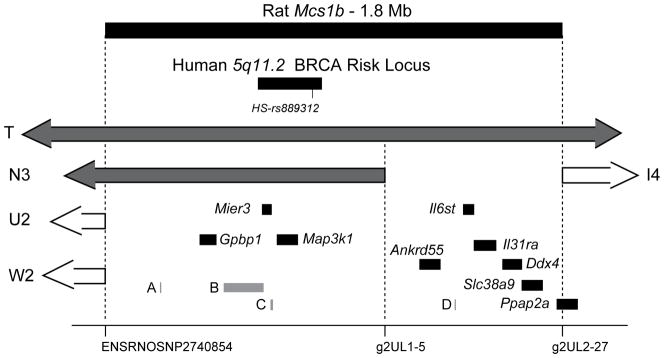
As shown in Fig. 3, rat Mcs1b was found to contain thirteen potential candidate gene transcripts as well as sequence orthologous to human 5q11.2, a GWAS-identified breast cancer risk associated allele marked by SNP rs889312 [2]. To prioritize potential candidates we resequenced conserved protein coding open reading frames (ORFs) that were within the 1.8 Mb interval that delimited Mcs1b, and based on RT-PCR gel electrophoresis were expressed in mammary glands of susceptible WF and Mcs1b resistant females (lines N3 and T). Transcripts from Gpbp1, Map3k1, Mier3, Ankrd55, Il6st, Il31ra, Ddx4, Slc38a9, and Ppap2a genes were detected in mammary gland total RNA pools from each genotype by RT-PCR. No genetic variants were identified between susceptible WF and Mcs1b resistant genotype ORFs or 3′ UTRs for these transcripts. Nucleotide sequences were submitted to NLM-NCBI.
Figure 3. Rat Mcs1b contains the ortholog of GWAS identified human 5q11.2 breast cancer risk allele marked by SNP rs889312 and multiple human/rat conserved transcripts.
Genetic markers used to map Mcs1b to this region are marked on the x-axis. Ends of relevant congenic lines are included for orientation. Potential candidate breast cancer susceptibility gene transcripts mapping to rat Mcs1b are shown as filled black bars that represent exonic and intronic DNA. Gene names are flanking 5′-UTRs of respective transcripts. Elements shown as gray bars and designated as A, B, C, or D correspond to predicted Actbl2, ENSRNOG00000013098, C5orf35, and ENSRNOG00000034909, respectively.
Four of the Mcs1b potential candidate genes were predicted transcripts (gray bars in Fig. 3). Rat Actbl2 (A in Fig. 3) is a psuedogene located outside rat genomic sequence orthologous to the human 5q11.2 haplotype block that associates with breast cancer risk. Predicted transcript ENSRNOG00000013098 (B in Fig. 3) was listed on the Ensembl genome browser [40]. We found no evidence by RT-PCR of a transcript from Actbl2 or ENSRNOG00000013098 in total RNA samples from multiple susceptible and Mcs1b resistant mammary glands or in rat mixed tissue total-RNA samples that included embryo, brain, testes, ovary, thymus, spleen, and liver. Since cDNA was not attainable and Actbl2 was predicted to be a single-exon we sequenced genomic DNA spanning this predicted psuedogene and found no sequence differences between WF and COP alleles.
A rat orthologous transcript (C in Fig. 3) to predicted C5ORF35 was not present in any total RNA samples tested from various rat tissues. We successfully amplified C5ORF35 from human thymus, spleen, and ovary, but not human breast tissue cDNA (Supplementary Fig. S1). However, in an Oncomine [41] search we found that other groups have reported detection of C5ORF35 in human breast carcinoma and non-diseased breast tissue. We noted that the annotated 5′- and 3′-UTRs of human C5ORF35 are poorly conserved between humans and rodents (Fig. S1); therefore, we concluded that C5ORF35 is a human, but not a rat transcript.
A predicted small nuclear RNA at rat Chr 2:43765811-43765918:1 named U6 or ENSRNOG00000034909 (D in Fig. 3) is estimated to be 108 bp on the forward strand. We noted that ENSRNOG00000034909 sequence aligned to multiple regions of the rat genome using both NCBI/BLAST and UCSC/BLAT [42, 43]. Thus, because of the highly repetitive nature of the sequence we were unable to design specific probes to determine if this predicted single exon gene was transcribed from rat Mcs1b.
Mcs1b Potential Candidate Gene Transcript Levels
Rat Mcs1b did not contain any protein coding genetic variation between Mcs1b susceptible and resistant alleles; therefore, rat Mcs1b may contain variation in one or more non-protein-coding regulatory elements that differentially control gene expression between mammary cancer susceptible and resistant genotypes. To test this hypothesis we measured mammary gland transcript levels of genes located at Mcs1b in 12-week old virgin female rats that were exposed to DMBA at 50–55 d to induce mammary carcinogenesis and age matched controls without DMBA. We focused on mammary gland transcript levels due to the mammary gland autonomous nature of Mcs1b. Twelve-week old animals were used because this age is after acute DMBA-toxicity and before frank mammary carcinomas are detectable.
Potential candidate gene transcript levels between Mcs1b genotype and DMBA exposure were analyzed by two-way ANOVA (Table 3). Effect of Mcs1b genotype was statistically significant for Gpbp1, Mier3, Map3k1, and Il6st. There was a significant effect of DMBA exposure on Map3k1 transcript levels. The interaction between Mcs1b genotype and DMBA exposure approached statistical significance for Map3k1. When mammary cancer susceptible and Mcs1b resistant genotypes were compared by exposure (with DMBA or without) mammary gland transcript levels were significantly different between susceptible and Mcs1b resistant females that were not carcinogen-induced for Gpbp1, Mier3, and Map3k1. Transcript levels of Gpbp1 and Map3k1 were not different between genotypes when DMBA-exposed females were evaluated. Significant expression differences between susceptible and Mcs1b resistant genotypes were sustained only for Mier3 when females given DMBA were compared between genotypes. We did not observe statistically significant differences in transcript levels of Ankrd55, Il31ra, Ddx4, Slc38a9, or Ppap2a between susceptible and Mcs1b resistant genotype mammary glands with DMBA or without.
Table 3.
Analysis and Statistics of Mcs1b Potential Candidate Gene Mammary Gland Transcript Levels in Mcs1b Resistant and Susceptible Genotypes at Twelve Weeks of Age
| Two-Way ANOVA F-Test P-Values
|
Log2 Target/Rplp2 Mean ± SD (n)
|
||||||
|---|---|---|---|---|---|---|---|
| Target | Mcs1b Genotype | Exposure | G X E | Exposure | Susceptible | Mcs1b Resistant | P value* |
| Gpbp1 | 0.0101 | 0.2090 | 0.6422 | Control | 0.586 ± 0.600 (34) | 0.044 ± 0.734 (29) | 0.0020 |
| DMBA | 0.281 ± 1.309 (45) | −0.097 ± 1.246 (42) | 0.1716 | ||||
| Mier3 | 0.0023 | 0.7911 | 0.6682 | Control | 0.115 ± 0.594 (34) | −0.522 ± 1.278 (34) | 0.0104 |
| DMBA | 0.154 ± 1.557 (45) | −0.688 ± 1.943 (48) | 0.0240 | ||||
| Map3k1 | 0.0002 | 0.0003 | 0.0588 | Control | −0.092 ± 0.818 (34) | −0.725 ± 0.767 (32) | 0.0019 |
| DMBA | 0.105 ± 0.564 (47) | −0.104 ± 0.651 (45) | 0.1036 | ||||
| Ankrd55 | 0.4694 | 0.2025 | 0.9019 | Control | −0.691 ± 0.678 (24) | −0.826 ± 1.108 (22) | 0.6180 |
| DMBA | −0.377 ± 1.296 (17) | −0.567 ± 1.006 (22) | 0.6090 | ||||
| Il6st | 0.0199 | 0.1744 | 0.8435 | Control | −0.066 ± 0.755 (36) | −0.418 ± 1.054 (33) | 0.1137 |
| DMBA | 0.189 ± 1.006 (44) | −0.227 ± 1.181 (48) | 0.0734 | ||||
| Il31ra | 0.2869 | 0.8674 | 0.9928 | Control | −0.331 ± 1.072 (24) | −0.559 ± 0.761 (23) | 0.4072 |
| DMBA | −0.368 ± 0.942 (20) | −0.592 ± 1.159 (23) | 0.4949 | ||||
| Ddx4 | 0.0555 | 0.5442 | 0.4045 | Control | −0.107 ± 0.983 (36) | −0.359 ± 0.911 (33) | 0.2748 |
| DMBA | −0.055 ± 1.122 (18) | −0.690 ± 1.575 (17) | 0.1769 | ||||
| Slc38a9 | 0.1008 | 0.3929 | 0.9730 | Control | −0.285 ± 0.600 (24) | −0.575 ± 0.681 (23) | 0.1284 |
| DMBA | −0.144 ± 0.970 (20) | −0.422 ± 0.954 (23) | 0.3499 | ||||
| Ppap2a | 0.3918 | 0.8314 | 0.5788 | Control | −0.385 ± 0.632 (24) | −0.447 ± 0.765 (23) | 0.7629 |
| DMBA | −0.315 ± 1.357 (20) | −0.605 ± 1.029 (23) | 0.4315 | ||||
Abbreviations: n, number of females; G, genotype; E, exposure; DMBA, 7,12-dimethylbenz[a]anthracene
Fisher’s PLSD test P values from comparing susceptible and Mcs1b resistant genotypes by exposure
Mammary gland transcript levels were lower in Mcs1b resistant genotype females for all genes with a significant difference between genotypes. Mier3 mean transcript levels were approximately 4.5-fold lower in Mcs1b resistant compared to susceptible genotype mammary glands whether animals were exposed to DMBA or not (Table 3). Thus, exposure to mammary carcinogen had no appreciable effect on Mier3 differences between susceptible and Mcs1b resistant genotype females. No significant differences in Mier3 transcript levels were detected between Mcs1b resistant and susceptible genotypes in spleen, thymus, ovary, or brain tissues (Supplemental Fig. S2). This suggests that Mier3 transcript level differences between Mcs1b alleles may be specific to mammary gland tissue.
A loss in statistical significance between DMBA-exposed susceptible and Mcs1b resistant females for Map3k1 was due to a statistically significant (P = 0.0003) increase in mean level of Map3k1 in the Mcs1b resistant genotype females with DMBA compared to age-matched controls of the same genotype without DMBA (Table 3). Map3k1 levels were not different (P = 0.2038) between susceptible WF mammary glands with DMBA or without.
Effect of Mcs1b Genotype on Body Weight
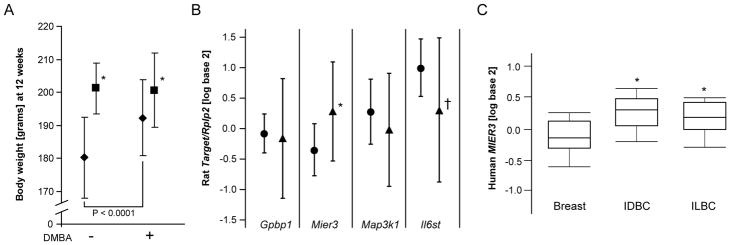
Travis et al. detected a significant association between human breast cancer risk associated SNP rs889312 and stature in women [44]. To determine if rat Mcs1b might also exhibit pleiotropy we analyzed rat body weight, which is information we routinely collect and relevant because body weight is genetically correlated to stature in humans [45]. Significant effects of Mcs1b genotype (P < 0.0001) and DMBA exposure (P = 0.0014) on body weight at 12 weeks of age were detected (Fig. 4A). The interaction between Mcs1b genotype and DMBA exposure was also significant (P = 0.0004). Females with the Mcs1b resistant genotype had mean ± SD body weights of 200 ± 11 grams with DMBA (n = 47) and 201 ± 7.7 grams without (n = 33), which were not significantly different (P = 0.7880). Comparatively, mammary cancer susceptible females had higher (P < 0.0001) mean ± SD body weight at 192 ± 11 grams with DMBA (n = 45) than unexposed susceptible females (n=34) who had a mean ± SD body weight of 180 ± 12 grams.
Figure 4. Rat Mcs1b resistant genotype is associated with higher body weight and rat/human Mier3/MIER3 levels are higher in rat mammary and human breast carcinomas.
Panel A: Lower body weight at 12 weeks of age was observed in mammary carcinoma susceptible (diamonds) compared to Mcs1b resistant females (squares) with DMBA and without (P < 0.0001 and P = 0.0007, respectively). Body weight was significantly higher in susceptible females that received DMBA compared to without (P < 0.0001). Panel B: Rat Mier3 transcript levels were significantly higher in DMBA-induced mammary carcinomas (triangles) compared to non-diseased mammary gland tissue (circles, P = 0.0120). Mean ± SD are graphed for each variable. Panel C: Human MIER3 was significantly higher in breast carcinomas compared to pathologically normal breast tissues. Oncomine (www.oncomine.org, reference 41 of main text) was used to query The Cancer Genome Atlas (cancergenome.nih.gov) gene expression database. Shown are box plots of log2 median centered MIER3 transcript levels for invasive ductal breast carcinomas (IDBC, n=392) and invasive lobular breast carcinomas (ILBC, n=36) compared to pathologically normal breast tissues (Breast, n=61). *P < 0.05. †P = 0.0569.
Higher Mier3 Transcript Levels in Mammary Carcinomas
Transcript levels of Mier3, Il6st, Gpbp1, and Map3k1 in DMBA-induced mammary carcinomas that developed in susceptible (n=28) and Mcs1b resistant genotype (n=25) mammary glands were measured by QPCR to determine if there was an effect of Mcs1b genotype on levels of any of these transcripts in mammary carcinoma tissue. These genes were evaluated because significant effects of Mcs1b genotype on mammary gland transcript levels of these genes were detected (Table 3). Further, Il6st was included because it had been reported to be higher in rat mammary carcinoma compared to mammary gland tissues [46]. We collected total RNA from DMBA-induced mammary carcinomas (n= 1 or 2 per rat) and adjacent non-diseased mammary gland tissue from 21–23 week old females (n= 6 per genotype). There were no statistically significant differences in mammary carcinoma transcript levels between Mcs1b genotypes for any of the four genes tested. However, as shown in Fig. 4B, Mier3 transcript levels were significantly higher (1.8-fold) in mammary carcinomas compared to non-diseased mammary tissue. We also observed that Il6st was potentially different between mammary carcinomas and non-diseased mammary glands; however, this comparison did not meet our statistical significance criterion (Fig. 4B).
Oncomine [41] was used to query The Cancer Genome Atlas (cancergenome.nih.gov) gene expression database to find that levels of human MIER3 were, respectively, 1.33 and 1.20 fold higher in invasive ductal (n = 392) and invasive lobular (n = 36) breast carcinoma samples compared to pathologically normal breast tissues (n = 61) (P = 2.8 × 10−13, ductal; P = 6.3 × 10−4,lobular; t-tests, Fig. 4C). Thus, both human/rat MIER3/Mier3 levels are higher in breast/mammary carcinoma compared to non-diseased breast/mammary tissues.
Localization of Human MIER3 Protein to Nuclei
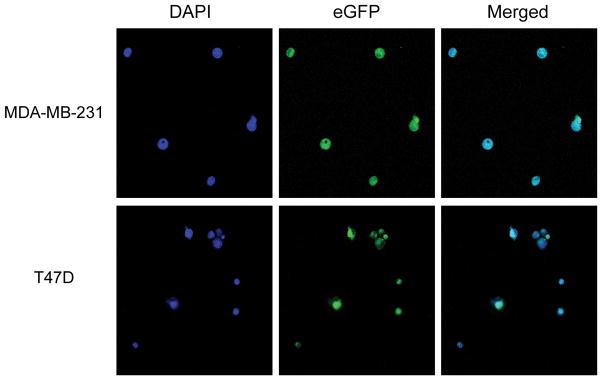
MIER3 encodes an uncharacterized member of the mesoderm induction early response family of proteins. The gene product of family member MIER1 is a transcription factor targeted to the nucleus [47]. Therefore, we determined if MIER3 may also encode a transcription factor targeted to the nucleus. We cloned a human MIER3 ORF into an enhanced green fluorescent protein (eGFP) expression vector and transiently transfected this expression vector into MDA-MB-231 and T47D breast cancer cell lines. In vitro expression of MIER3 linked to eGFP resulted in green fluorescence in distinct foci compared to eGFP alone (Supplementary Fig. S3). We confirmed that foci were nuclei by determining that MIER3-eGFP co-localized with DAPI-staining in both cell lines (Fig. 5).
Figure 5. Ectopically-expressed eGFP-MIER3 localizes to nuclei in breast cancer cell lines.
Plasmid expression vectors containing eGFP_MIER3 ORFs were transiently transfected into MDA-MB-231 or T47D breast cancer cell lines. At 24 h cells were fixed in formalin and stained with DAPI to visualize nuclei. Confocal microscopy was used to visualize DAPI staining and green fluorescence. Green fluorescence from eGFP-MIER3 and DAPI nuclear staining co-localized in both cell lines (Merged).
Discussion
Rat mammary carcinoma susceptibility, like human breast cancer risk, is complex as both are controlled by multiple susceptibility alleles and environmental factors. We have mapped rat Mcs1b to a 1.8 Mb region of rat chromosome 2 using multiple congenic lines. We found that rat Mcs1b is highly relevant to human breast cancer susceptibility as it contains genomic sequence orthologous to a low-penetrance breast cancer risk allele at human chromosome 5q11.2. This human susceptibility allele was first reported by Easton et al. in the first population-based breast cancer risk GWAS [2]. Human 5q11.2 has been confirmed to strongly associate with breast cancer risk in multiple independent studies of European- and Asian-descent populations [34–39]. This is the first report of a rodent complex disease susceptibility QTL with a GWAS-identified concordant human ortholog that had a probability of association below a stringent significance level of P ≤ 10−7, which is widely deemed to be required for genome-wide studies.
An experimental organism with a segregating concordant susceptibility allele implies that functional genetic studies may translate directly to human biology and disease. For example, Gould and colleagues reported that rat Mcs5a, a Wistar Kyoto (WKY) strain resistance QTL that is concordant to human MCS5A, acted in a non-mammary cell-autonomous manner that involves immune cells [48]. Here, we used rat genetic lines to show that Mcs1b controls mammary cancer susceptibility by an undetermined mechanism that is autonomous to mammary gland tissue. While our result is in agreement with previous work that concluded a majority, but not all, of the COP rat strain resistance to mammary cancer is mammary gland autonomous [49]; it further highlights that the WKY and COP strains may achieve mammary carcinoma resistance thru different genetically determined cellular and molecular mechanisms. Mechanisms that are likely genetically determined in humans.
Most common genetic variation associated with human complex disease susceptibility appears to be located in non-protein-coding DNA. Since we found no genetic variation between susceptible and resistant allele Mcs1b ORFs, we conclude that Mcs1b is likely a noncoding gene regulatory element(s), such as a transcription factor binding site or noncoding RNA. This would be similar to the hypothesized identity of the human 5q11.2 breast cancer risk associated element. Human polymorphisms that are contained in public databases and highly correlated with human 5q11.2 breast cancer risk associated SNP rs889312 are in non-protein-coding DNA. There are no known noncoding RNAs in either the human or rat ortholog; therefore, another type of gene regulatory element is likely responsible for or associated to susceptibility differences.
Our studies suggest that MIER3 is a strong candidate breast cancer susceptibility gene at human 5q11.2. We have identified Mier3 as a strong Mcs1b candidate gene in this study based on different Mier3 mammary gland transcript levels between susceptible and Mcs1b resistant genotypes. Lower levels of Mier3 in Mcs1b resistant genotype females were genetically determined and not dependent on the induction of mammary carcinogenesis by DMBA. We also found Mier3 levels to be significantly lower in non-diseased rat mammary tissue compared to mammary carcinoma. Further, we queried The Cancer Genome Atlas gene expression database and noted that human MIER3 levels were higher in both ductal and lobular breast carcinomas compared to breast tissue.
MIER3 or mesoderm induction early response 1, family member 3 (GenBank ref|NM_152622) is an uncharacterized gene. We determined that MIER3 localized to the nucleus. Human and rat MIER3/Mier3 (GenBank ref| NP_689835.3 and NP_001161472.1) gene products share 93% amino acid sequence identity, and human MIER3 and MIER1 (GenBank ref|NP_001071172.1) have 54% identical amino acids based on BLAST [42]. MIER1 physically interacts with estrogen receptor alpha, Sp1, and Creb-binding protein [50–52]. MIER1 contains one, while MIER3 has two conserved LXXLL sequences, which is a motif that facilitates nuclear hormone receptor interactions [53]. A potential functional difference between MIER1 and MIER3 may be that a difference in the number of LXXLL motifs between them results in physical interactions with different nuclear hormone receptors [54].
In addition to MIER3, MAP3K1 and C5ORF35 reside within the human 5q11.2 haplotype block that associates with breast cancer risk. Even though there are no published studies in support, MAP3K1 is often considered the candidate breast cancer susceptibility gene at 5q11.2 due to its location within the breast cancer risk associated haplotype block and known function as a serine/threonine kinase. In our rat studies, Map3k1 was differentially expressed between susceptible and Mcs1b resistant congenic rats that had not been induced to undergo mammary carcinogenesis; however, mammary glands that had been exposed to mammary carcinogen did not show a difference in Map3k1 levels between Mc1b alleles. An interesting result in our study with respect to Map3k1, which may have important implications for human studies of potential genotype-environment interactions, is exposure to mammary carcinogen resulted in increased mammary gland Map3k1 levels for the Mcs1b resistant, but not the susceptible genotype. We found no evidence of a rat orthologous transcript to human C5ORF35 in multiple rat tissues. Further, exonic elements of C5ORF35 have not been conserved in the rat. Therefore, we conclude that MAP3K1 and C5ORF35 are not as likely as MIER3 to be breast cancer susceptibility genes.
We noted that both rat Mcs1b and human 5q11.2 exhibit pleiotropy. Travis et al. reported that carriers of the increased risk allele at human 5q11.2 were significantly shorter in height than non-carriers [44]. In our study, high risk female rats had lower body weight than Mcs1b resistant females. We noted on the Rat Genome Database that there is a predicted rat body weight QTL named Bw1 that overlaps Mcs1b and is associated with mesenteric body fat amount [55]. Both human and rat study results are counter intuitive as one might expect taller women and heavier rats to be at greater cancer risk. Thus, it is important to note that, as expected with low-penetrance alleles, the quantitative difference between the means for each human genotype were subtle with overlapping distributions. Mean height difference was 7 mm between non-carriers and carriers of the increased risk allele. In our study, we analyzed only body weight, and not specific components of body weight, such as bone density or fat amounts. Thus, better descriptive traits would likely be more informative. It is notable that the pleitropic effects of these alleles opens the possibility that other experimental organisms, approaches, and study designs without focus on breast or mammary cancer may be useful to functionally characterize breast cancer risk associated genetic variation at 5q11.2.
In conclusion, rat Mcs1b is mammary gland autonomous allele and a non-protein-coding genetic element that is orthologous to the GWAS-identified human 5q11.2 breast cancer susceptibility locus. We propose that MIER3 is a strong candidate breast cancer susceptibility gene.
Supplementary Material
Acknowledgments
NIH grants R01CA137052 (DJS), P30ES014443 (DJS), T32ES011564 (ADD), and T35ES014559-06 (MDV); and Competitive Enhancement and Intramural Research Incentive Grants from the University of Louisville Office of the Executive Vice President for Research (DJS).
The authors thank Jason Willer, Jane Williams, Thomas Ray, Kendall Prater, and Deepa Patel for contributing technical assistance. This work was supported by National Institutes of Health grants CA137052 and ES014443, and partially supported by Competitive Enhancement and Intramural Research Incentive Grants from the University of Louisville Office of the Executive Vice President for Research. ADD and MDV were, respectively, supported by National Institutes of Health Training Grants T32ES011564 and T35ES014559-06.
Footnotes
Potential conflicts of interest: None.
References
- 1.Samuelson DJ, Hesselson SE, Aperavich BA, Zan Y, Haag JD, Trentham-Dietz A, Hampton JM, Mau B, Chen KS, Baynes C, et al. Rat Mcs5a is a compound quantitative trait locus with orthologous human loci that associate with breast cancer risk. Proc Natl Acad Sci U S A. 2007;104(15):6299–6304. doi: 10.1073/pnas.0701687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, Masson G, Jakobsdottir M, Thorlacius S, Helgason A, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 4.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng W, Long J, Gao Y-T, Li C, Zheng Y, Xiang Y-B, Wen W, Levy S, Deming SL, Haines JL, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(3):324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, Hankinson SE, Hutchinson A, Wang Z, Yu K, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41(5):579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, Jakobsdottir M, Bergthorsson JT, Gudmundsson J, Aben KK, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40(6):703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 8.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, Morrison J, Maranian M, Pooley KA, Luben R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(5):585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mavaddat N, Dunning AM, Ponder BAJ, Easton DF, Pharoah PD. Common Genetic Variation in Candidate Genes and Susceptibility to Subtypes of Breast Cancer. Cancer Epidemiol Biomarkers Prev. 2009;18(1):255–259. doi: 10.1158/1055-9965.EPI-08-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42(6):504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, Healey S, Morrison J, Kartsonaki C, Lesnick T, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42(10):885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Q, Long J, Lu W, Qu S, Wen W, Kang D, Lee J-Y, Chen K, Shen H, Shen C-Y, et al. Genome-wide association study identifies breast cancer risk variant at 10q21.2: results from the Asia Breast Cancer Consortium. Human Molecular Genetics. 2011;20(24):4991–4999. doi: 10.1093/hmg/ddr405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, Walker K, Zelenika D, Gut I, Heath S, Palles C, et al. Novel Breast Cancer Susceptibility Locus at 9q31.2: Results of a Genome-Wide Association Study. Journal of the National Cancer Institute. 2011;103(5):425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 14.Blackburn AC. Rats: gnawing through the barriers to understanding genetic susceptibility and breast cancer. Breast cancer research : BCR. 2011;13(5):112. doi: 10.1186/bcr2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gould MN. The Utility of Comparative Genetics to Inform Breast Cancer Prevention Strategies. Genetics. 2009;183(2):409–412. doi: 10.1534/genetics.109.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russo J, Tait L, Russo IH. Susceptibility of the mammary gland to carcinogenesis. III. The cell of origin of rat mammary carcinoma. Am J Pathol. 1983;113(1):50–66. [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan VC. Chemosuppression of breast cancer with long-term tamoxifen therapy. Prev Med. 1991;20(1):3–14. doi: 10.1016/0091-7435(91)90002-l. [DOI] [PubMed] [Google Scholar]
- 18.Stoll BA. Breast cancer and the western diet: role of fatty acids and antioxidant vitamins. Eur J Cancer. 1998;34(12):1852–1856. doi: 10.1016/s0959-8049(98)00204-4. [DOI] [PubMed] [Google Scholar]
- 19.Russo J, Russo IH. Atlas and histologic classification of tumors of the rat mammary gland. J Mammary Gland Biol Neoplasia. 2000;5(2):187–200. doi: 10.1023/a:1026443305758. [DOI] [PubMed] [Google Scholar]
- 20.Nagao M, Ushijima T, Watanabe N, Okochi E, Ochiai M, Nakagama H, Sugimura T. Studies on mammary carcinogenesis induced by a heterocyclic amine, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, in mice and rats. Environ Mol Mutagen. 2002;39(2-3):158–164. doi: 10.1002/em.10047. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins S, Rowell C, Wang J, Lamartiniere CA. Prenatal TCDD exposure predisposes for mammary cancer in rats. Reprod Toxicol. 2007;23(3):391–396. doi: 10.1016/j.reprotox.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarbl H. Toxicogenomic Analyses of Genetic Susceptibility to Mammary Gland Carcinogenesis in Rodents: Implications for Human Breast Cancer. Breast Disease. 2007;28(1):87–105. doi: 10.3233/bd-2007-28109. [DOI] [PubMed] [Google Scholar]
- 23.Gould MN, Wang B, Moore CJ. Modulation of Mammary Carcinogenesis by Enhancer and Suppressor Genes. New York and Basel: Marcel Dekker, Inc; 1989. [Google Scholar]
- 24.Dunning WF, Curtis MR, Segaloff A. Strain differences in response to estrone and the induction of mammary gland, adrenal, and bladder cancer in rats. Cancer Res. 1953;13(2):147–152. [PubMed] [Google Scholar]
- 25.Isaacs JT. Genetic control of resistance to chemically induced mammary adenocarcinogenesis in the rat. Cancer Res. 1986;46(8):3958–3963. [PubMed] [Google Scholar]
- 26.Moore CJ, Tricomi WA, Gould MN. Comparison of 7,12-dimethylbenz[a]anthracene metabolism and DNA binding in mammary epithelial cells from three rat strains with differing susceptibilities to mammary carcinogenesis. Carcinogenesis. 1988;9(11):2099–2102. doi: 10.1093/carcin/9.11.2099. [DOI] [PubMed] [Google Scholar]
- 27.Gould KA, Tochacek M, Schaffer BS, Reindl TM, Murrin CR, Lachel CM, VanderWoude EA, Pennington KL, Flood LA, Bynote KK, et al. Genetic determination of susceptibility to estrogen-induced mammary cancer in the ACI rat: mapping of Emca1 and Emca2 to chromosomes 5 and 18. Genetics. 2004;168(4):2113–2125. doi: 10.1534/genetics.104.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shull JD. The rat oncogenome: comparative genetics and genomics of rat models of mammary carcinogenesis. Breast Dis. 2007;28:69–86. doi: 10.3233/bd-2007-28108. [DOI] [PubMed] [Google Scholar]
- 29.Szpirer C, Szpirer J. Mammary cancer susceptibility: human genes and rodent models. Mamm Genome. 2007;18(12):817–831. doi: 10.1007/s00335-007-9073-x. [DOI] [PubMed] [Google Scholar]
- 30.Moore CJ, Bachhuber AJ, Gould MN. Relationship of mammary tumor susceptibility, mammary cell-mediated mutagenesis, and metabolism of polycyclic aromatic hydrocarbons in four types of rats. J Natl Cancer Inst. 1983;70(4):777–784. [PubMed] [Google Scholar]
- 31.Blackburn AC, Jerry DJ. Map making in the 21st century: charting breast cancer susceptibility pathways in rodent models. Journal of Mammary Gland Biology and Neoplasia. 2011;16(1):57–64. doi: 10.1007/s10911-011-9201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders J, Haag JD, Samuelson DJ. Physical Confirmation and Mapping of Overlapping Rat Mammary Carcinoma Susceptibility QTLs, Mcs2 and Mcs6. PLoS ONE. 2011;6(5):e19891. doi: 10.1371/journal.pone.0019891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haag JD, Shepel LA, Kolman BD, Monson DM, Benton ME, Watts KT, Waller JL, Lopez-Guajardo CC, Samuelson DJ, Gould MN. Congenic rats reveal three independent Copenhagen alleles within the Mcs1 quantitative trait locus that confer resistance to mammary cancer. Cancer Res. 2003;63(18):5808–5812. [PubMed] [Google Scholar]
- 34.Antoniou AC, Spurdle AB, Sinilnikova OM, Healey S, Pooley KA, Schmutzler RK, Versmold B, Engel C, Meindl A, Arnold N, et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet. 2008;82(4):937–948. doi: 10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, Bojesen SE, Nordestgaard BG, Axelsson CK, Arias JI, et al. Heterogeneity of Breast Cancer Associations with Five Susceptibility Loci by Clinical and Pathological Characteristics. PLoS Genetics. 2008;4(4):e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng W, Wen W, Gao YT, Shyr Y, Zheng Y, Long J, Li G, Li C, Gu K, Cai Q, et al. Genetic and clinical predictors for breast cancer risk assessment and stratification among Chinese women. Journal of the National Cancer Institute. 2010;102(13):972–981. doi: 10.1093/jnci/djq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han W, Woo JH, Yu JH, Lee MJ, Moon HG, Kang D, Noh DY. Common genetic variants associated with breast cancer in Korean women and differential susceptibility according to intrinsic subtype. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011;20(5):793–798. doi: 10.1158/1055-9965.EPI-10-1282. [DOI] [PubMed] [Google Scholar]
- 38.Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, Apicella C, Smith LD, Hammet F, Southey MC, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Human Molecular Genetics. 2011;20(16):3289–3303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campa D, Kaaks R, Le Marchand L, Haiman CA, Travis RC, Berg CD, Buring JE, Chanock SJ, Diver WR, Dostal L, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. Journal of the National Cancer Institute. 2011;103(16):1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Research. 2011;39(suppl 1):D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Research. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Travis RC, Reeves GK, Green J, Bull D, Tipper SJ, Baker K, Beral V, Peto R, Bell J, Zelenika D, et al. Gene-environment interactions in 7610 women with breast cancer: prospective evidence from the Million Women Study. Lancet. 2010;375(9732):2143–2151. doi: 10.1016/S0140-6736(10)60636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Czerwinski SA, Lee M, Choh AC, Wurzbacher K, Demerath EW, Towne B, Siervogel RM. Genetic factors in physical growth and development and their relationship to subsequent health outcomes. Am J Hum Biol. 2007;19(5):684–691. doi: 10.1002/ajhb.20663. [DOI] [PubMed] [Google Scholar]
- 46.Qiu C, Yu M, Shan L, Snyderwine EG. Allelic imbalance and altered expression of genes in chromosome 2q11-2q16 from rat mammary gland carcinomas induced by 2-amino-1-methyl-6-phenylimidazo[lsqb]4,5-b[rsqb]pyridine. Oncogene. 2003;22(8):1253–1260. doi: 10.1038/sj.onc.1206233. [DOI] [PubMed] [Google Scholar]
- 47.Paterno GD, Li Y, Luchman HA, Ryan PJ, Gillespie LL. cDNA Cloning of a Novel, Developmentally Regulated Immediate Early Gene Activated by Fibroblast Growth Factor and Encoding a Nuclear Protein. Journal of Biological Chemistry. 1997;272(41):25591–25595. doi: 10.1074/jbc.272.41.25591. [DOI] [PubMed] [Google Scholar]
- 48.Smits B, Sharma D, Samuelson D, Woditschka S, Mau B, Haag J, Gould M. The non-protein coding breast cancer susceptibility locus Mcs5a acts in a non-mammary cell-autonomous fashion through the immune system and modulates T-cell homeostasis and functions. Breast Cancer Research. 2011;13(4):R81. doi: 10.1186/bcr2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang R, Haag JD, Gould MN. Site of expression and biological function of the rat mammary carcinoma suppressor gene. Carcinogenesis. 1990;11(10):1765–1770. doi: 10.1093/carcin/11.10.1765. [DOI] [PubMed] [Google Scholar]
- 50.McCarthy PL, Mercer FC, Savicky MWJ, Carter BA, Paterno GD, Gillespie LL. Changes in subcellular localisation of MI-ER1[alpha], a novel oestrogen receptor-[alpha] interacting protein, is associated with breast cancer progression. Br J Cancer. 2008;99(4):639–646. doi: 10.1038/sj.bjc.6604518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blackmore T, Mercer C, Paterno G, Gillespie L. The transcriptional cofactor MIER1-beta negatively regulates histone acetyltransferase activity of the CREB-binding protein. BMC Research Notes. 2008;1(1):68. doi: 10.1186/1756-0500-1-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding Z, Gillespie LL, Mercer FC, Paterno GD. The SANT Domain of Human MI-ER1 Interacts with Sp1 to Interfere with GC Box Recognition and Repress Transcription from Its Own Promoter. Journal of Biological Chemistry. 2004;279(27):28009–28016. doi: 10.1074/jbc.M403793200. [DOI] [PubMed] [Google Scholar]
- 53.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387(6634):733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 54.McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, Inostroza J, Torchia J, Nolte RT, Assa-Munt N, et al. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes & Development. 1998;12(21):3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogino T, Wei S, Wei K, Moralejo DH, Kose H, Mizuno A, Shima K, Sasaki Y, Yamada T, Matsumoto K. Genetic evidence for obesity loci involved in the regulation of body fat distribution in obese type 2 diabetes rat, OLETF. Genomics. 2000;70(1):19–25. doi: 10.1006/geno.2000.6349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.