Abstract
Platinum-based drugs have been used to successfully treat diverse cancers for several decades. Cisplatin, the original compound of this class, cross-links DNA, resulting in cell cycle arrest and cell death via apoptosis. Cisplatin is effective against several tumor types, yet it exhibits toxic side effects and tumors often develop resistance. To mitigate these liabilities while maintaining potency, we generated a library of non-classical platinum-acridine hybrid agents and assessed their mechanisms of action using a validated genome-wide screening approach in Saccharomyces cerevisiae and in the distantly related yeast Schizosaccharomyces pombe. Chemogenomic profiles from both S. cerevisiae and S. pombe demonstrate that several of the platinum-acridines damage DNA differently than cisplatin based on their requirement for distinct modules of DNA repair.
Platinum-based drugs are extremely successful cancer chemotherapeutics. The widely used agent cisplatin (cis-diamminedichloroplatinum(II)) acts by forming cross-links in the major groove of DNA which inhibit DNA synthesis and lead to apoptosis(1). Cisplatin is effective against testicular, ovarian, bladder and cervical tumors but it manifests dose-limiting toxicities on the nervous and renal systems. Equally troubling is the frequency at which resistance develops during treatment(2). In light of these limitations, attention has focused on developing platinum-based agents with improved activity in notoriously cisplatin-resistant cancers.
Diverse designs for new platinum-based anticancer drugs have focused on modifying substituents surrounding the cisplatin core. Clinical data shows that these compounds offer certain advantages over cisplatin, yet because their mechanism of action is similar to that of cisplatin at the nuclear level, they represent an incremental improvement(3). Alternative approaches to platinum-based drug design, in contrast, do not adhere to all of the structural dictates of cisplatin(4). For example, a non-classical trinuclear platinum agent, BBR3464 (Novuspharma/Cell Therapeutics), damages DNA in a radically different manner than cisplatin and showed efficacy against colorectal, pancreatic and ovarian cancer through phase II clinical trials(5).
Recently, a class of promising platinum-acridine conjugates was developed by combining the DNA-damaging features of a platinum complex with the DNA-intercalating properties of acridine-based chromophores. The prototype of this class of hybrid agents, PT-ACRAMTU(EN) ([PtCl(en)(ACRAMTU)] dinitrate salt, ACRAMTU = 1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea, en = ethane-1,2-diamine), shows a dose-response similar or superior to cispla-tin in several solid tumor cell lines(6–8). In the DNA adducts produced by PT-ACRAMTU(EN), the metal forms a single coordinative bond with the nucleobase nitrogen while the ACRAMTU moiety intercalates into the base-pair step adjacent to the site of platination(9). This results in a novel mechanism of DNA damage distinct from the DNA cross-linking characteristic of cispla-tin. Based on these promising initial results, additional platinum-acridine compounds were synthesized with a focus on modifications to the prototypical PT-ACRAMTU(EN) structure (Fig. 1)(10–12). Structure-activity relationship studies revealed PT-ACRAMTU(EN) analogues with up to 500-fold higher potency in non-small cell lung cancer (NSCLC) cells when compared to cisplatin. The promise offered by the platinum-acridines is clear, yet, the cellular mechanism(s) of these dual-mode agents remains largely uncharacterized.
Figure 1.
Chemical structures of the platinum-acridine compounds and carriers screened.
Here we use a validated cell-based chemical genomic assay to examine a library of platinum-acridine hybrid agents to characterize their cellular mechanism(s) of action and evaluate their potential as anti-cancer agents. We have previously shown that the Saccharomyces cerevisiae gene deletion collection is a powerful means to dissect the detailed mechanisms of action of functionally diverse DNA-damaging agents(13) by screening these agents against the complete pool of ~4,800 barcoded non-essential deletion strains. In this study we apply a similar approach in both S. cerevisiae and the distantly related fission yeast Schizosaccharomyces pombe to dissect the mechanism(s) of action of these novel platinum-acridine conjugates. The results offer insight into the relative role of each structural modification on the genome-wide response and provide mechanistic insight into the relationship between compound structure and biological activity.
RESULTS AND DISCUSSION
Platinum-acridines derived from the prototype PT-ACRAMTU(EN) were developed using a modular synthetic approach. The compounds tested in this study can be grouped into three categories: platinum-free carriers (2 derivatives), platinating-intercalating hybrid agents (7 derivatives) and bisintercalators (4 derivatives) (Fig. 1). Each genome-wide screen produces a fitness profile that consists of a numerical value for each deletion strain’s fitness (relative growth with and without drug treatment) (Supplementary Dataset 1). To compare genome-wide profiles between compounds, we used a rank-based similarity scoring algorithm developed to account for non-biological “batch”-based systematic effects (14). With this algorithm, every gene deletion strain within a profile is distributed into sections, or “buckets,” where strains that are most influenced by the chemical compound (and therefore have the highest fitness defect scores) comprise one bucket, the next set of strains are distributed into a second bucket, and so forth. Once all strains are divided into buckets, a scoring matrix is constructed and used to evaluate the similarity between profiles (see Methods). This approach effectively minimizes the influence of “batch” effects that can overwhelm the biological signal. Using the resulting profile similarity scores, compounds with similar mechanisms cluster together, with experimental replicates being the most similar (Fig. 2).
Figure 2.
Two-dimensional hierarchical clustering of all 14 compounds based on the profile similarity scores of their genome-wide S. cerevisiae profiles. Red indicates high similarity between compound profiles and grey indicates low similarity. Profiles from experiments repeated under the same conditions cluster together with few outliers, indicating the reproducibility and resolution of the genome-wide assay. The dendrogram reveals structure-function relationships between compounds.
Clustering of all S. cerevisiae profiles revealed that, of all the platinum-acridines, PT-ACRAMTU(EN) induces a cellular response most similar to cisplatin, further suggesting that the most significantly sensitive strains represent those deleted for genes involved in DNA-damage repair. PT-ACRAMTU(PN), PT-ATUCA and PT-AMIDINE(EN) also grouped together with cis-platin and PT-ACRAMTU(EN). The platinum-free carriers and the 7 other platinum-acridine compounds clustered separately. Notably, PT-ACRAMTU(BPY), which was previously examined for its DNA-damaging properties(11) clusters with the platinum-free carriers, suggesting that it does not generate fitness defects in S. cerevisiae. The individual genome-wide strain sensitivities for each compound are discussed in detail below.
Cisplatin was profiled to serve as a benchmark for the platinum-acridines. We previously demonstrated that the cellular response to cisplatin requires genes with roles in nucleotide excision repair (NER), homologous recombination repair (HRR), post-replication repair (PRR) and translesion synthesis (TLS)(13). In this study, strains deleted for genes in NER (RAD1, RAD2, RAD4, RAD10 and RAD14), HRR (RAD51, RAD52, RAD54, RAD55 and RAD57) and PRR (RAD5 and RAD18) were among the top 30 most cisplatin-sensitive strains. Strains deleted for PSO2, an NER gene involved in cross-link repair, and genes involved in TLS (REV1, REV3 and REV7) also ranked highly; these were previously found to be the principal differences in the response to well-characterized cross-linking and non-cross-linking agents(13).
PT-ACRAMTU(EN) (Fig. 1) forms monofunctional-intercalative adducts in both grooves of double-stranded DNA but does not cross-link nucleobases(15), therefore we anticipated its ge-nome-wide profile would be distinct from cisplatin. Contrary to this expectation, the DNA repair modules important for resistance to PT-ACRAMTU(EN) are most similar to those of cisplatin. The top 30 strains are deleted for genes in NER (RAD1, RAD2, RAD4, RAD10 and RAD14), HRR (RAD51, RAD54 and XRS2) and PRR (RAD5 and RAD18) (Fig. 3) with NER and PRR being the most sensitive modules (Fig. 4a). One major difference from cisplatin is that deletion of PSO2 did not sensitize this strain to PT-ACRAMTU(EN), suggesting that the monoadducts formed are not recognized by cross-link-specific repair mechanisms. In addition, genes essential for TLS were not found in the PT-ACRAMTU(EN) profile, as would be predicted for a non-cross-linking compound. The relative ranking of strains from these five DNA-damage response (DDR) modules are shown in Figure 4a for those platinum-acridines whose profiles suggest they exert their cellular effects via damaging DNA. Significantly sensitive strains were tested in monoculture for compound sensitivity using fitness assays and spot dilutions with a 77% hit confirmation rate (Table S1, S2). To summarize the cellular responses captured by S. cerevisiae profiles, we performed gene set enrichment analysis (GSEA) to identify biological processes significantly enriched amongst genes whose deletions induce compound-sensitivity, and visualized the results as a network (Fig. S1). The current state of S. pombe gene annotation is too sparse to permit a similar analysis.
Figure 3.
S. cerevisiaeand S. pombe profiles for the platinum-acridine compounds that require DNA-damage response pathways. The top 30 sensitive strains are marked and labeled. Each graph represents the average of at least 3 replicate sensitivity screens in S. cerevisiae or 2 replicates in S. pombe. The x-axis represents gene names in alphabetical order and the y-axis is the log2 ratio of barcode intensity for drug vs control. Red indicates essential genes, blue indicates non-essential genes and purple indicates marked genes. Strains that often appear as hits in yeast chemogenomic assays are listed in Table S3.
Figure 4.
Four platinum-acridine compounds damage DNA, interfere with cell cycle progression and disrupt mitochondrial function. a) Relative importance of DNA-damage repair modules in the resistance to the DNA-damaging platinum compounds in S. cerevisiae and S. pombe. Each bar represents the median rank for genes in each of the DNA repair modules listed in the top 30 or top 250 most sensitive strains. The DNA-repair modules in S. cerevisiae were defined as follows: cross-linking genes (PSO2), NER (RAD1, RAD2, RAD4, RAD10, RAD14), PRR (RAD5, RAD6, RAD18), TLS (REV1, REV3, REV7), HRR (RAD51, RAD52, RAD54, RAD55, RAD57, RAD59), stalled replication fork repair (MUS81, MMS4). The DNA-repair modules in S. pombe were defined as follows: NER (RHP14, RHP41, RHP42, RAD13, RAD16, SWI10), PRR (RHP6, RHP18, RAD8), TLS (REV1, REV3, REV7), HRR (RHP51, RHP54, RHP55, RHP57, RAD22, RTI1), stalled replication fork repair (MUS81). b) DNA content analysis pro-files. Haploid BY4741 cells were synchronized at G1 prior to the addition of compound at IC50. Samples were taken every 30 min after compound addition. The positions of the 1N and 2N DNA content peaks are indicated. c) Mitochondrial function is disrupted by platinum-acridine compounds. Wildtype BY4743 cells were grown in the presence of four platinum-acridine compounds under fermenting (blue) or respiring (red) conditions. The concentration to inhibit growth by 20% (IC20) is plotted.
Three of the seven platinum-intercalating hybrid derivatives of PT-ACRAMTU(EN) elicit responses from DDR genes. The bidentate nonleaving group in PT-ACRAMTU(EN) was replaced with various diamines to yield PT-ACRAMTU(PN), PT-ACRAMTU(BPY) and PT-ACRAMTU(TMEDA) (Fig. 1). In previously reported in vitro DNA polymerase stop assays, PT-ACRAMTU(PN) and PT-ACRAMTU(BPY) were shown to produce DNA adducts(11). Clustering of genome-wide profiles revealed that PT-ACRAMTU(PN) groups with PT-ACRAMTU(EN) and cisplatin while PT-ACRAMTU(BPY) and PT-ACRAMTU(TMEDA) cluster separately (Fig. 2). The genome-wide response to PT-ACRAMTU(PN) differed from that of PT-ACRAMTU(EN); strains deleted for genes involved in NER ranked lower in sensitivity, while strains deleted for genes involved in HRR and PRR were more sensitive. Although PT-ACRAMTU(BPY) damages DNA, we did not uncover strains deleted for DDR genes as sensitive in our screens, consistent with previously reported model studies which showed that the trans-labilizing effect of the bipyridine (BPY) ligand renders the DNA adducts formed by this compound kinetically labile(11). Instead, PT-ACRAMTU(BPY) elicited responses for genes involved in RNA metabolic processes. PT-ACRAMTU(TMEDA), which does not bind to DNA, did not elicit responses from any known DDR genes.
To tune the DNA sequence specificity of the monofunctional platinum moiety, the carrier group of PT-ACRAMTU(EN) was modified to produce PT-ATUCA, which features a carboxamide group attached at the 4-position of the acridine chromophore. Despite this drastic modification, the fitness profile for PT-ATUCA was similar to that of PT-ACRAMTU(EN); strains deleted for genes involved in NER, HRR and PRR were the most sensitive (Fig. 3), with NER being the most important pathway for response to this compound (Fig. 4a). To generate PT-AMIDINE(EN) (the first member of this class able to slow NSCLC tumor growth in a murine xenograft(12)) the thiourea linkage was replaced with an amidine group. This seemingly minor modification resulted in greatly accelerated platinum-DNA binding(16) and reduced reactivity with cysteine sulphur, a contributor to cisplatin toxicity and tumor resistance(17). The fitness profile for this promising lead compound featured sensitive strains deleted for genes involved in NER, HRR and PRR. A distinct feature of this profile is that two genes essential for stalled-fork repair, MUS81 and MMS4, appeared in the top 15 most sensitive strains (Fig. 3). Stalled-fork repair is known to be important for the cellular response to most platinum agents, yet only for PT-AMIDINE(EN) do these deletion strains rank so highly (Fig. 4a). PT-ACR49NME2, where the intercalator group is modified with a DNA threading 4-carboxamide group, showed no significant gene enrichment, consistent with its low levels of DNA-binding and modest activity against cultured cells(18).
Platinum bisintercalators were generated by replacing the chloro leaving group in PT-ACRAMTU(EN) or PT-ACR49NME2 with an ACRAMTU or ACR49NME2 moiety, respectively, yielding C-PT-BIS(ACRAMTU) and C-PT-BIS(ACR49NME2) (Fig. 1). These four derivatives bind to DNA in a reversible manner by simultaneously inserting their two acridine moieties into the base stack(10). Likely because these compounds lack a suitable leaving group and therefore do not form permanent adducts with DNA, we did not identify genes involved in DNA repair pathways in our screens. Instead, for C-PT-BIS(ACRAMTU) and C-PT-BIS(ACR49NME2) the most sensitive strains are those deleted for genes involved in vesicle-mediated transport (AKR1, ARF1, ARL3, CHC1, MON2, PEP3, PMR1, RVS161, SUR4, SYS1, VPS15, VPS16, VPS33, VPS35, YPK1, YPT31) suggesting that transport and/or detoxification of these compounds is required for cellular resistance. The trans-isomers of these compounds, T-PT-BIS(ACRAMTU) and T-PT-BIS(ACR49NME2), did not yield any significantly sensitive hits in the screen, which may indicate limited uptake of the bulky compounds by the cells.
The recent availability of a genome-wide deletion set of S. pombe mutants allowed us to perform, for the first time, comparative chemogenomic characterization of a set of compounds in pooled culture. We used S. pombe to examine the mechanisms of cisplatin and the four compounds (PT-ACRAMTU(EN), PT-ACRAMTU(PN), PT-ATUCA, and PT-AMIDINE(EN)) that reveal DNA-damaging effects in S. cerevisiae. In genome-wide screens of S. pombe, resistance to cisplatin showed a requirement for NER, HRR and PRR genes (Fig. 4a), similar to that seen in S. cerevisiae. We observed that the unique requirement for TLS (rev3) for cisplatin resistance was also conserved in S. pombe (Fig. 4a). The S. pombe deletion collection is missing 20% of genes as deletion strains, including the Δpso2 strain, so its role in cisplatin resistance remains to be tested. The top 250 sensitive strains in the PT-ACRAMTU(EN) and PT-ACRAMTU(PN) profiles included those deleted for genes in NER, HRR and PRR (Fig. 3, 4a). In contrast to S. cere-visiae, HRR (rad22) ranked higher than other repair modules in response to PT-ACRAMTU(EN) (Fig. 4a). Furthermore, the requirement for intact PRR was less prominent than for S. cerevisiae, while the HRR requirement for resistance to PT-ACRAMTU(PN) treatment was conserved. Similar to S. cerevisiae, tolerance to PT-ATUCA required NER, HRR and PRR and the ranking of DDR genes was not as high as for the other compounds. PT-AMIDINE(EN) induced distinct profiles in S. pombe and S. cerevisiae; in S. pombe, only those strains deleted for NER and PRR genes were required for resistance. The most striking difference between the profiles from these two organisms is that S. pombe does not seem to require the stalled-fork repair gene mus81 for resistance to either cisplatin or the four DNA-damaging platinum-acridine agents. Selected strains from our genome-wide S. pombe screens were tested for their sensitivity in monoculture with a 35% confirmation rate (Table S1).
In light of the results from both yeast, we asked if these agents perturb cell cycle progression. We used flow cytometry to measure DNA content in synchronized wild-type haploid S. cere-visiae cells that were released into each of PT-ACRAMTU(EN), PT-ACRAMTU(PN), PT-ATUCA and PT-AMIDINE(EN). The compounds PT-ACRAMTU(EN), PT-ACRAMTU(PN) and PT-ATUCA treatment slowed progression through the cell cycle (Fig. 4b). Cells treated with PT-ATUCA accumulated in G1 and S phases, with only a small population of cells in G2/M at 60 minutes. Cells grown in PT-AMIDINE(EN) initially accumulated in S phase and only progress through to G2/M after 90 minutes. Cells remained arrested in G2/M and were unable to divide and re-enter the G1 phase of the subsequent cell cycle. These cells show a distinct metaphase arrest as a result of PT-AMIDINE(EN) treatment (Fig. S2).
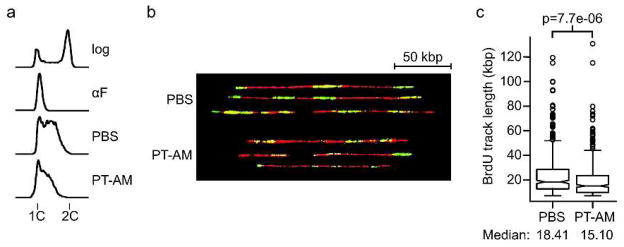
The observation that the S. cerevisiae profile of PT-AMIDINE(EN) shows an enrichment of genes involved in the replication of stalled DNA replication forks, and that the cell cycle data shows slowed progression through S-phase and then accumulation at G2/M, motivated us to further examine the effect of this compound on DNA replication. To examine DNA replication fork progression directly, we performed molecular combing using synchronized S. cerevisiae cells. These cells, which are genetically modified to contain several copies of the Herpes Simplex Virus thymidine kinase gene, were released from G1/S synchrony into medium containing the thy-midine analogue BrdU and PT-AMIDINE(EN) at an IC50 or into vehicle (PBS) for 30 min to al-low BrdU incorporation into newly-synthesized DNA. Genomic DNA was prepared and DNA fibers were combed on silanized coverslips(19). Replication fork progression was examined by measuring the length of BrdU tracks in the fibers. In cells treated with PT-AMIDINE(EN) the nascent DNA tracks, as indicated by BrdU labeling, were significantly shorter compared to cells treated with the vehicle (Fig. 5). This observation is consistent with PT-AMIDINE(EN) interfering with replication fork progression in vivo.
Figure 5.
PT-AMIDINE(EN) treated cells show defects in replication fork progression by DNA combing. Logarithmically growing cultures of S. cerevisiae cells (log) were arrested in G1 with α-factor (αF) and released in the presence of 400μg/mL BrdU and either PBS or PT-AMIDINE(EN) (PT-AM) at IC50. Samples were collected at 30 min and used in subsequent analysis. a) DNA content analysis. Samples were fixed and DNA contents were analyzed using flow cytometry. The positions of cells with 1C and 2C DNA contents are indicated. b) DNA combing. Representative chromosome fibers used for replication fork progression analysis. The image is assembled from fibers on different micrographs following extraction of fibers from the non-fiber background using Adobe Photoshop. A 50 kbp scale bar is indicated in the upper right corner. c) Distributions of BrdU track lengths in PBS or PT-AMIDINE(EN) treated cells, presented as a boxplot. Median BrdU track lengths are shown. The p-value was determined using a two-tailed Mann-Whitney U test.
We further explored the cellular effects of these compounds by examining mitochondrial function and integrity following treatment. Mitochondria are often subjected to high levels of DNA damage when treated with DNA-targeting drugs. Furthermore, mitochondria are an attractive target for cancer therapy due to their role in cellular energetics and apoptosis, and their derangement in different cancers. To examine whether the four DNA-damaging platinum-acridines interfere with mitochondrial function, wild-type S. cerevisiae cells were grown in medium that requires mitochondrial respiration (YPGE) or in a fermenting medium (YPD). In all cases, the inhibitory dose of each platinum-acridine on the cells was much lower in YPGE than YPD (Fig. 4c) indicating that lack of functional mitochondria sensitizes cells to these agents. To examine whether these compounds affect mitochondrial morphology, compound-treated cells were stained with Mitotracker Red CMXRos. For each compound tested, mitochondrial morphology changed from a filamentous network to clumped vesicles (Fig. S3). Together, these results indicate that the four DNA-damaging platinum-acridines affect mitochondrial function.
In this study, we use a validated genome-wide screen to characterize novel anti-cancer therapeutics and explore their structure-activity relationships in an unbiased manner. The utility of S. cerevisiae gene deletion studies for DNA-damaging agents has been exemplified by several published studies that provide a comprehensive view of DNA repair mechanisms(13, 20, 21). The results from our screens add to these reference datasets by providing a global view of the cellular response to novel agents and new insight into the activities previously attributed to these platinum-acridines including their ability to bind to DNA and their lack of a classical cross-linking mechanism for producing DNA-damage(6, 11, 12, 15). By classifying the DNA repair modules that were required for resistance to four of the platinum compounds, we revealed new details about their potential mechanisms at the nuclear level. The response from genes involved in NER strongly suggests that adducts generated by PT-ACRAMTU(EN) primarily lead to helical distortions in the DNA, while modification of the non-leaving group to produce PT-ACRAMTU(PN)(11) yielded a compound that appears to damage DNA via DNA double-strand breaks, a more severe form of DNA damage produced by either cisplatin or PT-ACRAMTU(EN). A distinctive pattern of resistance was seen in the profile of PT-AMIDINE(EN). Specifically, we found a requirement for genes involved in stalled-fork replication repair, a process which can be inhibited by DNA-damaging compounds with very different modes of action, such as alkylation, topoisomerase poisoning and ribonucleotide reductase inhibition. In addition, PT-AMIDINE(EN), the most DNA-reactive compound in this series, causes cells to initially accumulate in S phase, a delay consistent with PT-AMIDINE(EN) preventing or disrupting DNA synthesis, as demonstrated in NCI-H460 lung cancer cells(22). While the three other DNA-damaging hybrid compounds featured here also slow progression of cells through S phase, the cells are able to complete the cell cycle. Our molecular combing observations suggest that the S phase delay observed with PT-AMIDINE(EN) treatment is due, at least in part, to replication fork stalling at DNA adducts formed by this compound. In this scenario, the resumption of DNA synthesis without repair would lead to the observed cell cycle arrest at G2/M due to metaphase arrest and eventually lead to cell death. The hypothesis that severe and irreparable lesions caused by PT-AMIDINE(EN) are bypassed by DNA repair machinery frames future mechanistic studies. In addition, the mitochondrial disruption caused by each of these compounds present them as interesting candidate chemotherapeutics(23). For example, a minor modification to the PT-ACRAMTU(EN) structure produced a compound with a cytotoxicity profile far superior to that of current platinum drugs(12). Such pharmacophores should be of considerable interest for future clinical development.
In addition to revealing differential requirements for diverse DNA repair genes, our screens also highlight that for several platinum-acridines, the mechanism by which they interfere with cellular fitness does not (primarily) involve DDR pathways. For example, the moderately cytotoxic PT-ACRAMTU(BPY) has shown some degree of coordinative binding with DNA in vitro, yet its fitness profile did not suggest that DNA damage results. Similar results were observed for the bis(ACRAMTU) compounds, which enter cells and inhibit cancer cell proliferation at micromo-lar-to-submicromolar concentrations(24). However, the genome-wide profiles of C-PT-BIS(ACRAMTU) and C-PT-BIS(ACR49NME2) suggest that these highly charged compounds are actively detoxified by the cell and that this response is the primary means of cellular resistance. We also confirmed that bisintercalators likely do not form permanent DNA adducts based on our observation that DNA repair deficient strains are not sensitive to these analogues.
The genomes of S. pombe and S. cerevisiae share many features, however their evolutionary divergence allows for complementary functional genomic studies, particularly for examining conserved biological processes such as DNA-damage response pathways and the cell cycle, which are well-characterized in both yeast species(25). Our genome-wide interrogation of S. pombe showed an array of shared and distinct cellular responses to DNA-reactive agents. The hypersensitivity of translesion repair mutants to cisplatin treatment in both yeasts emphasizes the importance of this pathway to repair DNA cross-links, while its absence in the platinum-acridine profiles further highlights the different mechanism by which they function. The PT-AMIDINE(EN) profiles in S. cerevisiae and S. pombe were similar, however, the absence of HRR deletion strains as hits in the S. pombe screen, alludes to the presence of alternative mechanisms for resistance against PT-AMIDINE(EN)-induced double-strand breaks. Additional studies using the fission yeast deletion collection will initiate a productive cycle of gene annotation, in the same manner in the accumulated genome-wide data have greatly increased our understanding of gene function in budding yeast (for review see (26)). The current study demonstrates the power of chemogenomic screening in delineating structure–activity relationships in a new class of DNA-targeted anticancer agents. The requirement for DNA-repair modules for survival of evolutionary divergent yeast cells treated with the platinating-intercalation pharmacophore unequivocally demonstrates that DNA is the principal target of these agents and gives confidence that these results can be applied in human studies. Unlike NSCLC, several cancers may be relatively insensitive to platinum-acridines. To overcome resistance in these tissues, structural designs are currently pursued which feature platinum-acridines as cytotoxic “warheads” conjugated to molecules that modulate DNA repair and/or elicit pro-apoptotic responses. Comparative chemoge-nomics will be an invaluable tool for studying these novel multifunctional entities.
METHODS
Reagents
Cisplatin was purchased from Sigma-Aldrich. All other compounds screened were generated according to published procedures: ACRAMTU, PT-ACRAMTU(EN)(6), ACRAMGU(27), PT-ACRAMTU(PN/TMEDA/BPY)(11), PT-AMIDINE(EN)(12), PT-ACR49NME2(18), PT-ATUCA(28), C/T-PTBISACR, C/T-PTBISACR49NME2(24).
Yeast strains and media
The S. cerevisiae deletion collection(29) and the S. pombe deletion collection(30) were used for this study. S. cerevisiae BY4741 and BY4743 strains were used for small-scale analyses. S. cerevisiae cells were grown in YPD (Yeast extract/Peptone/Dextrose) and S. pombe cells were grown in YES (Yeast Extract with Supplements) at 30°C.
Deletion pool growth and chip experiments
Chemical genomic screens of the homozygous and heterozygous S. cerevisiae deletions pools were performed as described(31). The same protocol was used for the S. pombe deletion pool. The concentrations of the compounds screened were determined using dose-response analysis on a wild-type strain. The platinum-acridine com-pounds were applied at a concentration that inhibits wildtype growth by 15% (200 μM for most compounds; Fig. S4). Genomic DNA preparation, PCR and chip hybridization were performed for both yeast as described(31) with the following change: S. pombe samples were hybridized to the GMAP microarray(32).
Data analysis (all microarray data has been deposited with Array Express)
Using the GeneChip Operating Software (Affymetrix), intensity values for the probes on the chip were ex-tracted. For each strain, fitness defect ratios were calculated as the log2 of the ratio between the mean signal intensities from the control and the compound chips. The larger ratio means greater sensitivity of the strain to the compound. Complete screening data for all compounds can be found here: http://chemogenomics.med.utoronto.ca/supplemental/platinum.
Comparing genome-wide profiles
The algorithm we use for comparing genome-wide profiles (14) is based on ranking of strain sensitivities and comparing a large number of full-genome profiles. Because the scale of fitness defect values of a profile can vary between profiles, a rank-based correlation of these values is advantageous. We address this challenge by scoring each profile as a series of sections, dividing each profile’s gene scores into sections, or “buckets,” with bucket sizes modified according to significance. The smallest size bucket (0.05% of all genes) contains the most significant genes (here, genes with higher fitness defect scores) and the bucket sizes increase exponentially such that the larger size buckets contain the least significant genes, meaning those with the lowest fitness defect scores. The bucket number and sizes were the same for all profiles.
Once the genes of each profile are divided into buckets, we use a scoring matrix for scoring similarity between profiles. The scoring matrix is formulated in decreasing scores while taking into account the bucket location of each gene. When comparing profiles, the scoring matrix yields the score of Si, j to a gene located in bucket i and bucket j in each of the profiles com-pared. The scoring matrix follows these constraints:
Runs on a variety of datasets shows that the algorithm surpasses batch effects that are common in large-scale array-based experiments.
Two-dimensional hierarchical clustering was conducted using the R statistical software package with Euclidean distance metric and the complete linkage agglomerative method.
DNA content analysis
Wildtype S. cerevisiae BY4741 cells at an OD600 0.1 were arrested in G1 using 2 μg mL−1 α-factor for 2.5 h at 23 °C. Cells were released from the arrest and grown for an additional 2 h in the presence of compound at an IC50 at 23 °C. Samples were collected at half-hour time points, fixed in 70% (v/v) ethanol and analyzed by flow cytometry as described(33).
Molecular combing
S. cerevisiae E1670 (MATa ade2-1 trp1-1 can1–100 his3–11,15 leu2–3,112 RAD5+ GAL psi+ ura3::URA3/GPD-TK(7x)) cultures at an OD600 = 0.25 were arrested in G1 by addition of 2.5 μM α-factor for 75 min at 23°C, followed by an additional 1 μM α-factor for 75 min. 400 μg mL−1 BrdU was added to each culture 15 minutes prior to compound addition. Cultures were divided and released from G1 by addition of 100 μg mL−1 pronase (Sigma) and either PBS or PT-AMIDINE(EN) (350nM). Samples were collected after 30 min and incubated for 10 min in 0.1% (w/v) sodium azide on ice. Parallel samples were fixed in ethanol and ana-lysed by flow cytometry, as described above. Cultures were pelleted, re-suspended in SCE buffer (1 M sorbitol, 100 mM sodium citrate pH = 8.5, 10 mM EDTA pH = 8.0, 0.126% (v/v) β-mercaptoethanol, 10 U mL−1 zymolase) and cast into 1% low melt agarose plugs to a final con-centration of 3 × 108 cells mL−1. Processing of DNA plugs and subsequent DNA combing and detection with anti-BrdU and anti-DNA antibodies was performed as described(34). DNA fibers were imaged using an Axiovert inverted microscope (Carl Zeiss) with a 63x objective. Individual coverslips were blinded prior to image acquisition. Images were processed to maximize signal intensity and fluorescent tracks measured in Adobe Photoshop. Approximately 175 tracks were measured per sample and track lengths were converted from pixels to kbp using a conversion factor based on combing λ DNA(19). Experiments were repeated twice and data from independent experiments pooled. The distribution of track lengths was plotted as a boxplot and the two-tailed Mann-Whitney U test was used to compare the distributions of track lengths.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported in part by a grant from the US National Institutes of Health, CA101880 (to UB) and from the NHGRI to CN and GG and from the Canadian Cancer Society (#020380) to GG and CN and from the Canadian Cancer Society (#020254) to GWB. AYL was supported by the Charles H. Best fellowship.
We thank T. Sing for suggestions on cell cycle analysis and M. Gebbia for experimental assistance.
Footnotes
Author Contributions
KC-O oversaw all data collection and wrote the manuscript. KTS performed the chemogenomic screens. ZM synthesized the compounds. DS, AYL, LEH analyzed the data. DG and GB performed and analyzed the combing experiments. UB, GG, and CN supervised all aspects of the project and CN developed the initial concept.
Supporting Information. Supplementary datasets, figures, tables and methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 2.Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutat Res. 2001;478:23–43. doi: 10.1016/s0027-5107(01)00141-5. [DOI] [PubMed] [Google Scholar]
- 3.Wong E, Giandomenico CM. Current status of platinum-based antitumor drugs. Chemical reviews. 1999;99:2451–2466. doi: 10.1021/cr980420v. [DOI] [PubMed] [Google Scholar]
- 4.Cleare MJ, Hoeschele JD. Studies on the antitumor activity of group VIII transition metal complexes. Part I. Platinum (II) complexes. Bioinorganic Chemistry. 1973;2:187–210. [Google Scholar]
- 5.Manzotti C, Pratesi G, Menta E, Di Domenico R, Cavalletti E, Fiebig HH, Kelland LR, Farrell N, Polizzi D, Supino R, Pezzoni G, Zunino F. BBR 3464: a novel triplatinum complex, exhibiting a preclinical profile of antitumor efficacy different from cisplatin. Clin Cancer Res. 2000;6:2626–2634. [PubMed] [Google Scholar]
- 6.Martins ET, Baruah H, Kramarczyk J, Saluta G, Day CS, Kucera GL, Bierbach U. Design, synthesis, and biological activity of a novel non-cisplatin-type platinum-acridine pharmacophore. Journal of medicinal chemistry. 2001;44:4492–4496. doi: 10.1021/jm010293m. [DOI] [PubMed] [Google Scholar]
- 7.Hess SM, Anderson JG, Bierbach U. A non-crosslinking platinum-acridine hybrid agent shows enhanced cytotoxicity compared to clinical BCNU and cisplatin in glioblastoma cells. Bioorganic & medicinal chemistry letters. 2005;15:443–446. doi: 10.1016/j.bmcl.2004.10.049. [DOI] [PubMed] [Google Scholar]
- 8.Hess SM, Mounce AM, Sequeira RC, Augustus TM, Ackley MC, Bierbach U. Platinum-acridinylthiourea conjugates show cell line-specific cytotoxic enhancement in H460 lung carcinoma cells compared to cisplatin. Cancer chemotherapy and pharmacology. 2005;56:337–343. doi: 10.1007/s00280-004-0987-7. [DOI] [PubMed] [Google Scholar]
- 9.Baruah H, Wright MW, Bierbach U. Solution structural study of a DNA duplex containing the guanine-N7 adduct formed by a cytotoxic platinum-acridine hybrid agent. Biochemistry. 2005;44:6059–6070. doi: 10.1021/bi050021b. [DOI] [PubMed] [Google Scholar]
- 10.Augustus TM, Anderson J, Hess SM, Bierbach U. Bis(acridinylthiourea)platinum(II) complexes: synthesis, DNA affinity, and biological activity in glioblastoma cells. Bioorganic & medicinal chemistry letters. 2003;13:855–858. doi: 10.1016/s0960-894x(02)01078-8. [DOI] [PubMed] [Google Scholar]
- 11.Guddneppanavar R, Choudhury JR, Kheradi AR, Steen BD, Saluta G, Kucera GL, Day CS, Bierbach U. Effect of the diamine nonleaving group in platinum-acridinylthiourea conjugates on DNA damage and cytotoxicity. Journal of medicinal chemistry. 2007;50:2259–2263. doi: 10.1021/jm0614376. [DOI] [PubMed] [Google Scholar]
- 12.Ma Z, Choudhury JR, Wright MW, Day CS, Saluta G, Kucera GL, Bierbach U. A non-cross-linking platinum-acridine agent with potent activity in non-small-cell lung cancer. Journal of medicinal chemistry. 2008;51:7574–7580. doi: 10.1021/jm800900g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 2005;1:e24. doi: 10.1371/journal.pgen.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shabtai D, Giaever G, Nislow C. An algorithm for chemical genomic profiling that minimizes batch effects: Bucket Evaluations. BMC Bioinformatics. 2012 doi: 10.1186/1471-2105-13-245. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Budiman ME, Alexander RW, Bierbach U. Unique base-step recognition by a platinum-acridinylthiourea conjugate leads to a DNA damage profile complementary to that of the anticancer drug cisplatin. Biochemistry. 2004;43:8560–8567. doi: 10.1021/bi049415d. [DOI] [PubMed] [Google Scholar]
- 16.Choudhury JR, Rao L, Bierbach U. Rates of intercalator-driven platination of DNA determined by a restriction enzyme cleavage inhibition assay. J Biol Inorg Chem. 2011;16:373–380. doi: 10.1007/s00775-010-0733-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Z, Rao L, Bierbach U. Replacement of a thiourea-S with an amidine-NH donor group in a platinum-acridine antitumor compound reduces the metal’s reactivity with cysteine sulfur. Journal of medicinal chemistry. 2009;52:3424–3427. doi: 10.1021/jm900451y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guddneppanavar R, Saluta G, Kucera GL, Bierbach U. Synthesis, biological activity, and DNA-damage profile of platinum-threading intercalator conjugates designed to target adenine. Journal of medicinal chemistry. 2006;49:3204–3214. doi: 10.1021/jm060035v. [DOI] [PubMed] [Google Scholar]
- 19.Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 20.St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, Fiedler D, Dutkowski J, Guenole A, van Attikum H, Shokat KM, Kolodner RD, Huh WK, Aebersold R, Keogh MC, Krogan NJ, Ideker T. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyre CL, Saluta G, Kute TE, Kucera GL, Bierbach U. Inhibition of DNA Synthesis by a Platinum–Acridine Hybrid Agent Leads to Potent Cell Kill in Nonsmall Cell Lung Cancer. ACS Medicinal Chemistry Letters. 2011;2:870–874. doi: 10.1021/ml2001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galluzzi L, Larochette N, Zamzami N, Kroemer G. Mitochondria as therapeutic targets for cancer chemotherapy. Oncogene. 2006;25:4812–4830. doi: 10.1038/sj.onc.1209598. [DOI] [PubMed] [Google Scholar]
- 24.Choudhury JR, Guddneppanavar R, Saluta G, Kucera GL, Bierbach U. Tuning the DNA conformational perturbations induced by cytotoxic platinum-acridine bisintercalators: effect of metal cis/trans isomerism and DNA threading groups. J Med Chem. 2008;51:3069–3072. doi: 10.1021/jm8003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsburg SL. The yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe: models for cell biology research. Gravit Space Biol Bull. 2005;18:3–9. [PubMed] [Google Scholar]
- 26.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Z, Day CS, Bierbach U. Unexpected reactivity of the 9-aminoacridine chromophore in guanidylation reactions. J Org Chem. 2007;72:5387–5390. doi: 10.1021/jo0705972. [DOI] [PubMed] [Google Scholar]
- 28.Ma Z, Saluta G, Kucera GL, Bierbach U. Effect of linkage geometry on biological activity in thiourea- and guanidine-substituted acridines and platinum-acridines. Bioorganic & medicinal chemistry letters. 2008;18:3799–3801. doi: 10.1016/j.bmcl.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 30.Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, Han S, Jeffery L, Baek ST, Lee H, Shim YS, Lee M, Kim L, Heo KS, Noh EJ, Lee AR, Jang YJ, Chung KS, Choi SJ, Park JY, Park Y, Kim HM, Park SK, Park HJ, Kang EJ, Kim HB, Kang HS, Park HM, Kim K, Song K, Song KB, Nurse P, Hoe KL. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierce SE, Davis RW, Nislow C, Giaever G. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat Protoc. 2007;2:2958–2974. doi: 10.1038/nprot.2007.427. [DOI] [PubMed] [Google Scholar]
- 32.Ketela T, Heisler LE, Brown KR, Ammar R, Kasimer D, Surendra A, Ericson E, Blakely K, Karamboulas D, Smith AM, Durbic T, Arnoldo A, Cheung-Ong K, Koh JL, Gopal S, Cowley GS, Yang X, Grenier JK, Giaever G, Root DE, Moffat J, Nislow C. A comprehensive platform for highly multiplexed mammalian functional genetic screens. BMC Genomics. 2011;12:213. doi: 10.1186/1471-2164-12-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellay J, Atluri G, Sing TL, Toufighi K, Costanzo M, Ribeiro PS, Pandey G, Baller J, VanderSluis B, Michaut M, Han S, Kim P, Brown GW, Andrews BJ, Boone C, Kumar V, Myers CL. Putting genetic interactions in context through a global modular decomposition. Genome Res. 2011;21:1375–1387. doi: 10.1101/gr.117176.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, O’Donnell L, Durocher D, Brown GW. RMI1 Promotes DNA Replication Fork Progression and Recovery from Replication Fork Stress. Mol Cell Biol. 2012 doi: 10.1128/MCB.00255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Dongen S. A cluster algorithm for graphs. Technical Report INS-R0010. National Research Institute for Mathematics and Computer Science in the Netherlands, Amsterdam. 2000:1386–3681. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.