Abstract
Purpose
The molecular epidemiology of most EGFR and KRAS mutations in lung cancer remains unclear.
Experimental Design
We genotyped 3026 lung adenocarcinomas for the major EGFR (exon 19 deletions and L858R) and KRAS (G12, G13) mutations and examined correlations with demographic, clinical and smoking history data.
Results
EGFR mutations were found in 43% of never smokers (NS) and in 11% of smokers. KRAS mutations occurred in 34% of smokers and in 6% of NS. In patients with smoking histories up to 10 pack-years, EGFR predominated over KRAS. Among former smokers with lung cancer, multivariate analysis showed that, independent of pack-years, increasing smoking-free years raise the likelihood of EGFR mutation. NS were more likely than smokers to have KRAS G>A transition mutation (mostly G12D) (58% vs. 20%, p=0.0001). KRAS G12C, the most common G>T transversion mutation in smokers, was more frequent in women (p=0.007) and these women were younger than men with the same mutation (median 65 vs. 69, p=0.0008) and had smoked less.
Conclusions
The distinct types of KRAS mutations in smokers vs. NS suggest that most KRAS-mutant lung cancers in NS are not due to secondhand smoke exposure. The higher frequency of KRAS G12C in women, their younger age, and lesser smoking history together support a heightened susceptibility to tobacco carcinogens.
Keywords: lung cancer, tobacco, EGFR, KRAS, molecular epidemiology
Introduction
EGFR and KRAS mutations are present in almost 50% of lung adenocarcinomas in Caucasian patients. More than 90% of EGFR mutations are small in frame deletions in exon 19 and L858R missense mutation in exon 21.1 These mutations are associated with responsiveness to tyrosine kinase inhibitors (TKI) therapy. 2–4 EGFR mutations are more frequently found in women, Asians, and in never smokers.5,6 There is an inverse relationship between duration and intensity of cigarette smoking and frequency of EGFR mutations suggesting that smoking history has predictive value for presence of EGFR mutations.7,8
Although KRAS mutations were identified in lung cancer more than two decades ago, 9,10 the clinical importance of KRAS mutation status became apparent only relatively recently, as lung adenocarcinomas harboring KRAS mutations were found to show lack of response to EGFR TKI therapy.11,12 KRAS-mutated lung cancers are prognostically unfavorable when compared with EGFR-mutateds.13–16 In more than 95% of cases, KRAS missense mutations are found in codons 12 and 13.17 Unlike EGFR mutations, KRAS mutations show no sex predilection, are more frequent in white populations than Asians and most patients are former or current cigarette smokers.18,19 KRAS mutations known to be smoking-associated (G12C, G12V) are transversion mutations (G>T and G>C), whereas KRAS transitions mutations (G>A) are more common in lung adenocarcinomas from patients without any smoking history.20,21
Even though the distinctive distribution of EGFR and KRAS mutations in relation to ethnicity, sex and smoking history suggests that patient characteristics have a significant predictive value for the presence of these mutations, the etiology of most mutations arising in never smokers remains unknown. In the current study, we hypothesized that correlations between demographic, epidemiologic, and clinical data and types of EGFR and KRAS mutations could provide a better insight into specific etiology and/or biology of these mutations. Therefore, we took the advantage of our large clinical dataset and performed an in-depth retrospective analysis of more than 3000 consecutive lung adenocarcinoma cases subjected to routine testing for EGFR and KRAS mutations over a 5-year period.
Methods
Clinical Samples/Patients
From September 2004 to December 2009, 3026 lung adenocarcinomas (including 2 adenosquamous and one large cell carcinoma with adenocarcinoma component) were consecutively received and clinically tested for presence of EGFR exon 19 deletion and exon 21 L858R mutation. In January 2006, testing for KRAS mutations (codons 12 and 13) was introduced for all cases negative for EGFR mutation, and 2529 cases were received after that time. Cases with more than one tumor were included if: all the tumors were either mutation negative, harbored the same mutation, or if one tumor harbored EGFR or KRAS mutation and the other(s) was(were) mutation negative. Twenty three patients with more than one tumor harboring different KRAS or EGFR mutations were excluded from the study; some of these have been reported separately.22 Clinical samples submitted for molecular testing included surgically resected tumor samples, biopsies and cytology specimens. Clinical data were collected with the approval of Institutional Review Board (IRB) of Memorial Sloan-Kettering Cancer Center. Stage designated as IIIB/IV included stages IIIB, IV and multifocal bronchioloalveolar carcinoma (BAC). Smoking status was defined as never smokers (<100 lifetime cigarettes), former smokers (quit >1 year prior to diagnosis), or current smokers (still smoking, or quit <1 year prior to diagnosis). Pack-years of smoking was defined as (average number of cigarettes per day/20) × years smoking.
Mutation Detection
DNA was extracted using a kit (DNeasy, Qiagen) from frozen tumor tissue or formalin-fixed paraffin embedded tumor tissue. If necessary, manual microdissection of paraffin sections was done to ensure at least 50% tumor content. EGFR mutations were detected by sensitive PCR-specific assays as previously described.23 KRAS mutations were detected by PCR-sequencing of exon 2 as described.24 In limited volume tumor samples, presence of an exuberant inflammatory response or extensive fibrosis, PCR was performed with addition of locked nucleic acid (LNA) oligonucleotide to favor amplification of-mutated allele, if present.25
Statistical Analysis
Cases were divided into three groups based on mutation status: EGFR-mutated, KRAS-mutated or wild type for EGFR/KRAS. The associations were tested between the mutation groups and the demographic or clinical characteristics, and the smoking status using Fisher exact test or unpaired t-test. A p value <0.01 was considered significant. The Bonferroni method was used to control for family-wise error rate. Univariate and multivariate logistic regression analyses were used to test the association of smoking-free years and pack-years of smoking with EGFR and KRAS mutational status.
Nomogram Development and Validation
A nomogram was generated for the likelihood of EGFR mutation among Caucasian smokers based on the following logistic regression model: EGFR~β0+ β1smoke-free-years+ β2pack-years + β3gender + β4age+ β5age2. The quadratic term allows a U-shape pattern of the age association with the mutational outcome. All analyses were performed using the R package Design and Hmisc. An independent data set was used for validation26; specifically, we used 375 adenocarcinoma patients who were Caucasian smokers from the Boston cohort included in the study by Girard and colleagues 26 as the validation cohort.
Results
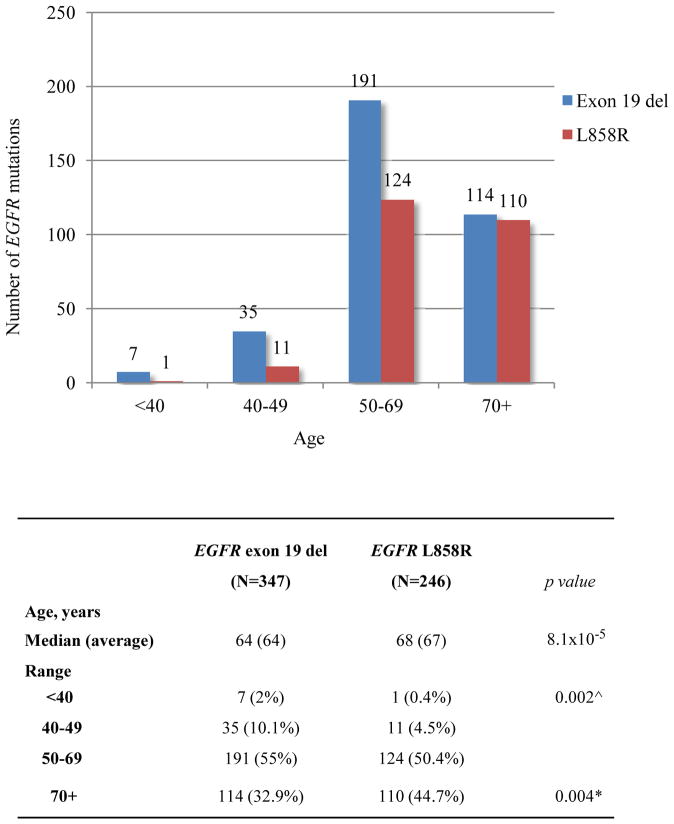
Table 1 summarizes characteristics of lung cancer patients with EGFR and KRAS mutations. Our lung adenocarcinoma patient population was predominantly female (1898/3026, 62.7%) and this was consistent in each year from 2006 to 2009 (1624/2620, 62%) reflecting the routine reflex EGFR/KRAS testing which was initiated in 200627. Only 13% of the cases (406/3026) were submitted for testing before 2006 and these showed a slightly higher female to male ratio (274/406; 67.5%) presumably reflecting some referral bias. Of 3026 cases tested clinically for the two major EGFR mutations, 593 (20%) were-mutated, including 347 exon 19 deletions (59%) and 246 L858R mutations (41%). Patients with EGFR L858R tended to be older than exon 19-mutateds (median age 68 vs. 64; p=8.1×10−5), reflected by an exon 19 del to L858R ratio of 3.5 under age 50 (p=0.002) and of 1.0 in patients 70 and over (p=0.004) (Figure 1). Men with EGFR mutations were more likely than women to present at late stage (i.e. IIIB/IV) of disease (118/170, 69% vs. 235/423, 56%; p=0.002), whereas women predominated at stage I (31% vs. 19%, p=0.004) (Supplementary Figure 1A). Tumors with EGFR L858R presented more often at stage I than tumors with exon 19 del (83/246, 34% vs. 82/347, 24%; p=0.009) (Supplementary Figure 1B). Testing of 2529 cases for KRAS mutations (codons 12 and 13) detected 670 (26%) mutations, including G12C (39%), G12V (21%), G12D (17%), G12A (11%), and other G12 and G13 mutations (12%). Although none of the EGFR-mutated tumors in the present clinical data set were tested for concomitant KRAS mutations, our more recent experience using multiplex genotyping by MALDI-TOF mass spectrometry (Sequenom) further confirm their mutually exclusive occurrence pattern.28 No significant differences in age or stage at presentation were noted between different subtypes of KRAS mutations (Supplementary Figure 2).
Table 1.
Demographic and clinical characteristics of patients according to the EGFR and KRAS mutation status
| All patients (N=3026) | EGFR mutations | All patients (N=2529) | KRAS mutations | |||||
|---|---|---|---|---|---|---|---|---|
| Patients with EGFR mutations (N=593) | All 3026 patients % (95% CI) | p value | Patients with KRAS mutations (N=670) | All 2529 patients % (95% CI) | p value | |||
| Sex | ||||||||
| Male | 1128 | 170 | 15.1 (13.1–17.3) | 0.0001 | 959 | 248 | 25.9 (23.2–28.7) | 0.58 |
| Female | 1898 | 423 | 22.3 (20.5–24.2) | 1570 | 422 | 26.9 (24.7–29.1) | ||
| Age, years | ||||||||
| Median (Average) | 66 (65) | 66 (65) | NA | 66 (65) | 66 (66) | NA | ||
| Range | 15–96 | 24–90 | NA | 15–96 | 30–88 | NA | ||
| Ethnicity/Race | ||||||||
| White | 2736 | 478 | 17.5 | NA | 2285 | 641 | 28.1 | NA |
| Asian/Pacific | 136 | 75 | 55.1 (46.8–63.2) | 0.0001 | 114 | 7 | 6.1 (2.8–12.4) | 0.0001 |
| Black | 77 | 16 | 20.8 (13.1–31.2) | 0.45 | 66 | 12 | 18.2 (10.6–29.3) | 0.09 |
| Asian/Indian | 28 | 14 | 50 (32.6–67.4) | 0.0001 | 23 | 0 | 0 | 0.0007 |
| Other/Unknown | 49 | 10 | 20.4 (11.3–33.8) | 41 | 10 | 24.4 (13.7–39.5) | ||
| Stage | ||||||||
| I | 902 | 165 | 18.3 (15.9–21.0) | 760 | 207 | 27.2 (24.2–30.5) | ||
| II | 188 | 35 | 18.6 (13.7–24.8) | 158 | 43 | 27.2 (20.9–34.7) | ||
| IIIA | 260 | 40 | 15.4 (11.5–20.3) | 210 | 54 | 25.7 (20.3–32.0) | ||
| IIIB&IV | 1676 | 353 | 21.1 (19.2–23.7) | 1401 | 366 | 26.1 (23.9–28.5) | ||
| Smoking History | ||||||||
| Never smokers | 828 | 352 | 42.5 (39.2–45.9) | NA | 669 | 43 | 6.4 (4.8–8.6) | NA |
| Former smokers | 1548 | 209 | 13.5 (11.9–15.3) | 0.0001 | 1297 | 419 | 32.3 (29.8–34.9) | 0.0001 |
| Current smokers | 650 | 32 | 4.9 (3.5–7.7) | 0.0001 | 563 | 208 | 36.9 (33.1–41.0) | 0.0001 |
P values compare the frequency of EGFR or KRAS mutations between men and women, between white patients and other ethnicities/races, and between never smokers and former and current smokers, respectively.
Figure 1.
Age distribution of EGFR exon 19 del and EGFR L858R. Patients with EGFR L858R mutant tumors presented at older age than those harboring EGFR exon 19 del (median age 68 vs. 64; p=8.1×10−5). Fisher exact test, P value <0.01 is considered significant.
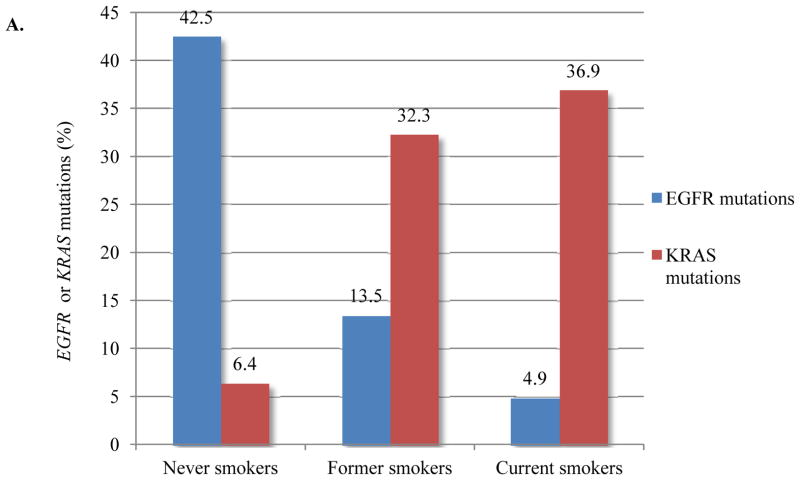
The positive and negative associations of KRAS and EGFR mutations, respectively, with smoking are well known but had not previously been analyzed in detail in a single large dataset. Figure 2 illustrates the frequency of EGFR and KRAS mutations in relation to smoking history and smoking pack-years. EGFR mutations were found in 352 of 828 (43%) of never smokers and in 241 of 2198 (11%) former and current smokers. There was no significant difference in frequency of EGFR exon 19 del vs. EGFR L858R relative to smoking pack-years (data not shown). KRAS mutations were found in 627/1860 (34%) of former and current smokers and in 43/669 (6%) of never smokers, the latter proportion being notably lower than in a smaller study from our center but within the confidence interval of the previously reported higher percentage21. While any smoking history significantly decreased the likelihood of EGFR mutations, no difference was noted among smokers with less than 10 pack-years smoking history. Furthermore, in smokers of more than 10 pack-years, EGFR mutations were 5-fold less likely to be found than in never smokers (p=0.0001). In contrast, the proportion of KRAS-mutated lung cancers was significantly higher in smokers with any smoking history than in never smokers; among smokers, we found 15 pack-years as a cut-point above which the likelihood of a lung cancer harboring KRAS mutations was 6-fold higher than in never smokers (p=0.0001). Notably, even in patients with up to 10 pack-years of smoking, tumors with EGFR mutations were still more common than those with KRAS mutations.
Figure 2.
(A) Frequency of EGFR and KRAS mutations by smoking history. (B) Frequency of EGFR and KRAS mutations by pack-years of smoking. In the range of up to 10 pack-years, tumors with EGFR mutations are still more common than KRAS mutations.
The effect of smoking and smoking-free period on the likelihood of EGFR mutation has been previously reported in Asian patients with lung adenocarcinoma 29,30, but the impact of these two smoking variables on the proportions of lung adenocarcinomas with either EGFR or KRAS mutations has not been previously investigated in a predominantly white patient population. Because smoking-free years and pack-years of smoking are partly dependent variables, we performed a multivariate logistic regression analysis to examine the effect of these two parameters in current and former Caucasian smokers. Interestingly, this showed that, among patients with lung cancer, smoking-free years change the likelihood of EGFR mutation but not that of KRAS mutation (Supplementary Table 1).
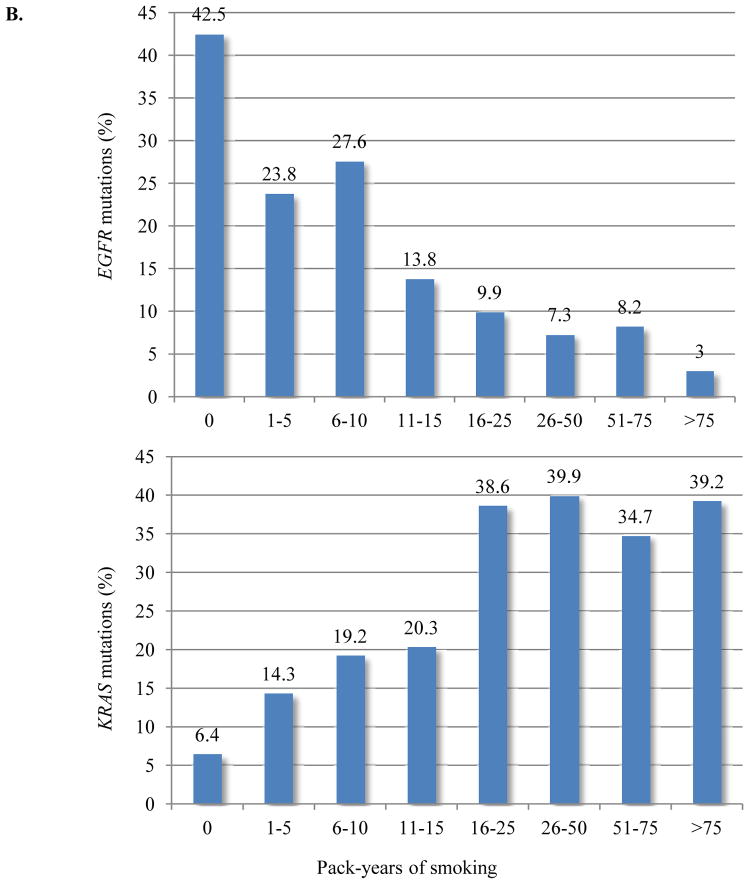
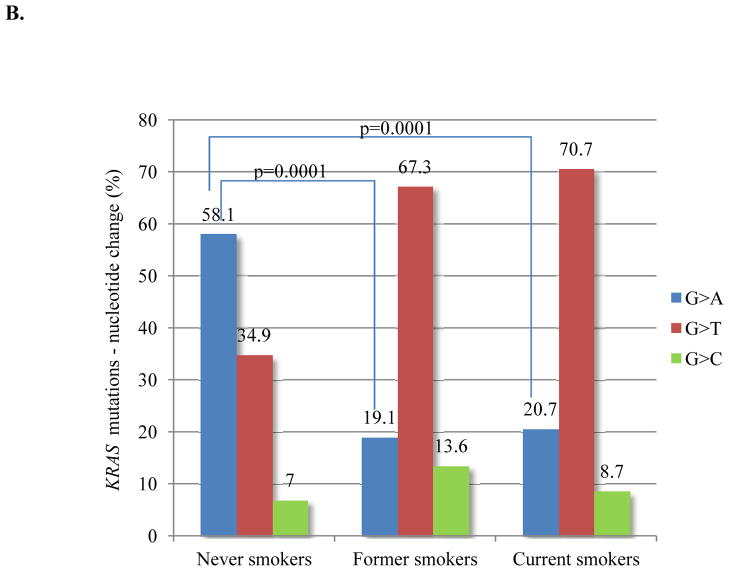
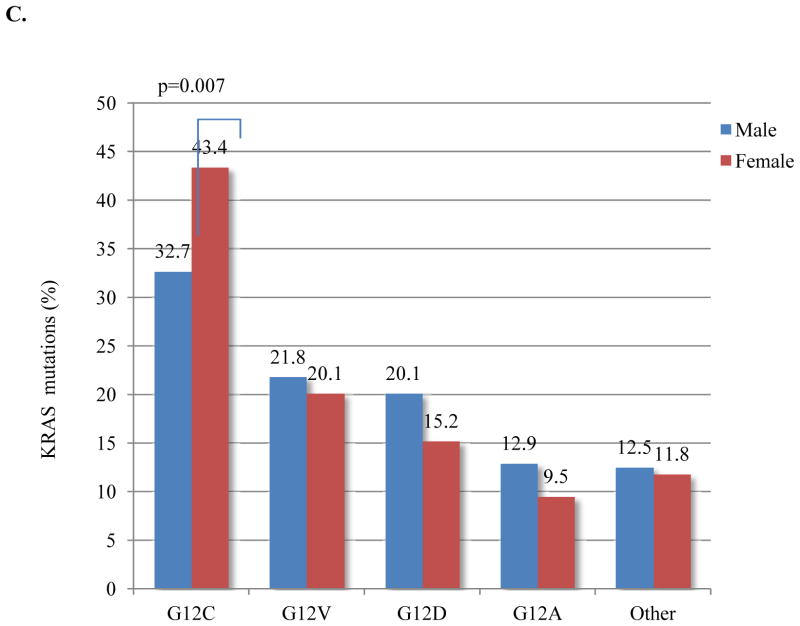
Given the variety of possible nucleotide substitutions leading to missense mutations of KRAS G12 and G13, we examined their association with smoking in this large dataset. Among never smokers, the most common KRAS mutation was G12D (56%), and G12C was the most frequent mutation among former and current smokers (41%) (Figure 3A). Never smokers were significantly more likely than former and current smokers to have G>A transition mutations (as in G12D) (58% vs. 19% vs. 21%; p=0.0001), whereas G>T transversion mutations (as in G12C), a typical change associated with tobacco carcinogens, was the most common nucleotide change in former and current smokers (67% and 71%, respectively) (Figure 3B). Compared to other KRAS mutations types, G12C was more frequent in women (p=0.007) (Figure 3C), who were also younger than men with the same mutation (median age 65 vs. 69; p=0.0008). Intriguingly, women with G>T transversions had smoked less (average 34 pack-years vs. 40 pack-years, p=0.001) (Supplementary Table 2) and were younger than men with the same nucleotide change (median age 64 vs. 67, p=0.006). As discussed below, this pattern of findings suggests an increased susceptibility to tobacco carcinogenesis in women.
Figure 3.
KRAS mutation type as a function of smoking history. (A) KRAS G12D is the most common mutation in never smokers and KRAS G12C is the most frequent mutation among former and current smokers. (B) Never smokers are significantly more likely to have G>A transition mutation (p<0.0001). G>T transversion is the most common nucleotide change in former and current smokers (p<0.0001). (C) KRAS G12C was relatively more frequent in women than in men (p=0.007). Fisher exact test, P value <0.01 is considered significant.
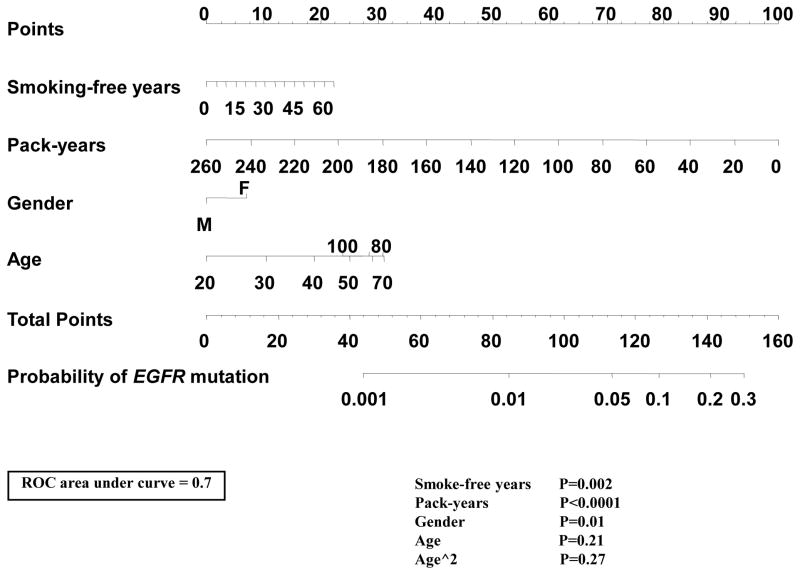
There is continuing interest in using clinical variables to prioritize EGFR mutation testing. Certain patient subsets, such as Asians and never-smokers are routinely tested, while other subsets, such as male Caucasian smokers, are considered of lower priority for testing. However, it is also becoming clear that these patient characteristics should not be used individually to exclude patients from testing, as shown in a recent analysis of a subset of the present data31. Given the significant associations of EGFR mutation with sex (p=0.01), pack-years of smoking (p<0.0001) and smoking-free years (p=0.002), we used these variables along with age to generate a nomogram to predict the EGFR status specifically in Caucasian smokers (current and former). We excluded Asians and never smokers from the nomogram dataset because it is generally agreed that patients in these groups should be tested regardless. The area under the receiver operating characteristic (ROC) curve was 0.70 (Figure 4). To validate the performance of our nomogram in an independent dataset, we used the Caucasian smokers from the Boston cohorts used in the study by Girard and colleagues.26 In this independent set of patients, our nomogram generated an area under the ROC curve for predicting EGFR status of 0.71 (Supplementary Figure 3). In the MSKCC training dataset (n=2078), 16 had a predicted probability of EGFR mutation of 1% or less, and none were EGFR-mutated, and 421 had a predicted probability of 0.05 or lower, of which 14 (3%) had EGFR mutations. In the Boston dataset (n=375) used for validation, ten patients had probability below 1%, one of which was EGFR-mutated, 145 had a probability below 5%, including ten (7%) EGFR-mutated cases. As discussed below, we view these results as indicating that, even in the context of a rigorously developed nomogram, clinical variables cannot be used to robustly identify patients with a negligible chance of harboring an EGFR-mutated lung cancer.
Figure 4.
Development of a nomogram including clinical variables and smoking history data for prediction of EGFR mutant status among Caucasian smokers (current or former). Mark the Smoking-free years on the axis and draw vertical line up to the Points axis to determine the number of points. Repeat the same for Pack-years, Gender and Age and sum the total points for all four variables. Plot the given number on the Total Points axis and draw a vertical line down to the Probability of EGFR mutation.
Discussion
To accurately and reliably determine the frequency of the major mutations in EGFR and KRAS in lung adenocarcinoma in relation to patient characteristics and different levels of smoking, a sufficiently large number of case subjects is necessary to provide statistical power for more detailed analyses. Here, we performed a retrospective analysis of our large clinical database of lung adenocarcinomas with established EGFR/KRAS mutation status. We found (1) distinct differences in sex, age and stage distribution of two most common types of EGFR mutations; (2) we determined the likelihood of EGFR and KRAS mutations by intensity and duration of smoking, and (3) evaluated the effects of smoking-free period on the proportions of EGFR and KRAS mutations in lung cancers arising in former smokers; (4) we designed a nomogram to predict presence of EGFR mutation in Caucasian smokers; (5) we noted a distinct distribution of types of KRAS mutations in smokers vs. never smokers; (6) we observed significant sex and age differences in the frequency of G12C as the most common smoking-related KRAS mutation.
EGFR exon 19 del was relatively more common than L858R mutation in younger patients. Notably, of eight patients below age 40 with EGFR-mutated lung adenocarcinoma, seven were EGFR exon 19 del. On the other hand, L858R occurred in a relatively older age distribution and the patients more often presented with stage I disease. These findings may suggest a potentially more aggressive natural history of adenocarcinomas with EGFR exon 19 del and relatively less aggressive one for L858R-mutateds. Differences between EGFR exon 19 del and L858R-mutateds have been reported in patients treated with TKI or chemotherapy. EGFR exon 19 deletions have been associated with better response to TKI and with a longer time-to-progression (TTP) and overall survival (OS) in advanced adenocarcinoma patients.32–35 However, the better clinical outcome of patients with EGFR exon 19 del compared to patients harboring EGFR L858R mutations remains controversial; two prospective randomized phase III trial studies did not confirm these observations. 36,37 A distinct age and stage distribution as well as differences in response to molecular targeted therapy may suggest subtle differences in biology and/or etiology for EGFR exon 19 del and L858R mutation.
Although typically seen in the absence of smoking history, a significant minority (11%) of former and current smokers harbor EGFR-mutated tumors, arguing against excluding smokers from EGFR testing. Moreover, among smokers with less than ten pack-years, EGFR mutations were more common than KRAS mutations. In a study of 265 lung adenocarcinomas, some of which are included in the current dataset, Pham et al. found significantly fewer EGFR mutations in people who smoked for more than 15 pack-years or stopped smoking less than 25 years ago compared with individuals who never smoked.7 Our extended dataset allowed for more accurate risk stratification by pack-years categories and showed, that any smoking history at or above one pack-year significantly decreased the likelihood of EGFR mutated tumors with no notable difference up to ten pack-years. Although our patient population was primarily Caucasian, the results appear generalizable, as a similar relationship of EGFR mutations to pack-years and smoke-free years has also been reported in Asian patients with lung cancer.29,30,38
As expected, most of the KRAS mutations were found among current and former smokers, and consistent with other studies 39, we identified 6% of never smokers with KRAS-mutated tumors. In our earlier study that included 102 KRAS-mutated tumors, 21 we failed to demonstrate predictive value of pack-years for the presence of KRAS mutations likely due to small number of cases. Here, we have shown that any smoking history significantly increases the likelihood of a KRAS mutation being found in the lung cancer. Smoking-free years provided additional value in predicting the likelihood of EGFR mutations but not that of KRAS mutations, independent of pack-years of smoking. These multivariate results suggest a model in which KRAS mutations occur at the time of smoking and lead to cancer eventually, explaining the lack of impact of smoke-free years. This is also supported by the observation that former and current smokers have similar proportions of KRAS-mutated lung cancers (Figure 2A). Overall, this further supports the notion that permanent DNA damage by tobacco carcinogens acquired at the time of smoking is the major source of most KRAS-mutated lung adenocarcinomas. Thus, the likelihood that a patient with lung cancer has a KRAS mutation is determined by pack-years of smoking and does not decrease significantly over time upon smoking cessation; in contrast, because overall lung cancer incidence decreases with increasing smoke-free years, the relative proportion of non-smoking associated cancers (represented by EGFR-mutated tumors) increases. Importantly, these data should not be misinterpreted as supporting a “protective” effect of smoking on the risk of EGFR-mutated lung adenocarcinoma.
Based on the need for efficient medical resource utilization and concerns regarding health care costs and possible treatment delays due to testing, there is continuing controversy regarding routine EGFR mutation testing in certain patient subsets perceived as having a low chance of EGFR mutation in their lung cancer, such as male Caucasian smokers. Using the readily available clinical parameters of age, sex, pack-years, and smoking-free years, we developed a nomogram to predict the likelihood of EGFR mutation in Caucasian current or former smokers with lung adenocarcinoma. We should note that a similar, recently published nomogram 26 differs in two important ways from the one we have developed. First, it includes never smokers, a group in which the value of EGFR testing is no longer in question. Secondly, it includes the histologic subtype of adenocarcinoma, which usually can only be properly analyzed in resection specimens, but decisions regarding EGFR testing often have to be made in advanced stage patients in whom the available small biopsies are sometimes suboptimal for histologic subtyping. The accuracy of our nomogram was 70% on the source dataset and 71% in an independent validation dataset. Based on clinical considerations (for instance, the fact that testing of ALK fusions present in only 3–5% of lung adenocarcinomas is now indicated to select patients for crizotinib),40 we deemed that only a probability of harboring an EGFR mutation of less than 1% was clinically negligible and therefore actionable in terms of bypassing EGFR testing. However, only a very small proportion of patients fall in this category, 0.8% is the source dataset and 2.7% in the validation dataset and the latter included one incorrect prediction (10% error rate). Overall, the 70–71% accuracy of nomogram prediction, along with the very low proportion of predictions below 1%, suggests that clinical variables cannot be used to robustly identify patients with a negligible chance of harboring an EGFR-mutated lung cancer. Nonetheless, our nomogram may still be helpful in situations where mutation analysis for EGFR is simply not possible and the clinical parameters and smoking history are used to direct the treatment decision.
In a previous smaller study, we showed that never smokers were significantly more likely than former or current smokers to have a KRAS transition mutation (G>A) rather than the transversion mutations known to be smoking-related (G>T or G>C).21 The much larger number of cases in the present series allowed us to robustly confirm these earlier findings as well as to detect sex and age differences in the frequency of the most common smoking-related G>T transversion mutation, KRAS G12C. These findings support the notion that most KRAS-mutant lung adenocarcinomas in never smokers are not likely to be caused by environmental (second hand) tobacco smoke, a potentially important observation in assessing the level of risk posed by such exposure.
Sex differences in sensitivity to tobacco smoke have been well documented.41 Zang and Wynder have reported that the odds ratios for major lung cancer types are consistently higher in women than in men at every level of exposure to cigarette smoke and that these differences cannot be explained by differences in baseline exposure, smoking history, or body size, but are likely due to a higher susceptibility to tobacco carcinogens in women.42 Computed tomographic screening data suggest that female smokers are almost twice as likely as male smokers to have a lung cancer detected in spite of lesser smoking histories.43 Consistent with our findings in KRAS, studies of the mutational spectrum of TP53 in relation to smoking and sex showed that cancers arising in female smokers had significantly more tobacco-related mutations (G>T transversions) than in male smokers.44,45 Therefore, taken together, the relatively higher percentage of the female patients with tumors containing KRAS G12C (due to G>T transversion), their younger age at diagnosis, and the fewer pack-years of smoking in women with this KRAS mutation, compared to men with the same KRAS mutation, provide yet another type of data supporting the hypothesis that women are more susceptible to tobacco carcinogens.
The apparent increased susceptibility of women to tobacco carcinogenesis may reflect constitutive differences in genes encoding tobacco carcinogen-metabolizing enzymes. For example, the cytochrome P450 phase I detoxifying enzyme CYP1A1 shows higher expression in the normal lung tissue of female smokers than male smokers.46 The most common polymorphism found in cytochrome P450 phase II detoxification enzymes is the GSTM1-null genotype, which is present in 40%–50% of the general population due to homozygosity for a deletion polymorphism and the impact of this GSTM1 genotype may be enhanced in female smokers.47
In summary, several observations emerge from this large analysis of the molecular epidemiology of EGFR and KRAS mutations in lung adenocarcinoma. Pack-years of smoking have a significant predictive value for the presence of EGFR and KRAS mutations and smoking-free years have additional predictive value for presence of EGFR mutations but not that of KRAS mutations. However, even in the context of a rigorously developed nomogram incorporating these clinical variables, it remains difficult to reliably identify a significant subset of smokers who would have an EGFR mutation likelihood of <1%, and therefore our data do not support excluding any subset of patients with lung adenocarcinoma from EGFR testing. Our results suggest a different etiology of KRAS mutations in smokers vs. never smokers and firmly support earlier observations of increased susceptibility to tobacco carcinogenesis in women. More broadly, our observations strengthen the notion that careful consideration of histologic subtypes (focusing on adenocarcinoma instead of mixing all lung cancer types) and molecular subtypes defined by distinct, non-overlapping driver mutations (EGFR, KRAS) can help to clarify epidemiologic associations that may otherwise remain elusive.48,49 This approach, which recognizes the possible etiologic diversity represented by different histologic and molecular subtypes, has recently been termed molecular pathological epidemiology.50
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
To clarify the molecular epidemiology of EGFR and KRAS mutations in lung adenocarcinoma, we examined tumor genotyping data in 3026 patients in relation to demographic, clinical and smoking history data. In addition to the expected reciprocal associations of EGFR and KRAS mutations with smoking history, this showed that 11% of smokers had EGFR-mutated tumors and 6% of never smokers had KRAS-mutated tumors. Pack-years of smoking were predictive for EGFR and KRAS mutations but even in the context of a nomogram, it is difficult to identify a significant subset of smokers with an EGFR mutation likelihood of <1%, and therefore our data do not support excluding any patient subset from EGFR testing. The distinct types of KRAS mutations in smokers vs. never smokers suggest that most KRAS-mutant lung cancers in never smokers are not due to secondhand smoke exposure. The higher frequency of KRAS G12C in women, their younger age, and lesser smoking history support a heightened susceptibility to tobacco carcinogens.
Acknowledgments
Financial support: NIH P01 CA129243 (to M.L., M.G.K.)
The authors wish to thank Justyna Sadowska, Jacklyn Casanova, and Lin Dong for excellent technical support. We also thank Dr Cameron Brennan for helpful discussions.
References
- 1.Pao W, Miller V. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005 Apr 10;23:2556–68. doi: 10.1200/JCO.2005.07.799. Epub 2005 Mar 14. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005 Mar 2;97:339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 6.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009 Aug;28(Suppl 1):S24–31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pham D, Kris MG, Riely GJ, et al. Use of cigarette smoking history to estimate the likelihood of mutations in epidermal growth factor receptor gene exons 19 and21 in lung adenocarcinomas. J Clin Oncol. 2006;24:1700–4. doi: 10.1200/JCO.2005.04.3224. [DOI] [PubMed] [Google Scholar]
- 8.Lee YJ, Shim HS, Kang YA, et al. Dose effect of cigarette smoking on frequency and spectrum of epidermal growth factor receptor gene mutations in Korean patients with non-small cell lung cancer. J Cancer Res Clin Oncol. 2010 Dec;136:1937–44. doi: 10.1007/s00432-010-0853-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos E, Martin-Zanca D, Reddy E, Pierotti MA, Della Porta G, Barbacid M. Malignant activation of a K-RAS oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–664. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 10.Rodenhuis S, van de Wetering ML, Mooi WJ, Evers SG, van Zandwijk N, Bos JL. Mutational activation of the K-ras oncogene: a possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med. 1987;317:929–35. doi: 10.1056/NEJM198710083171504. [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005 Jan;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005 Sep 1;23:5900–9. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 13.Graziano SL, Gamble GP, Newman NB, et al. Prognostic significance of K-ras codon 12 mutations in patients with resected stage I and II non-small-cell lung cancer. J Clin Oncol. 1999;17:668–75. doi: 10.1200/JCO.1999.17.2.668. [DOI] [PubMed] [Google Scholar]
- 14.Slebos R, Kibbelaar R, Dalesio O, et al. K-RAS oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–5. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- 15.Rosell R, Molina F, Moreno I, et al. Mutated K–ras gene analysis in a randomized trial of preoperative chemotherapy plus surgery versus surgery in stage IIIA non-small cell lung cancer. Lung Cancer. 1995;12(Suppl 1):S59–70. doi: 10.1016/0169-5002(95)00421-v. [DOI] [PubMed] [Google Scholar]
- 16.Johnson ML, Sima CS, Paik P, et al. Association of KRAS and EGFR Mutations with Survival in Patients with Advanced Lung Adenocarcinoma. J Clin Oncol. 2010;28:7s. doi: 10.1002/cncr.27730. (suppl; abstr 7541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes S, Clements J, Dawson E, et al. Cosmic 2005. Br J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buttitta F, Barassi F, Fresu G, et al. Mutational analysis of the HER2 gene in lung tumors from Caucasian patients: mutations are mainly present in adenocarcinomas with bronchioloalveolar features. Int J Cancer. 2006;119:2586–91. doi: 10.1002/ijc.22143. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki M, Shigematsu H, Iizasa T, et al. Exclusive mutation in epidermal growth factor receptor gene, HER-2, and KRAS, and synchronous methylation of nonsmall cell lung cancer. Cancer. 2006;106:2200–7. doi: 10.1002/cncr.21853. [DOI] [PubMed] [Google Scholar]
- 20.Ahrendt SA, Decker PA, Alawi EA, et al. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001;92:1525–30. doi: 10.1002/1097-0142(20010915)92:6<1525::aid-cncr1478>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 21.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girard N, Deshpande C, Azzoli CG, et al. Use of EGFR/KRAS mutation testing to define clonal relationships among multiple lung adenocarcinomas: comparison with clinical guidelines. Chest. 2010;137:46–52. doi: 10.1378/chest.09-0325. [DOI] [PubMed] [Google Scholar]
- 23.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005 Aug;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005 Jan;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011 Jan;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girard N, Sima CS, Jackman DM, et al. Nomogram to predict the presence of EGFR activating mutation in lung adenocarcinoma. Eur Respir J. 2012 Feb;39:366–72. doi: 10.1183/09031936.00010111. [DOI] [PubMed] [Google Scholar]
- 27.D’Angelo SP, Park B, Azzoli CG, et al. Reflex testing of resected stage I through III lung adenocarcinomas for EGFR and KRAS mutation: report on initial experience and clinical utility at a single center. J Thor Cardiovasc Surg. 2011;141:476–80. doi: 10.1016/j.jtcvs.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arcila ME, Lau C, Jhanwar J, et al. Comprehensive Analysis for Clinically Relevant Oncogenic Driver Mutations in 1131 Consecutive Lung Adenocarcinomas. Modern Pathology. 2011;24:404A, abstract 1721. [Google Scholar]
- 29.Sugio K, Uramoto H, Ono K, et al. Mutations within the tyrosine kinase domain of EGFR gene specifically occur in lung adenocarcinoma patients with a low exposure of tobacco smoking. Br J Cancer. 2006 Mar 27;94(6):896–903. doi: 10.1038/sj.bjc.6603040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuo K, Ito H, Yatabe Y, et al. Risk factors differ for non-small-cell lung cancers with and without EGFR mutation: assessment of smoking and sex by a case-control study in Japanese. Cancer Sci. 2007 Jan;98(1):96–101. doi: 10.1111/j.1349-7006.2006.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Angelo SP, Pietanza MC, Johnson ML, Riely GJ, Miller VA, Sima CS, Zakowski MF, Rusch VW, Ladanyi M, Kris MG. Incidence of EGFR exon 19 deletions and L858R mutations in tumor specimens from men and cigarette smokers with lung adenocarcinomas. J Clin Oncol. 2011;29:2066–70. doi: 10.1200/JCO.2010.32.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009 Sep 3;361:958–67. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 33.Jackman DM, Miller VA, Cioffredi LA, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009 Aug 15;15:5267–73. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Won YW, Han JY, Lee GK, et al. Comparison of clinical outcome of patients with non-small-cell lung cancer harbouring epidermal growth factor receptor exon 19 or exon 21 mutations. J Clin Pathol. 2011 Nov;64(11):947–52. doi: 10.1136/jclinpath-2011-200169. [DOI] [PubMed] [Google Scholar]
- 35.Kim DW, Lee SH, Lee JS, et al. A multicenter phase II study to evaluate the efficacy and safety of gefitinib as first-line treatment for Korean patients with advanced pulmonary adenocarcinoma harboring EGFR mutations. Lung Cancer. 2011 Jan;71:65–9. doi: 10.1016/j.lungcan.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010 Feb;11(2):121–8. doi: 10.1016/S1470-2045(09)70364-X. Epub 2009 Dec 18. [DOI] [PubMed] [Google Scholar]
- 37.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010 Jun 24;362(25):2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 38.Huang YS, Yang JJ, Zhang XC, et al. Impact of smoking status and pathologic type on epidermal growth factor receptor mutations in lung cancer. Chin Med J (Engl) 2011 Aug;124:2457–60. [PubMed] [Google Scholar]
- 39.Mao C, Qiu LX, Liao RY, et al. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: A meta-analysis of 22 studies. Lung Cancer. 2010 Sep;69:272–8. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 40.Riely GJ, Chaft JE, Ladanyi M, Kris MG. Incorporation of crizotinib into the NCCN Guidelines. J Natl Compr Canc Netw. 2011 Dec;9(12):1328–30. doi: 10.6004/jnccn.2011.0113. [DOI] [PubMed] [Google Scholar]
- 41.Kiyohara C, Ohno Y. Sex differences in lung cancer susceptibility: a review. Gend Med. 2010 Oct;7:381–401. doi: 10.1016/j.genm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Zang EA, Wynder EL. Differences in lung cancer risk between men and women: examination of the evidence. J Natl Cancer Inst. 1996;88:183–92. doi: 10.1093/jnci/88.3-4.183. [DOI] [PubMed] [Google Scholar]
- 43.International Early Lung Cancer Action Program Investigators. Women’s susceptibility to tobacco carcinogens and survival after diagnosis of lung cancer. JAMA. 2006;296(2):180–184. doi: 10.1001/jama.296.2.180. [DOI] [PubMed] [Google Scholar]
- 44.Bennett WP, Hussain SP, Vahakangas KH, et al. Molecular epidemiology of human cancer risk: gene-environment interactions and p53 mutation spectrum in human lung cancer. J Pathol. 1999 Jan;187(1):8–18. doi: 10.1002/(SICI)1096-9896(199901)187:1<8::AID-PATH232>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 45.Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat. 2003;21:229–39. doi: 10.1002/humu.10177. [DOI] [PubMed] [Google Scholar]
- 46.Uppstad H, Osnes GH, Cole KJ, Phillips DH, Haugen A, Mollerup S. Sex differences in susceptibility to PAHs is an intrinsic property of human lung adenocarcinoma cells. Lung Cancer. 2011 Mar;71:264–70. doi: 10.1016/j.lungcan.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Tang DL, Rundle A, Warburton D, et al. Associations between both genetic and environmental biomarkers and lung cancer: evidence of a greater risk of lung cancer in women smokers. Carcinogenesis. 1998;19:1949–53. doi: 10.1093/carcin/19.11.1949. [DOI] [PubMed] [Google Scholar]
- 48.Wakelee HA, Gomez SL, Chang ET. Sex differences in lung-cancer susceptibility: a smoke screen? Lancet Oncol. 2008 Jul;9:609–10. doi: 10.1016/S1470-2045(08)70162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gazdar AF, Thun MJ. Lung cancer, smoke exposure, and sex. J Clin Oncol. 2007 Feb 10;25:469–71. doi: 10.1200/JCO.2006.09.4623. [DOI] [PubMed] [Google Scholar]
- 50.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011 Mar;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.